Effects of Dufulin on Oxidative Stress and Metabolomic Profile of Tubifex
Abstract
:1. Introduction
2. Results and Discussion
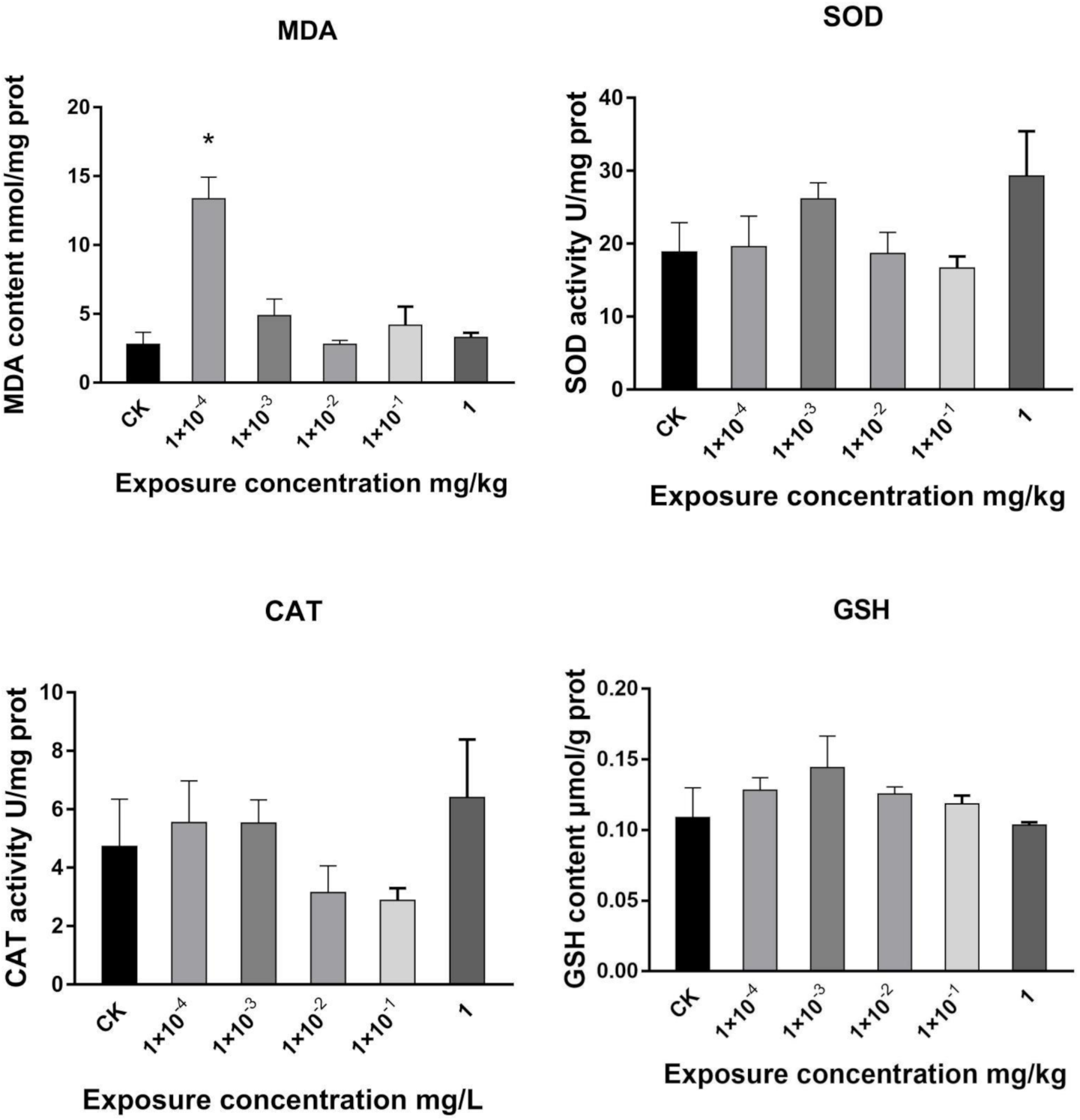
2.1. Effects of Dufulin on Oxidative Stress in Tubifex
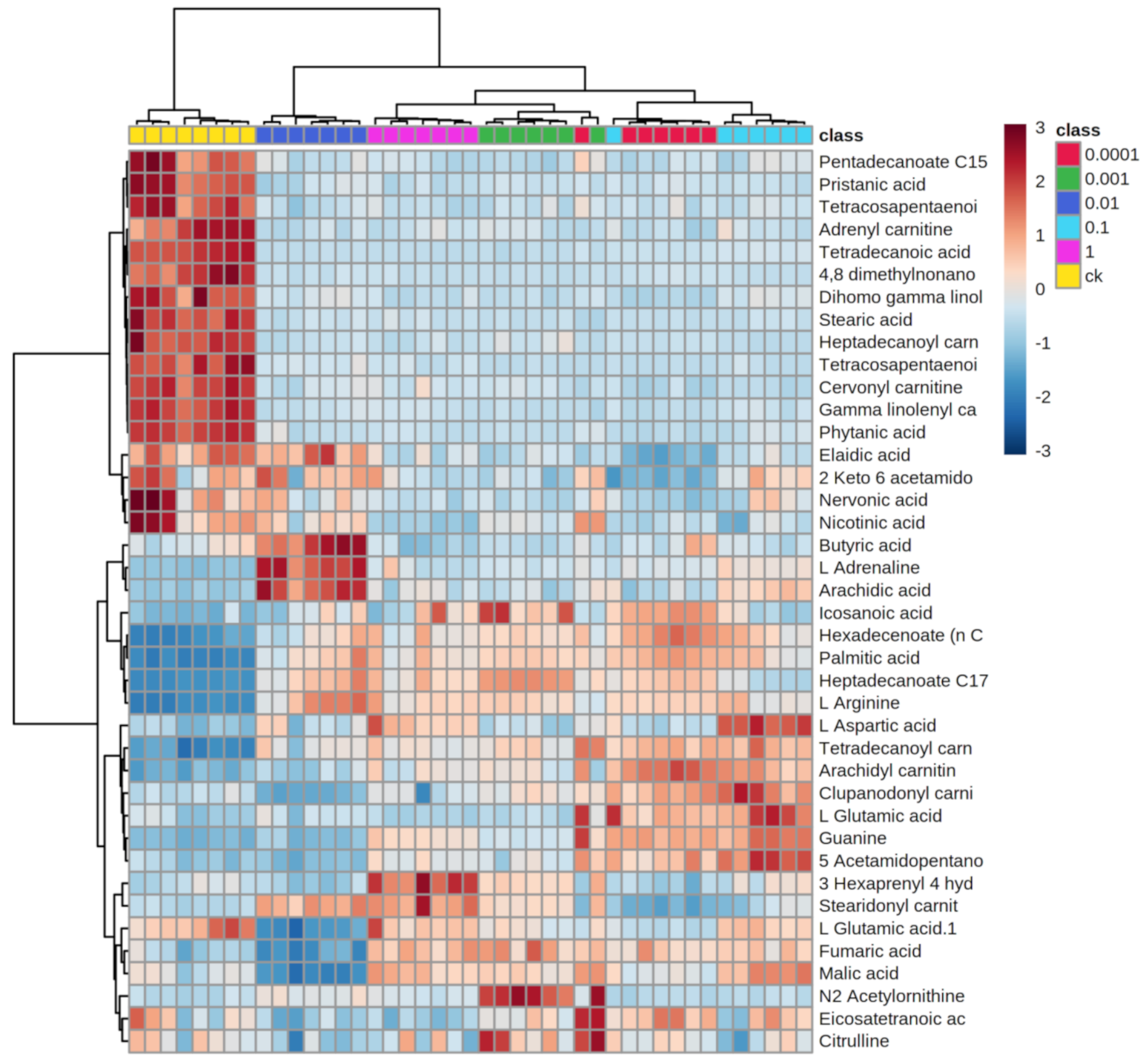
2.2. LC-MS-Based Metabolomic Data
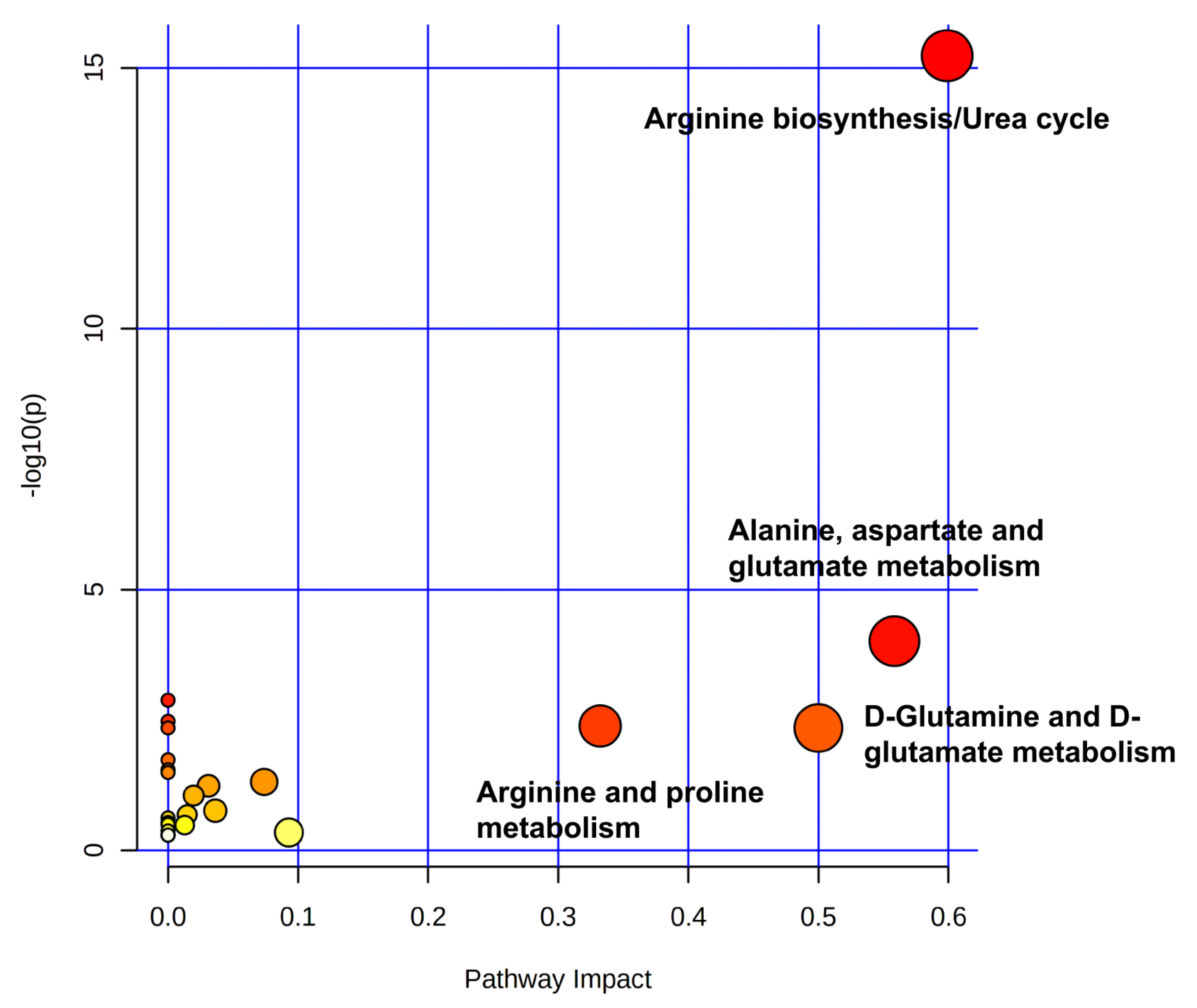
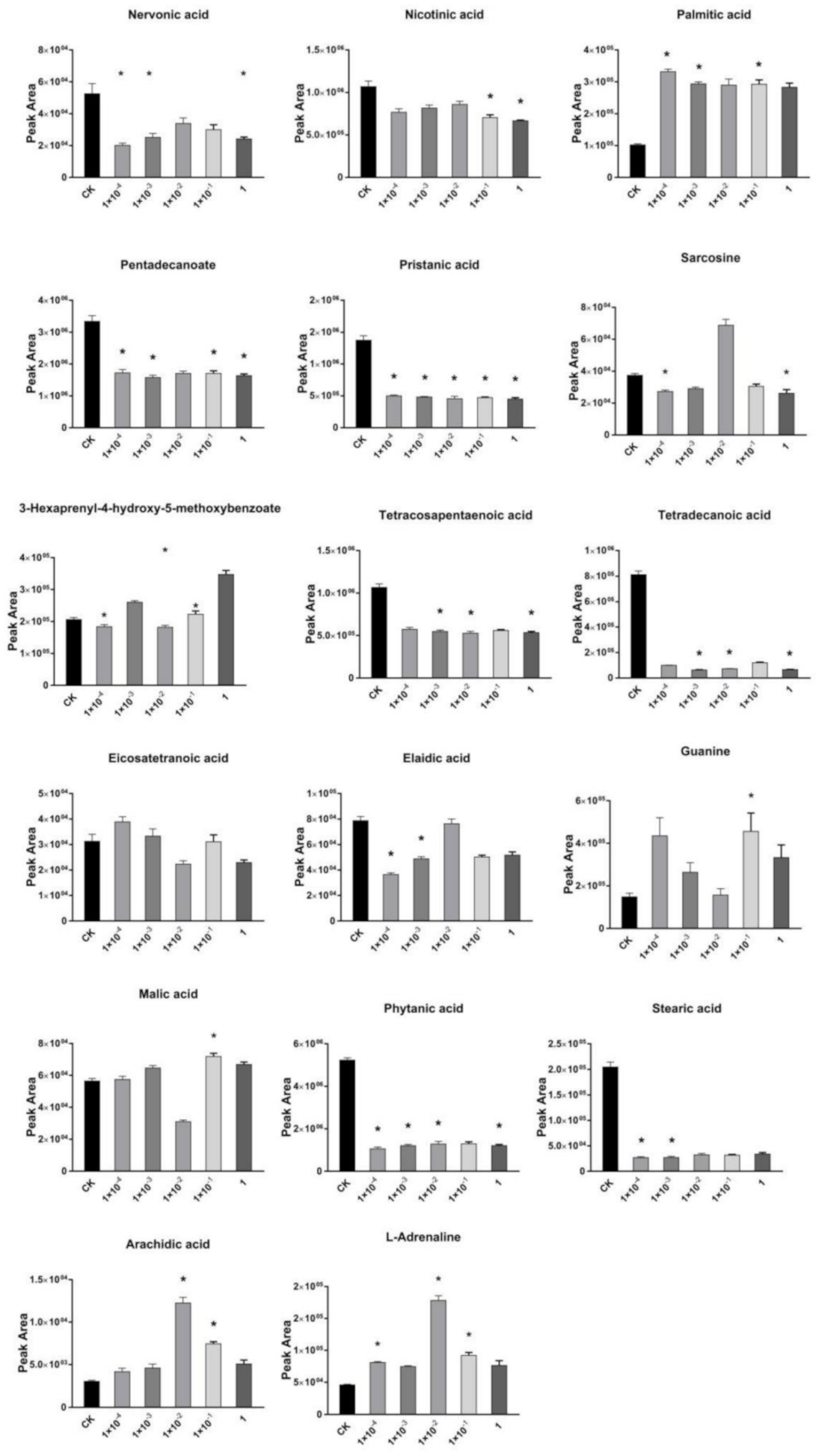
2.3. Effects of Dufulin on Tubifex Metabolism
2.4. Impact of Dufulin on Tubifex Urea Cycle
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Experimental Design
3.2.1. Growth of the Experimental Organism
3.2.2. Exposure of Dufulin on Tubifex
3.2.3. Sample Pretreatment for the Analysis of Oxidative Stress
3.2.4. Metabolomic Analysis of Tubifex by LC-Q-TOF
3.3. Biochemical Parameters Measurement
3.4. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooper, J.; Dobson, H. The benefits of pesticides to mankind and the environment. Crop. Prot. 2007, 26, 1337–1348. [Google Scholar] [CrossRef]
- Wilhoit, L. History of Pesticide Use Reporting in California. In Managing and Analyzing Pesticide Use Data for Pest Management, Environmental Monitoring, Public Health, and Public Policy; American Chemical Society: Washington, DC, USA, 2018; pp. 3–14. [Google Scholar]
- Zhang, K.-K.; Hu, D.-Y.; Zhu, H.-J.; Yang, J.-C.; Song, B.-A. Enantioselective Degradation of Dufulin in Four Types of Soil. J. Agric. Food Chem. 2014, 62, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Hao, G.; Pan, J.; Zhang, J.; Hu, D.; Song, B. Asymmetric Synthesis and Bioselective Activities of α-Amino-phosphonates Based on the Dufulin Motif. J. Agric. Food Chem. 2016, 64, 4207–4213. [Google Scholar] [CrossRef]
- Zhang, K.; Hu, D.; Zhu, H.; Yang, J.; Wu, J.; He, M.; Jin, L.; Yang, S.; Song, B. Enantioselective hydrolyzation and photolyzation of dufulin in water. Chem. Central J. 2013, 7, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Zeng, M.; Song, B.; Hou, C.; Hu, D.; Li, X.; Wang, Z.; Fan, H.; Bi, L.; Liu, J.; et al. Dufulin Activates HrBP1 to Produce Antiviral Responses in Tobacco. PLoS ONE 2012, 7, e37944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Xue, J.; Zhang, H.-M.; Yang, J.; Xie, L.; Chen, J.