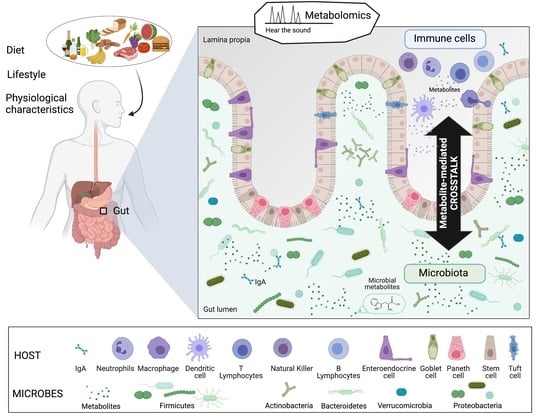
What We Know So Far about the Metabolite-Mediated Microbiota-Intestinal Immunity Dialogue and How to Hear the Sound of This Crosstalk
Abstract
1. Introduction
2. Microbiota-Derived Metabolites That Modulate Host Immunity in the Gut
2.1. Soluble Microbially-Derived Metabolites Affecting the Immune System
2.1.1. Short Chain Fatty Acids (Propionate, Butyrate, and Acetate)
2.1.2. Lactate
2.1.3. Succinate
2.1.4. B Vitamins
Vitamin B1
Vitamin B2
Vitamin B3
Vitamin B5
Vitamin B6
Vitamin B8
Vitamin B9 and B12
2.1.5. Amino Acids (AA) and AA-Derived Metabolites
Tryptophan
Taurine
p-Cresol and Its Derivatives
Histidine and Derivatives
Polyamines
D-Amino Acids
Gamma-Aminobutyric Acid
Quorum Sensing Molecules
2.1.6. Catecholamines
2.1.7. Cyclic-Dinucleotides (CDNs) and Cyclic-Trinucleotides (CTNs)
2.1.8. Inosine
2.1.9. Secondary Bile Acids
2.2. Microbial Membrane Metabolites Affecting the Immune System
2.2.1. Sphingolipids
2.2.2. Lipoteichoic Acids
3. Metabolites Derived from Immune Cells Affecting the Microbiota
3.1. Gut Microbiota Modulation upon Acute and Chronic Host Immune System Activation
3.2. The Antimicrobial Peptides
3.3. Immune Cells-Derived Metabolites and Metabolite Mimicry
4. Conclusions
5. Challenges and Perspectives: How to Hear the Sound from Metabolite-Mediated Microbiota-Host Immunity Crosstalk
- Improve the analytical tools to detect metabolites in the gut (depth of data acquisition, detection of chemicals of various nature, dynamic range of detection with a large order of magnitude of detection). Currently, ultra-high performance liquid chromatography-high resolution mass spectrometry (UHPLC-HRMS) variants represent the best methods to address these challenges, and, in particular, to increase metabolite detection coverage [315]. The main limitations of LC-MS are the limited structural information and the complexity of obtaining absolute quantification, especially in non-targeted approaches.
- Correctly assign the identity of a metabolite. The vast majority of information collected by metabolomics is “dark matter”, i.e., chemical signatures that remain uncharacterized. Therefore, new computational solutions to illuminating dark matter are needed [316]. Significant progress has been reported in exploring public data-rich libraries, finding chemicals and associated metadata, and applying molecular networking strategies to accelerate metabolite annotation, especially in the last few years [317,318,319,320,321]. Moreover, the development and maintenance of MS-based spectral databases, together with the increasing practice of sharing metabolomics data through these resources, play a key role in translating dark matter into biological knowledge [322]. Authentic chemical standards should be used to acquire both positive and negative mode MS/MS and MSn spectra to supplement spectral databases such as HMBD [323], NIST20 [324], METLIN [325], MZcloud [326], MassBank [327], ReSpect [328], and GNPS [329].
- Integrate multi-omics datasets to recover microbe-metabolite relationships by using statistical analysis. Linear or neural network methods estimate the conditional probability that each molecule is present, given the presence of a specific microorganism [330,331,332]. The mmmvec tool (https://github.com/biocore/mmvec; accessed on 15 January 2021), for instance, can reliably identify all of the experimentally determined P. aeruginosa-produced molecules of interest in the lung of cystic fibrosis patient chronically infected with P. aeruginosa [331].
- Use experimental models relevant to the concept of molecular dialogue to unveil the respective role of microbiota and/or immune cells in the production of specific metabolites of interest. In a context where the microbial and host metabolite relationships are not elucidated, this first work consists of a detailed experimental design, followed by a fundamental study on metabolites using specific pathological models (KO mice, mono-colonization of the microbiota with certain strains, synthetic biology strategy, GF and specific pathogen-free model, wilding mice [333], culturomic, metabolomic model with minimal microbiota, (reviewed in [334]) and in vitro models [335,336].