-P. Characterization of homologous and heterologous interactions between viroplasm proteins P6 and P9-1 of the fijivirus southern rice black-streaked dwarf virus. Arch. Virol. 2014, 160, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xie, X.; Gao, D.; Chen, K.; Chen, Z.; Jin, L.; Li, X.; Song, B. Dufulin Intervenes the Viroplasmic Proteins as the Mechanism of Action against Southern Rice Black-Streaked Dwarf Virus. J. Agric. Food Chem. 2019, 67, 11380–11387. [Google Scholar] [CrossRef]
- Fang, H.; Yu, Y.; Chu, X.; Wang, X.; Yang, X.; Yu, J. Degradation of chlorpyrifos in laboratory soil and its impact on soil microbial functional diversity. J. Environ. Sci. 2009, 21, 380–386. [Google Scholar] [CrossRef]
- Wang, H.Z.; Zuo, H.G.; Ding, Y.J.; Miao, S.S.; Jiang, C.; Yang, H. Biotic and abiotic degradation of pesticide Dufulin in soils. Environ. Sci. Pollut. Res. 2014, 21, 4331–4342. [Google Scholar] [CrossRef]
- Teng, M.; Zhou, Y.; Song, M.; Dong, K.; Chen, X.; Wang, C.; Bi, S.; Zhu, W. Chronic Toxic Effects of Flutolanil on the Liver of Zebrafish (Danio rerio). Chem. Res. Toxicol. 2019, 32, 995–1001. [Google Scholar] [CrossRef]
- van den Berg, F.; Holterman, H.J.; Leistra, M. Emission of Pesticides into the Air. Water Air Soil Pollut. 1999, 115, 195–218. [Google Scholar] [CrossRef]
- Jiang, W.; Gan, J. Importance of Fine Particles in Pesticide Runoff from Concrete Surfaces and Its Prediction. Environ. Sci. Technol. 2012, 46, 6028–6034. [Google Scholar] [CrossRef]
- Borja, A.; Tueros, I.; Belzunce, M.J.; Galparsoro, I.; Garmendia, J.M.; Revilla, M.; Solaun, O.; Valencia, V. Investigative monitoring within the European Water Framework Directive: A coastal blast furnace slag disposal, as an example. J. Environ. Monit. 2008, 10, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Camusso, M.; Galassi, S.; Vignati, D. Assessment of river Po sediment quality by micropollutant analysis. Water Res. 2002, 36, 2491–2504. [Google Scholar] [CrossRef]
- Lan, J.; Jia, J.; Liu, A.; Yu, Z.; Zhao, Z. Pollution levels of banned and non-banned pesticides in surface sediments from the East China Sea. Mar. Pollut. Bull. 2019, 139, 332–338. [Google Scholar] [CrossRef]
- Di, S.; Zhang, W.; Chen, L.; Zhou, Z.; Diao, J. Toxicokinetics and oxidative stress in Tubifex tubifex exposed to hexachlorocyclohexane isomers. RSC Adv. 2016, 6, 19016–19024. [Google Scholar] [CrossRef]
- Di, S.; Liu, R.; Cheng, C.; Chen, L.; Zhang, W.; Tian, Z.; Liu, C.; Zhou, Z.; Diao, J. Biomarkers in Tubifex tubifex for the metalaxyl and metalaxyl-M toxicity assessment in artificial sediment. Environ. Sci. Pollut. Res. 2016, 24, 3618–3625. [Google Scholar] [CrossRef]
- Lagauzère, S.; Terrail, R.; Bonzom, J.-M. Ecotoxicity of uranium to Tubifex tubifex worms (Annelida, Clitellata, Tubificidae) exposed to contaminated sediment. Ecotoxicol. Environ. Saf. 2009, 72, 527–537. [Google Scholar] [CrossRef]
- Wiederholm, T.; Wiederholm, A.-M.; Milbrink, G. Bulk sediment bioassays with five species of fresh-water oligochaetes. Water, Air, Soil Pollut. 1987, 36, 131–154. [Google Scholar] [CrossRef]
- Walker, J.G. Oxygen poisoning in the Annelid Tubifex tubifexi. Response to oxygen exposure. Biol. Bull. 1970, 138, 235–244. [Google Scholar] [CrossRef]
- Liu, C.; Wang, B.; Xu, P.; Liu, T.; Di, S.; Diao, J. Enantioselective Determination of Triazole Fungicide Epoxiconazole Bioaccumulation in Tubifex Based on HPLC-MS/MS. J. Agric. Food Chem. 2013, 62, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Oplinger, R.W.; Bartley, M.; Wagner, E.G. Culture of Tubifex tubifex: Effect of Feed Type, Ration, Temperature, and Density on Juvenile Recruitment, Production, and Adult Survival. N. Am. J. Aquac. 2011, 73, 68–75. [Google Scholar]
- Milbrink, G. Biological characterization of sediments by standardized tubificid bioassays. Hydrobiologia 1987, 155, 267–275. [Google Scholar] [CrossRef]
- Di, S.; Liu, T.; Diao, J.; Zhou, Z. Enantioselective Bioaccumulation and Degradation of Sediment-Associated Metalaxyl Enantiomers in Tubifex tubifex. J. Agric. Food Chem. 2013, 61, 4997–5002. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, P.; Hu, D.; Wang, S.; Zhang, Q.; Yu, Y.; Zeng, S. Stereoselective Bioaccumulation of Water and Soil-Associated Dufulin Enantiomers in Tubifex. J. Agric. Food Chem. 2017, 65, 8569–8577. [Google Scholar] [CrossRef]
- Wang, C.; Qian, Y.; Zhang, X.; Chen, F.; Zhang, Q.; Li, Z.; Zhao, M. A metabolomic study of fipronil for the anxiety-like behavior in zebrafish larvae at environmentally relevant levels. Environ. Pollut. 2016, 211, 252–258. [Google Scholar] [CrossRef]