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cavalier-Smith, T. Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis; The MIT Press: Cambridge, MA, USA, 1992; Volume 7, ISBN 0262519909. [Google Scholar]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; Fitzgerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Shi, Y.; Tyson, G.W.; Delong, E.F. Metatranscriptomics reveals unique microbial small RNAs in the oceans water column. Nature 2009, 459, 266–269. [Google Scholar] [CrossRef]
- Maron, P.A.; Ranjard, L.; Mougel, C.; Lemanceau, P. Metaproteomics: A new approach for studying functional microbial ecology. Microb. Ecol. 2007, 53, 486–493. [Google Scholar] [CrossRef]
- Francino, M.P. Early development of the gut microbiota and immune health. Pathogens 2014, 3, 769–790. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Dorrestein, P.C.; Mazmanian, S.K.; Knight, R. Finding the Missing Links among Metabolites, Microbes, and the Host. Immunity 2014, 40, 824–832. [Google Scholar] [CrossRef]
- Goodacre, R. Metabolomics of a superorganism. J. Nutr. 2007, 137, 259S–266S. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kibe, R.; Ooga, T.; Aiba, Y.; Kurihara, S.; Sawaki, E.; Koga, Y.; Benno, Y. Impact of intestinal microbiota on intestinal luminal metabolome. Sci. Rep. 2012, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef]
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Pomare, E.W.; Branch, H.W.J.; Naylor, C.P.E.; MacFarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef]
- Sivaprakasam, S.; Bhutia, Y.D.; Yang, S.; Ganapathy, V. Short-chain fatty acid transporters: Role in colonic homeostasis. In Comprehensive Physiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; Volume 8, pp. 299–314. [Google Scholar]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef]
- Schönfeld, P.; Wojtczak, L. Short- and medium-chain fatty acids in energy metabolism: The cellular perspective. J. Lipid Res. 2016, 57, 943–954. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Gerhauser, C. Impact of dietary gut microbial metabolites on the epigenome. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170359. [Google Scholar] [CrossRef]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef]
- Le Poul, E.; Loison, C.; Struyf, S.; Springael, J.Y.; Lannoy, V.; Decobecq, M.E.; Brezillon, S.; Dupriez, V.; Vassart, G.; Van Damme, J.; et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 2003, 278, 25481–25489. [Google Scholar] [CrossRef] [PubMed]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. GPFM 09A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A.R. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef]
- Sina, C.; Gavrilova, O.; Förster, M.; Till, A.; Derer, S.; Hildebrand, F.; Raabe, B.; Chalaris, A.; Scheller, J.; Rehmann, A.; et al. G Protein-Coupled Receptor 43 Is Essential for Neutrophil Recruitment during Intestinal Inflammation. J. Immunol. 2009, 183, 7514–7522. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Ryu, S.H.; Koues, O.I.; Collins, P.L.; Solnica-Krezel, L.; Pearce, E.J.; Pearce, E.L.; Oltz, E.M.; Stappenbeck, T.S. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 2016, 165, 1708–1720. [Google Scholar] [CrossRef]
- Kelly, C.J.; Zheng, L.; Campbell, E.L.; Saeedi, B.; Scholz, C.C.; Bayless, A.J.; Wilson, K.E.; Glover, L.E.; Kominsky, D.J.; Magnuson, A.; et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 2015, 17, 662–671. [Google Scholar] [CrossRef]
- Tedelind, S.; Westberg, F.; Kjerrulf, M.; Vidal, A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: A study with relevance to inflammatory bowel disease. World J. Gastroenterol. 2007, 13, 2826–2832. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; Deroos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Yang, W.; Yu, T.; Huang, X.; Bilotta, A.J.; Xu, L.; Lu, Y.; Sun, J.; Pan, F.; Zhou, J.; Zhang, W.; et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gurav, A.; Sivaprakasam, S.; Bhutia, Y.D.; Boettger, T.; Singh, N.; Ganapathy, V. Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem. J. 2015, 469, 267–278. [Google Scholar] [CrossRef]
- Hui, W.; Yu, D.; Cao, Z.; Zhao, X. Butyrate inhibit collagen-induced arthritis via Treg/IL-10/Th17 axis. Int. Immunopharmacol. 2019, 68, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, L.; Min, J.; Wang, J.; Wu, H.; Zeng, Y.; Chen, S.; Chu, Z. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell. Immunol. 2012, 277, 66–73. [Google Scholar] [CrossRef]
- Singh, N.; Thangaraju, M.; Prasad, P.D.; Martin, P.M.; Lambert, N.A.; Boettger, T.; Offermanns, S.; Ganapathy, V. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J. Biol. Chem. 2010, 285, 27601–27608. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445.e7. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Lutgendorff, F.; Phan, V.; Söderholm, J.D.; Sherman, P.M.; McKay, D.M. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm. Bowel Dis. 2010, 16, 1138–1148. [Google Scholar] [CrossRef]
- Venegas, D.P.; De La Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Mirmonsef, P.; Zariffard, M.R.; Gilbert, D.; Makinde, H.; Landay, A.L.; Spear, G.T. Short-Chain Fatty Acids Induce Pro-Inflammatory Cytokine Production Alone and in Combination with Toll-Like Receptor Ligands. Am. J. Reprod. Immunol. 2012, 67, 391–400. [Google Scholar] [CrossRef]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 1–15. [Google Scholar] [CrossRef]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406.e10. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015, 8, 80–93. [Google Scholar] [CrossRef]