- Aliferis, K.; Chrysayi-Tokousbalides, M. Metabolomics in pesticide research and development: Review and future perspec-tives. Metabolomics 2011, 7, 35–53. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Mass spectrometry-based metabolomics: Applications to biomarker and metabolic pathway research. Biomed. Chromatogr. 2016, 30, 7–12. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Connelly, J.; Lindon, J.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002, 1, 153–161. [Google Scholar] [CrossRef]
- Yao, C.; Sheng, J.; Yan, S.; Tian, S.; Meng, Z.; Zhou, Z.; Zhu, W. Enantioselectivity effects of imazethapyr enantiomers to metabolic responses in mice. Pestic. Biochem. Physiol. 2020, 168, 104619. [Google Scholar] [CrossRef]
- Li, J.; Wang, Q.; Zheng, Y.; Zhou, P.; Xu, X.; Liu, X.; Zhao, L.; Liu, H. Development of a mass spectrometry-based pseudotargeted metabolomics strategy to analyze hormone-stimulated gastric cancer cells. J. Pharm. Biomed. Anal. 2020, 180, 113041. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, X.; Qiu, J.; Zhang, L.; Zhang, Y.; Qiu, X.; Huang, Z.; Xu, W. Comprehensive Comparison on the Chemical Profile of Guang Chen Pi at Different Ripeness Stages Using Untargeted and Pseudotargeted Metabolomics. J. Agric. Food Chem. 2020, 68, 8483–8495. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, W.J.; Koal, T.; Wang, Y.; Kohl, M.; Enot, D.P.; Deigner, H.-P. Targeted Metabolomics for Biomarker Discovery. Angew. Chem. Int. Ed. 2010, 49, 5426–5445. [Google Scholar] [CrossRef]
- Zha, H.; Cai, Y.; Yin, Y.; Wang, Z.; Li, K.; Zhu, Z.-J. SWATHtoMRM: Development of High-Coverage Targeted Metabolomics Method Using SWATH Technology for Biomarker Discovery. Anal. Chem. 2018, 90, 4062–4070. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef]
- Evans, C.; Karnovsky, A.; Kovach, M.A.; Standiford, T.J.; Burant, C.F.; Stringer, K.A. Untargeted LC–MS Metabolomics of Bronchoalveolar Lavage Fluid Differentiates Acute Respiratory Distress Syndrome from Health. J. Proteome Res. 2014, 13, 640–649. [Google Scholar] [CrossRef]
- Gu, J.; Ji, C.; Yue, S.; Shu, D.; Su, F.; Zhang, Y.; Xie, Y.; Zhang, Y.; Liu, W.; Zhao, M. Enantioselective Effects of Metalaxyl Enantiomers in Adolescent Rat Metabolic Profiles Using NMR-Based Metabolomics. Environ. Sci. Technol. 2018, 52, 5438–5447. [Google Scholar] [CrossRef]