- Belenguer, A.; Duncan, S.H.; Holtrop, G.; Anderson, S.E.; Lobley, G.E.; Flint, H.J. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl. Environ. Microbiol. 2007, 73, 6526–6533. [Google Scholar] [CrossRef]
- Belenguer, A.; Holtrop, G.; Duncan, S.H.; Anderson, S.E.; Calder, A.G.; Flint, H.J.; Lobley, G.E. Rates of productionand utilization of lactate by microbial communities fromthe human colon. FEMS Microbiol. Ecol. 2011, 77, 107–119. [Google Scholar] [CrossRef]
- Iraporda, C.; Errea, A.; Romanin, D.E.; Cayet, D.; Pereyra, E.; Pignataro, O.; Sirard, J.C.; Garrote, G.L.; Abraham, A.G.; Rumbo, M. Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology 2015, 220, 1161–1169. [Google Scholar] [CrossRef]
- Hove, H.; Nordgaard-Andersen, I.; Mortensen, P.B. Faecal DL-lactate concentration in 100 gastrointestinal patients. Scand. J. Gastroenterol. 1994, 29, 255–259. [Google Scholar] [CrossRef]
- Vernia, P.; Caprilli, R.; Latella, G.; Barbetti, F.; Magliocca, F.M.; Cittadini, M. Fecal Lactate and Ulcerative Colitis. Gastroenterology 1988, 95, 1564–1568. [Google Scholar] [CrossRef]
- Wang, S.P.; Rubio, L.A.; Duncan, S.H.; Donachie, G.E.; Holtrop, G.; Lo, G.; Farquharson, F.M.; Wagner, J.; Parkhill, J.; Louis, P.; et al. Pivotal Roles for pH, Lactate, and Lactate-Utilizing Bacteria in the Stability of a Human Colonic Microbial Ecosystem. mSystems 2020, 5, 645–665. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Fan, J.; Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2021, 19, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.; Shanmugam, A.; Swafford, D.; Suryawanshi, A.; Bhattacharjee, P.; Hussein, M.S.; Koni, P.A.; Prasad, P.D.; Kurago, Z.B.; Thangaraju, M.; et al. GPR81, a Cell-Surface Receptor for Lactate, Regulates Intestinal Homeostasis and Protects Mice from Experimental Colitis. J. Immunol. 2018, ji1700604. [Google Scholar] [CrossRef] [PubMed]
- Errea, A.; Cayet, D.; Marchetti, P.; Tang, C.; Kluza, J.; Offermanns, S.; Sirard, J.-C.; Rumbo, M. Lactate Inhibits the Pro-Inflammatory Response and Metabolic Reprogramming in Murine Macrophages in a GPR81-Independent Manner. PLoS ONE 2016, 11, e0163694. [Google Scholar] [CrossRef] [PubMed]
- SHAN, T.; CHEN, S.; CHEN, X.; WU, T.; YANG, Y.; LI, S.; MA, J.; ZHAO, J.; LIN, W.; LI, W.; et al. M2-TAM subsets altered by lactic acid promote T-cell apoptosis through the PD-L1/PD-1 pathway. Oncol. Rep. 2020, 44, 1885–1894. [Google Scholar] [CrossRef] [PubMed]
- Dietl, K.; Renner, K.; Dettmer, K.; Timischl, B.; Eberhart, K.; Dorn, C.; Hellerbrand, C.; Kastenberger, M.; Kunz-Schughart, L.A.; Oefner, P.J.; et al. Lactic Acid and Acidification Inhibit TNF Secretion and Glycolysis of Human Monocytes. J. Immunol. 2010, 184, 1200–1209. [Google Scholar] [CrossRef] [PubMed]
- Haas, R.; Smith, J.; Rocher-Ros, V.; Nadkarni, S.; Montero-Melendez, T.; D’Acquisto, F.; Bland, E.J.; Bombardieri, M.; Pitzalis, C.; Perretti, M.; et al. Lactate regulates metabolic and proinflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 2015, 13, e1002202. [Google Scholar] [CrossRef] [PubMed]
- Angelin, A.; Gil-de-Gómez, L.; Dahiya, S.; Jiao, J.; Guo, L.; Levine, M.H.; Wang, Z.; Quinn, W.J.; Kopinski, P.K.; Wang, L.; et al. Foxp3 Reprograms T Cell Metabolism to Function in Low-Glucose, High-Lactate Environments. Cell Metab. 2017, 25, 1282–1293.e7. [Google Scholar] [CrossRef]
- Upadhyay, D.; Singh, A.; Das, P.; Mehtab, J.; Dattagupta, S.; Ahuja, V.; Makharia, G.K.; Jagannathan, N.R.; Sharma, U. Abnormalities in metabolic pathways in celiac disease investigated by the metabolic profiling of small intestinal mucosa, blood plasma and urine by NMR spectroscopy. NMR Biomed. 2020, 33, e4305. [Google Scholar] [CrossRef]
- Faith, J.J.; Ahern, P.P.; Ridaura, V.K.; Cheng, J.; Gordon, J.I. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 2014, 6, 220ra11. [Google Scholar] [CrossRef]
- Meijer-Severs, G.J.; Van Santen, E. Short-chain fatty acids and succinate in feces of healthy human volunteers and their correlation with anaerobe cultural counts. Scand. J. Gastroenterol. 1987, 22, 672–676. [Google Scholar] [CrossRef]
- Macias-Ceja, D.C.; Ortiz-Masiá, D.; Salvador, P.; Gisbert-Ferrándiz, L.; Hernández, C.; Hausmann, M.; Rogler, G.; Esplugues, J.V.; Hinojosa, J.; Alós, R.; et al. Succinate receptor mediates intestinal inflammation and fibrosis. Mucosal Immunol. 2019, 12, 178–187. [Google Scholar] [CrossRef]
- Littlewood-Evans, A.; Sarret, S.; Apfel, V.; Loesle, P.; Dawson, J.; Zhang, J.; Muller, A.; Tigani, B.; Kneuer, R.; Patel, S.; et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J. Exp. Med. 2016, 213, 1655–1662. [Google Scholar] [CrossRef]
- Pålsson-McDermott, E.M.; O’Neill, L.A.J. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020, 30, 300–314. [Google Scholar] [CrossRef]
- Rubic, T.; Lametschwandtner, G.; Jost, S.; Hinteregger, S.; Kund, J.; Carballido-Perrig, N.; Schwärzler, C.; Junt, T.; Voshol, H.; Meingassner, J.G.; et al. Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat. Immunol. 2008, 9, 1261–1269. [Google Scholar] [CrossRef]
- Saraiva, A.L.; Veras, F.P.; Peres, R.S.; Talbot, J.; De Lima, K.A.; Luiz, J.P.; Carballido, J.M.; Cunha, T.M.; Cunha, F.Q.; Ryffel, B.; et al. Succinate receptor deficiency attenuates arthritis by reducing dendritic cell traffic and expansion of Th17 cells in the lymph nodes. FASEB J. 2018, 32, 6550–6558. [Google Scholar] [CrossRef]
- Nadjsombati, M.S.; McGinty, J.W.; Lyons-Cohen, M.R.; Jaffe, J.B.; DiPeso, L.; Schneider, C.; Miller, C.N.; Pollack, J.L.; Nagana Gowda, G.A.; Fontana, M.F.; et al. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 2018, 49, 33–41.e7. [Google Scholar] [CrossRef]
- Schneider, C.; O’Leary, C.E.; von Moltke, J.; Liang, H.E.; Ang, Q.Y.; Turnbaugh, P.J.; Radhakrishnan, S.; Pellizzon, M.; Ma, A.; Locksley, R.M. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 2018, 174, 271–284.e14. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Ren, W.; Ohmoto, M.; Urban, J.F.; Matsumoto, I.; Margolskee, R.F.; Jiang, P. Activation of intestinal tuft cell-expressed sucnr1 triggers type 2 immunity in the mouse small intestine. Proc. Natl. Acad. Sci. USA 2018, 115, 5552–5557. [Google Scholar] [CrossRef]
- Jakobsdottir, G.; Xu, J.; Molin, G.; Ahrné, S.; Nyman, M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS ONE 2013, 8, e80476. [Google Scholar] [CrossRef] [PubMed]
- Ooi, M.; Nishiumi, S.; Yoshie, T.; Shiomi, Y.; Kohashi, M.; Fukunaga, K.; Nakamura, S.; Matsumoto, T.; Hatano, N.; Shinohara, M.; et al. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm. Res. 2011, 60, 831–840. [Google Scholar] [CrossRef]
- Connors, J.; Dawe, N.; Van Limbergen, J. The role of succinate in the regulation of intestinal inflammation. Nutrients 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Wolak, N.; Zawrotniak, M.; Gogol, M.; Kozik, A.; Rapala-Kozik, M. Vitamins B1, B2, B3 and B9—Occurrence, Biosynthesis Pathways and Functions in Human Nutrition. Mini-Rev. Med. Chem. 2016, 17, 1075–1111. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Ravcheev, D.; De Crécy-Lagard, V.; Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests cooperation among gut microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef] [PubMed]
- Putnam, E.E.; Goodman, A.L. B vitamin acquisition by gut commensal bacteria. PLoS Pathog. 2020, 16, e1008208. [Google Scholar] [CrossRef]
- Rodionov, D.A.; Arzamasov, A.A.; Khoroshkin, M.S.; Iablokov, S.N.; Leyn, S.A.; Peterson, S.N.; Novichkov, P.S.; Osterman, A.L. Micronutrient requirements and sharing capabilities of the human gut microbiome. Front. Microbiol. 2019, 10, 1316. [Google Scholar] [CrossRef]
- Radjabzadeh, D.; Boer, C.G.; Beth, S.A.; van der Wal, P.; Kiefte-De Jong, J.C.; Jansen, M.A.E.; Konstantinov, S.R.; Peppelenbosch, M.P.; Hays, J.P.; Jaddoe, V.W.V.; et al. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Costea, P.I.; Hildebrand, F.; Manimozhiyan, A.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; De Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2017, 3, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, K.; Hosomi, K.; Sawane, K.; Kunisawa, J. Metabolism of dietary and microbial vitamin b family in the regulation of host immunity. Front. Nutr. 2019, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Nabokina, S.M.; Said, H.M. A high-affinity and specific carrier-mediated mechanism for uptake of thiamine pyrophosphate by human colonic epithelial cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 2012, 303, G389–G395. [Google Scholar] [CrossRef] [PubMed]
- Frank, R.A.W.; Leeper, F.J.; Luisi, B.F. Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell. Mol. Life Sci. 2007, 64, 892–905. [Google Scholar] [CrossRef]
- Ashokkumar, B.; Kumar, J.S.; Hecht, G.A.; Said, H.M. Enteropathogenic Escherichia coli inhibits intestinal vitamin B1 (thiamin) uptake: Studies with human-derived intestinal epithelial Caco-2 cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 2009, 297, 825–833. [Google Scholar] [CrossRef]
- Subramanya, S.B.; Subramanian, V.S.; Said, H.M. Chronic alcohol consumption and intestinal thiamin absorption: Effects on physiological and molecular parameters of the uptake process. Am. J. Physiol.-Gastrointest. Liver Physiol. 2010, 299, G23–G31. [Google Scholar] [CrossRef] [PubMed]
- Seligmann, H.; Levi, R.; Konijn, A.M.; Prokocimer, M. Thiamine deficiency in patients with B-chronic lymphocytic leukaemia: A pilot study. Postgrad. Med. J. 2001, 77, 582–585. [Google Scholar] [CrossRef]
- Müri, R.M.; Von Overbeck, J.; Furrer, J.; Ballmer, P.E. Thiamin deficiency in HIV-positive patients: Evaluation by erythrocyte transketolase activity and thiamin pyrophosphate effect. Clin. Nutr. 1999, 18, 375–378. [Google Scholar] [CrossRef]
- Ottinger, C.A.; Honeyfield, D.C.; Densmore, C.L.; Iwanowicz, L.R. Impact of thiamine deficiency on T-cell dependent and T-cell independent antibody production in lake trout. J. Aquat. Anim. Health 2012, 24, 258–273. [Google Scholar] [CrossRef]
- de Moreno de LeBlanc, A.; Levit, R.; de Giori, G.S.; LeBlanc, J.G. Vitamin Producing Lactic Acid Bacteria as Complementary Treatments for Intestinal Inflammation. Antiinflamm. Antiallergy Agents Med. Chem. 2018, 17, 50–56. [Google Scholar] [CrossRef][Green Version]
- Li, L.; Krause, L.; Somerset, S. Associations between micronutrient intakes and gut microbiota in a group of adults with cystic fibrosis. Clin. Nutr. 2017, 36, 1097–1104. [Google Scholar] [CrossRef]
- Ghashut, R.A.; McMillan, D.C.; Kinsella, J.; Talwar, D. Erythrocyte concentrations of B1, B2, B6 but not plasma C and E are reliable indicators of nutrition status in the presence of systemic inflammation. Clin. Nutr. ESPEN 2017, 17, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Bishayi, B. Riboflavin along with antibiotics balances reactive oxygen species and inflammatory cytokines and controls Staphylococcus aureus infection by boosting murine macrophage function and regulates inflammation. J. Inflamm. 2016, 13, 1–21. [Google Scholar] [CrossRef]