- Coen, M.; Holmes, E.; Lindon, J.; Nicholson, J. NMR-Based Metabolic Profiling and Metabonomic Approaches to Problems in Molecular Toxicology. Chem. Res. Toxicol. 2008, 21, 9–27. [Google Scholar] [CrossRef]
- Charris-Molina, A.; Riquelme, G.; Burdisso, P.; Hoijemberg, P.A. Consecutive Queries to Assess Biological Correlation in NMR Metabolomics: Performance of Comprehensive Search of Multiplets over Typical 1D 1H NMR Database Search. J. Proteome Res. 2020, 19, 2977–2988. [Google Scholar] [CrossRef]
- Rutledge, A.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Fraisier-Vannier, O.; Chervin, J.; Cabanac, G.; Puech-Pages, V.; Fournier, S.; Durand, V.; Amiel, A.; André, O.; Benamar, O.A.; Dumas, B.; et al. MS-CleanR: A Feature-Filtering Workflow for Untargeted LC–MS Based Metabolomics. Anal. Chem. 2020, 92, 9971–9981. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.-Y.; Fairfull-Smith, K.E.; Morrow, B.J.; Lussini, V.; Kim, B.; Bondar, M.V.; Bottle, S.E.; Belfield, K.D. Two-Photon Fluorescence Microscopy Imaging of Cellular Oxidative Stress Using Profluorescent Nitroxides. J. Am. Chem. Soc. 2012, 134, 4721–4730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livingstone, D.R. Oxidative stress in aquatic organisms in relation to pollution and aquaculture. Rev. Med. Veteri-Naire 2003, 154, 427–430. [Google Scholar]
- Analytical Currents. Measuring oxidative damage. Anal. Chem. 1998, 70, 91A. [Google Scholar] [CrossRef]
- Gamon, L.F.; Wille, U. Oxidative Damage of Biomolecules by the Environmental Pollutants NO2• and NO3•. Acc. Chem. Res. 2016, 49, 2136–2145. [Google Scholar] [CrossRef]
- Stanicka, J.; Landry, W.; Cotter, T.G. Oxidative Stress Biomarkers and ROS Molecular Probes. In Oxidative Stress: Diagnostics, Prevention, and Therapy Volume 2; American Chemical Society: Washington, DC, USA, 2015; pp. 353–374. [Google Scholar]
- Sergiev, P.V.; Dontsova, O.A.; Berezkin, G.V. Theories of Aging: An Ever-Evolving Field. Acta Naturae 2015, 7, 9–18. [Google Scholar] [CrossRef]
- Tang, J.-Y.; Ou-Yang, F.; Hou, M.-F.; Huang, H.-W.; Wang, H.-R.; Li, K.-T.; Fayyaz, S.; Shu, C.-W.; Chang, H.-W. Oxidative stress-modulating drugs have preferential anticancer effects—involving the regulation of apoptosis, DNA damage, endoplasmic reticulum stress, autophagy, metabolism, and migration. Semin. Cancer Biol. 2019, 58, 109–117. [Google Scholar] [CrossRef]