- Mazur-Bialy, A.I.; Pocheć, E. Riboflavin reduces pro-inflammatory activation of adipocyte-macrophage co-culture. Potential application of vitamin B2 enrichment for attenuation of insulin resistance and metabolic syndrome development. Molecules 2016, 21, 1724. [Google Scholar] [CrossRef]
- Qureshi, A.A.; Tan, X.; Reis, J.C.; Badr, M.Z.; Papasian, C.J.; Morrison, D.C.; Qureshi, N. Suppression of nitric oxide induction and pro-inflammatory cytokines by novel proteasome inhibitors in various experimental models. Lipids Health Dis. 2011, 10, 1–25. [Google Scholar] [CrossRef]
- Tastan, C.; Karhan, E.; Zhou, W.; Fleming, E.; Voigt, A.Y.; Yao, X.; Wang, L.; Horne, M.; Placek, L.; Kozhaya, L.; et al. Tuning of human MAIT cell activation by commensal bacteria species and MR1-dependent T-cell presentation. Mucosal Immunol. 2018, 11, 1591–1605. [Google Scholar] [CrossRef]
- Kumar, V.; Ahmad, A. Role of MAIT cells in the immunopathogenesis of inflammatory diseases: New players in old game. Int. Rev. Immunol. 2018, 37, 90–110. [Google Scholar] [CrossRef]
- Von Martels, J.Z.H.; Bourgonje, A.R.; Klaassen, M.A.Y.; Alkhalifah, H.A.A.; Sadaghian Sadabad, M.; Vich Vila, A.; Gacesa, R.; Gabriëls, R.Y.; Steinert, R.E.; Jansen, B.H.; et al. Riboflavin Supplementation in Patients with Crohn’s Disease [the RISE-UP study]. J. Crohn’s Colitis 2020, 14, 595–607. [Google Scholar] [CrossRef]
- Mohedano, M.L.; Hernández-Recio, S.; Yépez, A.; Requena, T.; Martínez-Cuesta, M.C.; Peláez, C.; Russo, P.; LeBlanc, J.G.; Spano, G.; Aznar, R.; et al. Real-time detection of riboflavin production by Lactobacillus plantarum strains and tracking of their gastrointestinal survival and functionality in vitro and in vivo using mCherry labeling. Front. Microbiol. 2019, 10, 1748. [Google Scholar] [CrossRef]
- Shats, I.; Williams, J.G.; Liu, J.; Makarov, M.V.; Wu, X.; Lih, F.B.; Deterding, L.J.; Lim, C.; Xu, X.; Randall, T.A.; et al. Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway. Cell Metab. 2020, 31, 564–579.e7. [Google Scholar] [CrossRef]
- Cetina Biefer, H.R.; Vasudevan, A.; Elkhal, A. Aspects of tryptophan and nicotinamide adenine dinucleotide in immunity: A new twist in an old tale. Int. J. Tryptophan Res. 2017, 10, 117864691771349. [Google Scholar] [CrossRef]
- Rajman, L.; Chwalek, K.; Sinclair, D.A. Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence. Cell Metab. 2018, 27, 529–547. [Google Scholar] [CrossRef]
- Blacher, E.; Bashiardes, S.; Shapiro, H.; Rothschild, D.; Mor, U.; Dori-Bachash, M.; Kleimeyer, C.; Moresi, C.; Harnik, Y.; Zur, M.; et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 2019, 572, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Nakanishi, Y.; Murakami, S.; Aw, W.; Tsukimi, T.; Nozu, R.; Ueno, M.; Hioki, K.; Nakahigashi, K.; Hirayama, A.; et al. A Metabolomic-Based Evaluation of the Role of Commensal Microbiota throughout the Gastrointestinal Tract in Mice. Microorganisms 2018, 6, 101. [Google Scholar] [CrossRef]
- Montserrat- de la Paz, S.; Naranjo, M.C.; Lopez, S.; Abia, R.; Muriana, F.J.G.; Bermudez, B. Niacin and its metabolites as master regulators of macrophage activation. J. Nutr. Biochem. 2017, 39, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.; Zheng, D.; Zhang, L.; Ni, R.; Wang, G.; Fan, G.C.; Lu, Z.; Peng, T. Administration of nicotinamide riboside prevents oxidative stress and organ injury in sepsis. Free Radic. Biol. Med. 2018, 123, 125–137. [Google Scholar] [CrossRef]
- Ganji, S.H.; Kamanna, V.S.; Kashyap, M.L. Niacin decreases leukocyte myeloperoxidase: Mechanistic role of redox agents and Src/p38MAP kinase. Atherosclerosis 2014, 235, 554–561. [Google Scholar] [CrossRef]
- Ferreira, R.G.; Matsui, T.C.; Godin, A.M.; Gomides, L.F.; Pereira-Silva, P.E.M.; Duarte, I.D.G.; Menezes, G.; Coelho, M.M.; Klein, A. Neutrophil recruitment is inhibited by nicotinamide in experimental pleurisy in mice. Eur. J. Pharmacol. 2012, 685, 198–204. [Google Scholar] [CrossRef][Green Version]
- Bhatt, B.; Zeng, P.; Zhu, H.; Sivaprakasam, S.; Li, S.; Xiao, H.; Dong, L.; Shiao, P.; Kolhe, R.; Patel, N.; et al. Gpr109a Limits Microbiota-Induced IL-23 Production To Constrain ILC3-Mediated Colonic Inflammation. J. Immunol. 2018, 200, 2905–2914. [Google Scholar] [CrossRef]
- Nitto, T.; Onodera, K. The linkage between coenzyme A metabolism and inflammation: Roles of pantetheinase. J. Pharmacol. Sci. 2013, 123, 1–8. [Google Scholar] [CrossRef]
- He, W.; Hu, S.; Du, X.; Wen, Q.; Zhong, X.P.; Zhou, X.; Zhou, C.; Xiong, W.; Gao, Y.; Zhang, S.; et al. Vitamin B5 reduces bacterial growth via regulating innate immunity and adaptive immunity in mice infected with Mycobacterium tuberculosis. Front. Immunol. 2018, 9, 365. [Google Scholar] [CrossRef]
- Ghosal, A.; Lambrecht, N.; Subramanya, S.B.; Kapadia, R.; Said, H.M. Conditional knockout of the Slc5a6 gene in mouse intestine impairs biotin absorption. Am. J. Physiol.-Gastrointest. Liver Physiol. 2013, 304, G64–G71. [Google Scholar] [CrossRef]
- Sabui, S.; Kapadia, R.; Ghosal, A.; Schneider, M.; Lambrecht, N.W.G.; Said, H.M. Biotin and pantothenic acid oversupplementation to conditional SLC5A6 KO mice prevents the development of intestinal mucosal abnormalities and growth defects. Am. J. Physiol.-Cell Physiol. 2018, 315, C73–C79. [Google Scholar] [CrossRef]
- Spady, M. Vitamin B6 Deficiency. AJN, Am. J. Nurs. 1960, 60, 83. [Google Scholar] [CrossRef]
- Rosenberg, J.; Ischebeck, T.; Commichau, F.M. Vitamin B6 metabolism in microbes and approaches for fermentative production. Biotechnol. Adv. 2017, 35, 31–40. [Google Scholar] [CrossRef]
- Rosenberg, J.; Yeak, K.Y.C.; Commichau, F.M. A two-step evolutionary process establishes a non-native vitamin B6 pathway in Bacillus subtilis. Environ. Microbiol. 2018, 20, 156–168. [Google Scholar] [CrossRef]
- Nirmagustina, D.E.; Yang, Y.; Kumrungsee, T.; Yanaka, N.; Kato, N. Gender difference and dietary supplemental vitamin B6: Impact on colon luminal environment. J. Nutr. Sci. Vitaminol. 2018, 64, 116–128. [Google Scholar] [CrossRef]
- Zheng, X.; Feng, L.; Jiang, W.D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.Y.; Tang, L.; Tang, W.N.; Zhang, Y.A.; et al. Dietary pyridoxine deficiency reduced growth performance and impaired intestinal immune function associated with TOR and NF-κB signalling of young grass carp (Ctenopharyngodon idella). Fish Shellfish. Immunol. 2017, 70, 682–700. [Google Scholar] [CrossRef]
- Roubenoff, R.; Roubenoff, R.A.; Selhub, J.; Nadeau, M.R.; Cannon, J.G.; Freeman, L.M.; Dinarello, C.A.; Rosenberg, I.H. Abnormal vitamin b6 status in rheumatoid cachexia association with spontaneous tumor necrosis factor α production and markers of inflammation. Arthritis Rheum. 1995, 38, 105–109. [Google Scholar] [CrossRef]
- Chiang, E.P.I.; Selhub, J.; Bagley, P.J.; Dallal, G.; Roubenoff, R. Pyridoxine supplementation corrects vitamin B6 deficiency but does not improve inflammation in patients with rheumatoid arthritis. Arthritis Res. Ther. 2005, 7, R1404. [Google Scholar] [CrossRef]
- Molina-López, J.; Florea, D.; Quintero-Osso, B.; de la Cruz, A.P.; Rodríguez-Elvira, M.; del Pozo, E.P. Pyridoxal-5′-phosphate deficiency is associated with hyperhomocysteinemia regardless of antioxidant, thiamine, riboflavin, cobalamine, and folate status in critically ill patients. Clin. Nutr. 2016, 35, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Dusitanond, P.; Eikelboom, J.W.; Hankey, G.J.; Thom, J.; Gilmore, G.; Loh, K.; Yi, Q.; Klijn, C.J.M.; Langton, P.; Van Bockxmeer, F.M.; et al. Homocysteine-lowering treatment with folic acid, cobalamin, and pyridoxine does not reduce blood markers of inflammation, endothelial dysfunction, or hypercoagulability in patients with previous transient ischemic attack or stroke: A randomized substudy of the Vitatops. Stroke 2005, 36, 144–146. [Google Scholar] [CrossRef]
- Selhub, J.; Byun, A.; Liu, Z.; Mason, J.B.; Bronson, R.T.; Crott, J.W. Dietary vitamin B6 intake modulates colonic inflammation in the IL10-/- model of inflammatory bowel disease. J. Nutr. Biochem. 2013, 24, 2138–2143. [Google Scholar] [CrossRef]
- Zhang, P.; Tsuchiya, K.; Kinoshita, T.; Kushiyama, H.; Suidasari, S.; Hatakeyama, M.; Imura, H.; Kat, N.; Suda, T. Vitamin B6 Prevents IL-1β Protein Production by Inhibiting NLRP3 Inflammasome Activation. J. Biol. Chem. 2016, 291, 24517–24527. [Google Scholar] [CrossRef]
- Qian, B.; Shen, S.; Zhang, J.; Jing, P. Effects of Vitamin B6 Deficiency on the Composition and Functional Potential of T Cell Populations. J. Immunol. Res. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, C.; Kurohane, K.; Imai, Y. High dose dietary pyridoxine induces T-helper type 1 polarization and decreases contact hypersensitivity response to fluorescein isothiocyanate in mice. Biol. Pharm. Bull. 2012, 35, 532–538. [Google Scholar] [CrossRef]
- Katunuma, N.; Matsui, A.; Endo, K.; Hanba, J.; Sato, A.; Nakano, M.; Yuto, Y.; Tada, Y.; Asao, T.; Himeno, K.; et al. Inhibition of intracellular cathepsin activities and suppression of immune responses mediated by helper T lymphocyte type-2 by peroral or intraperitoneal administration of vitamin B6. Biochem. Biophys. Res. Commun. 2000, 272, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Kumrungsee, T.; Zhang, P.; Chartkul, M.; Yanaka, N.; Kato, N. Potential Role of Vitamin B6 in Ameliorating the Severity of COVID-19 and Its Complications. Front. Nutr. 2020, 7, 562051. [Google Scholar] [CrossRef]
- León-Del-Río, A. Biotin in metabolism, gene expression, and human disease. J. Inherit. Metab. Dis. 2019, 42, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Kuroishi, T. Regulation of immunological and inflammatory functions by biotin. Can. J. Physiol. Pharmacol. 2015, 93, 1091–1096. [Google Scholar] [CrossRef]
- Skupsky, J.; Sabui, S.; Hwang, M.; Nakasaki, M.; Cahalan, M.D.; Said, H.M. Biotin Supplementation Ameliorates Murine Colitis by Preventing NF-κB Activation. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 557–567. [Google Scholar] [CrossRef]
- Elahi, A.; Sabui, S.; Narasappa, N.N.; Agrawal, S.; Lambrecht, N.W.; Agrawal, A.; Said, H.M. Biotin Deficiency Induces Th1- and Th17-Mediated Proinflammatory Responses in Human CD4 + T Lymphocytes via Activation of the mTOR Signaling Pathway. J. Immunol. 2018, 200, 2563–2570. [Google Scholar] [CrossRef] [PubMed]
- Valentini, L.; Pinto, A.; Bourdel-Marchasson, I.; Ostan, R.; Brigidi, P.; Turroni, S.; Hrelia, S.; Hrelia, P.; Bereswill, S.; Fischer, A.; et al. Impact of personalized diet and probiotic supplementation on inflammation, nutritional parameters and intestinal microbiota—The “RISTOMED project”: Randomized controlled trial in healthy older people. Clin. Nutr. 2015, 34, 593–602. [Google Scholar] [CrossRef]
- Morscher, R.J.; Ducker, G.S.; Li, S.H.J.; Mayer, J.A.; Gitai, Z.; Sperl, W.; Rabinowitz, J.D. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 2018, 554, 128–132. [Google Scholar] [CrossRef]
- Kapadia, C.R. Vitamin B12 in health and disease. Part I—Inherited disorders of function, absorption, and transport. Gastroenterologist 1995, 3, 329–344. [Google Scholar]
- Kok, D.E.; Steegenga, W.T.; McKay, J.A. Folate and epigenetics: Why we should not forget bacterial biosynthesis. Epigenomics 2018, 10, 1147–1150. [Google Scholar] [CrossRef]