- Prieto, A.I.; Pichardo, S.; Jos, A.; Moreno, I.; Cameán, A.M. Time-dependent oxidative stress responses after acute exposure to toxic cyanobacterial cells containing microcystins in tilapia fish (Oreochromis niloticus) under laboratory conditions. Aquat. Toxicol. 2007, 84, 337–345. [Google Scholar] [CrossRef]
- Liu, T.; Diao, J.; Di, S.; Zhou, Z. Stereoselective Bioaccumulation and Metabolite Formation of Triadimefon in Tubifex tubifex. Environ. Sci. Technol. 2014, 48, 6687–6693. [Google Scholar] [CrossRef]
- Abdollahi, M.; Ranjbar, A.; Shadnia, S.; Nikfar, S.; Rezaiee, A. Pesticides and oxidative stress: A review. Med Sci. Monit. 2004, 10, RA141–RA147. [Google Scholar]
- Janero, D.R. Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free. Radic. Biol. Med. 1990, 9, 515–540. [Google Scholar] [CrossRef]
- Shin, S.-K.; Cho, H.-W.; Song, S.-E.; Im, S.-S.; Bae, J.-H.; Song, D.-K. Oxidative stress resulting from the removal of endogenous catalase induces obesity by promoting hyperplasia and hypertrophy of white adipocytes. Redox Biol. 2020, 37, 101749. [Google Scholar] [CrossRef]
- Scott, M.; Meshnick, S.; Eaton, J. Superoxide dismutase-rich bacteria. Paradoxical increase in oxidant toxicity. J. Biol. Chem. 1987, 262, 3640–3645. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, L.; Tan, D.; Sun, W.; Ke, Q.; Yue, X.; Bai, B. S-desulfurization: A different covalent modification mechanism from persulfidation by GSH. Free. Radic. Biol. Med. 2021, 167, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.C.; Watt, A.P.; Nicoll-Griffith, D.A.; Baillie, T.A. Drug−Protein Adducts: An Industry Perspective on Minimizing the Potential for Drug Bioactivation in Drug Discovery and Development. Chem. Res. Toxicol. 2004, 17, 3–16. [Google Scholar] [CrossRef]
- Rinaldi, R.; Eliasson, E.; Swedmark, S.; Morgenstern, R. Reactive Intermediates and The Dynamics of Glutathione Transferases. Drug Metab. Dispos. 2002, 30, 1053–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beronius, A.; Vandenberg, L.N. Using systematic reviews for hazard and risk assessment of endocrine disrupting chemicals. Rev. Endocr. Metab. Disord. 2015, 16, 273–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakata, S.; Fujita, R. Self-Propelled Motion of Camphor Disk on Nervonic Acid Molecular Layer and Its Dependence on Phase Transition. J. Phys. Chem. B 2020, 124, 5525–5529. [Google Scholar] [CrossRef]
- Tu, X.; Wan, J.; Xie, Y.; Wei, F.; Quek, S.; Lv, X.; Du, L.; Chen, H. Lipid analysis of three special nervonic acid resources in China. Oil Crop. Sci. 2020, 5, 180–186. [Google Scholar] [CrossRef]
- Watkins, P.A. Fatty acid activation. Prog. Lipid Res. 1997, 36, 55–83. [Google Scholar] [CrossRef]
- Spiekerkoetter, U.; Duran, M. Mitochondrial Fatty Acid Oxidation Disorders, in Physician’s Guide to the Diagnosis, Treatment, and Follow-Up of Inherited Metabolic Diseases; Blau, N., Duran, M., Gibson, K.M., Dionisi-Vici, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 247–264. [Google Scholar]
- Houten, S.M.; Wanders, R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 2010, 33, 469–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta Bioenerg. 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Li, J.-M.; Li, L.-Y.; Qin, X.; Ning, L.-J.; Lu, D.-L.; Zhang, M.-L.; Wang, X.; Du, Z.-Y. Systemic regulation of L-carnitine in nutritional metabolism in zebrafish, Danio rerio. Sci. Rep. 2017, 7, 40815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epstein, F.H.; Moncada, S.; Higgs, A. The L-Arginine-Nitric Oxide Pathway. N. Engl. J. Med. 1993, 329, 2002–2012. [Google Scholar] [CrossRef]
- Beaumier, L.; Castillo, L.; Yu, Y.M.; Ajami, A.M.; Young, V.R. Arginine: New and exciting developments for an “old” amino acid. Biomed. Environ. Sci. 1996, 9, 296–315. [Google Scholar]
- Cabello, J.; Basílio, C.; Prajoux, V. Kinetic properties of erythrocyte- and liver arginase. Biochim. Biophys. Acta 1961, 48, 148–152. [Google Scholar] [CrossRef]
- Morris, J.S. Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 2002, 22, 87–105. [Google Scholar] [CrossRef]
- Garrard, L.J.; Mathis, J.M.; Raushel, F.M. Substrate-induced inactivation of argininosuccinate lyase by mono-fluorofumarate and difluorofumarate. Biochemistry 1983, 22, 3729–3735. [Google Scholar] [CrossRef]