- Bermingham, A.; Derrick, J.P. The folic acid biosynthesis pathway in bacteria: Evaluation of potential for antibacterial drug discovery. BioEssays 2002, 24, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.H.; Barg, H.; Warren, M.; Jahn, D. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 2002, 58, 275–285. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Kang, J.; Zhang, D. Microbial production of vitamin B12: A review and future perspectives. Microb. Cell Fact. 2017, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, T.B.; O’Neill, M.P.; Webb, S.A.R.; Ho, K.M. Inflammation, vitamin deficiencies and organ failure in critically ill patients. Anaesth. Intensive Care 2009, 37, 740–747. [Google Scholar] [CrossRef]
- Tal, S.; Shavit, Y.; Stern, F.; Malnick, S. Association between vitamin B12 levels and mortality in hospitalized older adults. J. Am. Geriatr. Soc. 2010, 58, 523–526. [Google Scholar] [CrossRef]
- Abe, I.; Shirato, K.; Hashizume, Y.; Mitsuhashi, R.; Kobayashi, A.; Shiono, C.; Sato, S.; Tachiyashiki, K.; Imaizumi, K. Folate-deficiency induced cell-specific changes in the distribution of lymphocytes and granulocytes in rats. Environ. Health Prev. Med. 2013, 18, 78–84. [Google Scholar] [CrossRef]
- Courtemanche, C.; Huang, A.C.; Elson-Schwab, I.; Kerry, N.; Ng, B.Y.; Ames, B.N. Folate deficiency and ionizing radiation cause DNA breaks in primary human lymphocytes: A comparison. FASEB J. 2004, 18, 209–211. [Google Scholar] [CrossRef]
- Socha-Banasiak, A.; Kamer, B.; Gach, A.; Wysocka, U.; Jakubowski, L.; Głowacka, E.; Czkwianianc, E. Folate status, regulatory T cells and MTHFR C677T polymorphism study in allergic children. Adv. Med. Sci. 2016, 61, 300–305. [Google Scholar] [CrossRef]
- Kunisawa, J.; Hashimoto, E.; Ishikawa, I.; Kiyono, H. A pivotal role of vitamin B9 in the maintenance of regulatory T cells in vitro and in vivo. PLoS ONE 2012, 7, e32094. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Hilgenbrink, A.R.; Matteson, E.L.; Lockwood, M.B.; Cheng, J.X.; Low, P.S. A functional folate receptor is induced during macrophage activation and can be used to target drugs to activated macrophages. Blood 2009, 113, 438–446. [Google Scholar] [CrossRef]
- Hilgendorf, I.; Swirski, F.K. Folate Receptor: A Macrophage “Achilles’ Heel”? J. Am. Heart Assoc. 2012, 1, e004036. [Google Scholar] [CrossRef]
- Poh, S.; Putt, K.S.; Low, P.S. Folate-Targeted Dendrimers Selectively Accumulate at Sites of Inflammation in Mouse Models of Ulcerative Colitis and Atherosclerosis. Biomacromolecules 2017, 18, 3082–3088. [Google Scholar] [CrossRef] [PubMed]
- Spiller, R. Serotonin, inflammation, and IBS: Fitting the jigsaw together? J. Pediatric Gastroenterol. Nutr. 2007, 45, S115–S119. [Google Scholar] [CrossRef] [PubMed]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef]
- Marcobal, A.; Kashyap, P.C.; Nelson, T.A.; Aronov, P.A.; Donia, M.S.; Spormann, A.; Fischbach, M.A.; Sonnenburg, J.L. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013, 7, 1933–1943. [Google Scholar] [CrossRef]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; DeLuca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, L.; Wu, T.; Li, Y.; Zhou, X.; Ruan, Z. Indole-3-propionic Acid Improved the Intestinal Barrier by Enhancing Epithelial Barrier and Mucus Barrier. J. Agric. Food Chem. 2021, 69, 1487–1495. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef] [PubMed]
- Marsland, B.J. Regulating inflammation with microbial metabolites. Nat. Med. 2016, 22, 581–583. [Google Scholar] [CrossRef]
- Huxtable, R.J. Physiological actions of taurine. Physiol. Rev. 1992, 72, 101–164. [Google Scholar] [CrossRef] [PubMed]
- Sturman, J.A. Taurine in development. Physiol. Rev. 1993, 73, 119–148. [Google Scholar] [CrossRef]
- Trachtman, H.; Barbour, R.; Sturman, J.A.; Finberg, L. Taurine and osmoregulation: Taurine is a cerebral osmoprotective molecule in chronic hypernatremic dehydration. Pediatr. Res. 1988, 23, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.F. Hepatology: A Textbook of Liver Disease. Arch. Intern. Med. 1983, 143, 2238. [Google Scholar] [CrossRef]
- Hardison, W.G.M. Hepatic Taurine Concentration and Dietary Taurine as Regulators of Bile Acid Conjugation with Taurine. Gastroenterology 1978, 75, 71–75. [Google Scholar] [CrossRef]
- Sjövall, J. Dietary Glycine and Taurine on Bile Acid Conjugation in Man. Bile Acids and Steroids 75. Proc. Soc. Exp. Biol. Med. 1959, 100, 676–678. [Google Scholar] [CrossRef]
- Fang, H.; Meng, F.; Piao, F.; Jin, B.; Li, M.; Li, W. Effect of Taurine on Intestinal Microbiota and Immune Cells in Peyer’s Patches of Immunosuppressive Mice. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2019; Volume 1155, pp. 13–24. [Google Scholar]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef] [PubMed]
- Aronov, P.A.; Luo, F.J.G.; Plummer, N.S.; Quan, Z.; Holmes, S.; Hostetter, T.H.; Meyer, T.W. Colonic contribution to uremic solutes. J. Am. Soc. Nephrol. 2011, 22, 1769–1776. [Google Scholar] [CrossRef]
- Saito, Y.; Sato, T.; Nomoto, K.; Tsuji, H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol. Ecol. 2018, 94, 125. [Google Scholar] [CrossRef]
- Gryp, T.; Vanholder, R.; Vaneechoutte, M.; Glorieux, G. P-Cresyl Sulfate. Toxins 2017, 9, 52. [Google Scholar] [CrossRef]
- Dalrymple, L.S.; Go, A.S. Epidemiology of acute infections among patients with chronic kidney disease. Clin. J. Am. Soc. Nephrol. 2008, 3, 1487–1493. [Google Scholar] [CrossRef]