- Bishop, S.H.; Campbell, J.W. Arginine and urea biosynthesis in the earthworm Lumbricus terrestris. Comp. Biochem. Physiol. 1965, 15, 51–71. [Google Scholar] [CrossRef]
- Wang, F.S.; Goh, D.L.M.; Ong, H.T. Urea cycle disorder presenting as bilateral mesial temporal sclerosis—An un-usual cause of seizures: A case report and review of the literature. J. Med Case Rep. 2018, 12, 208. [Google Scholar]
- Wang, B.; Jha, P. A Case of Atypical Adult Presentation of Urea Cycle Disorder. WMJ Off. Publ. State Med. Soc. Wis. 2019, 118, 98–100. [Google Scholar]
- Du, D.; Gu, H.; Djukovic, D.; Bettcher, L.; Gong, M.; Zheng, W.; Hu, L.; Zhang, X.; Zhang, R.; Wang, D.; et al. Multiplatform Metabolomics Investigation of Antiadipogenic Effects on 3T3-L1 Adipocytes by a Potent Diarylheptanoid. J. Proteome Res. 2018, 17, 2092–2101. [Google Scholar] [CrossRef]
- Wei, Y.; Jasbi, P.; Shi, X.; Turner, C.; Hrovat, J.; Liu, L.; Rabena, Y.; Porter, P.; Gu, H. Early Breast Cancer Detection Using Untargeted and Targeted Metabolomics. J. Proteome Res. 2021. [Google Scholar] [CrossRef]
- Peng, Y.; Hong, J.; Raftery, D.; Xia, Q.; Du, D. Metabolomic-based clinical studies and murine models for acute pancreatitis disease: A review. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166123. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, J.; Raftery, D. Five Easy Metrics of Data Quality for LC–MS-Based Global Metabolomics. Anal. Chem. 2020, 92, 12925–12933. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Zhu, Y.; Yang, J.; Zhu, W.; Zhou, Z.; Zhang, R. Effects of Dufulin on Oxidative Stress and Metabolomic Profile of Tubifex. Metabolites 2021, 11, 381. https://doi.org/10.3390/metabo11060381
Yu Y, Zhu Y, Yang J, Zhu W, Zhou Z, Zhang R. Effects of Dufulin on Oxidative Stress and Metabolomic Profile of Tubifex. Metabolites. 2021; 11(6):381. https://doi.org/10.3390/metabo11060381
Chicago/Turabian StyleYu, Yile, Yuxin Zhu, Jing Yang, Wentao Zhu, Zhiqiang Zhou, and Renke Zhang. 2021. "Effects of Dufulin on Oxidative Stress and Metabolomic Profile of Tubifex" Metabolites 11, no. 6: 381. https://doi.org/10.3390/metabo11060381
APA StyleYu, Y., Zhu, Y., Yang, J., Zhu, W., Zhou, Z., & Zhang, R. (2021). Effects of Dufulin on Oxidative Stress and Metabolomic Profile of Tubifex. Metabolites, 11(6), 381. https://doi.org/10.3390/metabo11060381