- Azevedo, M.L.V.; Bonan, N.B.; Dias, G.; Brehm, F.; Steiner, T.M.; Souza, W.M.; Stinghen, A.E.M.; Barreto, F.C.; Elifio-Esposito, S.; Pecoits-Filho, R.; et al. p-Cresyl sulfate affects the oxidative burst, phagocytosis process, and antigen presentation of monocyte-derived macrophages. Toxicol. Lett. 2016, 263, 1–5. [Google Scholar] [CrossRef]
- Shiba, T.; Makino, I.; Sasaki, T.; Fukuhara, Y.; Kawakami, K.; Kato, I.; Kobayashi, T. p-Cresyl sulfate decreases peripheral B cells in mice with adenine-induced renal dysfunction. Toxicol. Appl. Pharmacol. 2018, 342, 50–59. [Google Scholar] [CrossRef]
- Shiba, T.; Kawakami, K.; Sasaki, T.; Makino, I.; Kato, I.; Kobayashi, T.; Uchida, K.; Kaneko, K. Effects of intestinal bacteria-derived p-cresyl sulfate on Th1-type immune response in vivo and in vitro. Toxicol. Appl. Pharmacol. 2014, 274, 191–199. [Google Scholar] [CrossRef]
- Vanholder, R.; Bammens, B.; De Loor, H.; Glorieux, G.; Meijers, B.; Schepers, E.; Massy, Z.; Evenepoel, P. Warning: The unfortunate end of p-cresol as a uraemic toxin. Nephrol. Dial. Transplant. 2011, 26, 1464–1467. [Google Scholar] [CrossRef] [PubMed]
- Sudo, N. Biogenic Amines: Signals Between Commensal Microbiota and Gut Physiology. Front. Endocrinol. 2019, 10, 504. [Google Scholar] [CrossRef] [PubMed]
- Jutel, M.; Watanabe, T.; Klunker, S.; Akdis, M.; Thomet, O.A.R.; Malolepszy, J.; Zak-Nejmark, T.; Koga, R.; Kobayashi, T.; Blaser, K.; et al. Histamine regulates T-cell and antibody responses by differential expression of H1 and H2 receptors. Nature 2001, 413, 420–425. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Tsutsui, H.; Tominaga, K.; Hoshino, K.; Okamura, H.; Akira, S.; Paul, W.E.; Nakanishi, K. IL-18, although antiallergic when administered with IL-12, stimulates IL-4 and histamine release by basophils. Proc. Natl. Acad. Sci. USA 1999, 96, 13962–13966. [Google Scholar] [CrossRef]
- BEAVER, M.H.; WOSTMANN, B.S. Histamine and 5-Hydroxytryptamine in the Intestinal Tract of Germ-Free Animals, Animals Harbouring One Microbial Species and Conventional Animals. Br. J. Pharmacol. Chemother. 1962, 19, 385–393. [Google Scholar] [CrossRef]
- Barcik, W.; Pugin, B.; Westermann, P.; Perez, N.R.; Ferstl, R.; Wawrzyniak, M.; Smolinska, S.; Jutel, M.; Hessel, E.M.; Michalovich, D.; et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J. Allergy Clin. Immunol. 2016, 138, 1491–1494.e7. [Google Scholar] [CrossRef] [PubMed]
- Caron, G.; Delneste, Y.; Roelandts, E.; Duez, C.; Herbault, N.; Magistrelli, G.; Bonnefoy, J.-Y.; Pestel, J.; Jeannin, P. Histamine Induces CD86 Expression and Chemokine Production by Human Immature Dendritic Cells. J. Immunol. 2001, 166, 6000–6006. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Young, H.A.; Spitzer, J.H.; Visintin, A.; Segal, D.M. Histamine regulates cytokine production in maturing dendritic cells, resulting in altered T cell polarization. J. Clin. Investig. 2001, 108, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Van Der Pouw Kraan, T.C.T.M.; Snijders, A.; Boeije, L.C.M.; De Groot, E.R.; Alewijnse, A.E.; Leurs, R.; Aarden, L.A. Histamine inhibits the production of interleukin-12 through interaction with H2 receptors. J. Clin. Investig. 1998, 102, 1866–1873. [Google Scholar] [CrossRef]
- Noubade, R.; Milligan, G.; Zachary, J.F.; Blankenhorn, E.P.; Del Rio, R.; Rincon, M.; Teuscher, C. Histamine receptor H1 is required for TCR-mediated p38 MAPK activation and optimal IFN-γ production in mice. J. Clin. Investig. 2007, 117, 3507–3518. [Google Scholar] [CrossRef]
- Barcik, W.; Pugin, B.; Brescó, M.S.; Westermann, P.; Rinaldi, A.; Groeger, D.; Van Elst, D.; Sokolowska, M.; Krawczyk, K.; Frei, R.; et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy Eur. J. Allergy Clin. Immunol. 2019, 74, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 2010, 42, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Milovic, V. Polyamines in the gut lumen: Bioavailability and biodistribution. Eur. J. Gastroenterol. Hepatol. 2001, 13, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Molina, B.; Queipo-Ortuño, M.I.; Lambertos, A.; Tinahones, F.J.; Peñafiel, R. Dietary and gut microbiota polyamines in obesity- And age-related diseases. Front. Nutr. 2019, 6, 1–15. [Google Scholar] [CrossRef]
- Nakamura, A.; Ooga, T.; Matsumoto, M. Intestinal luminal putrescine is produced by collective biosynthetic pathways of the commensal microbiome. Gut Microbes 2019, 10, 159–171. [Google Scholar] [CrossRef]
- Matsumoto, M.; Benno, Y. The relationship between microbiota and polyamine concentration in the human intestine: A pilot study. Microbiol. Immunol. 2007, 51, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Borovikova, L.V.; Wang, H.; Metz, C.; Tracey, K.J. Spermine inhibition of monocyte activation and inflammation. Mol. Med. 1999, 5, 595–605. [Google Scholar] [CrossRef]
- Zhang, M.; Caragine, T.; Wang, H.; Cohen, P.S.; Botchkina, G.; Soda, K.; Bianchi, M.; Ulrich, P.; Cerami, A.; Sherry, B.; et al. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: A counterregulatory mechanism that restrains the immune response. J. Exp. Med. 1997, 185, 1759–1768. [Google Scholar] [CrossRef]
- Zhu, S.; Ashok, M.; Li, J.; Li, W.; Yang, H.; Wang, P.; Tracey, K.J.; Sama, A.E.; Wang, H. Spermine protects mice against lethal sepsis partly by attenuating surrogate inflammatory markers. Mol. Med. 2009, 15, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Soda, K.; Dobashi, Y.; Kano, Y.; Tsujinaka, S.; Konishi, F. Polyamine-rich food decreases age-associated pathology and mortality in aged mice. Exp. Gerontol. 2009, 44, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Kibe, R.; Kurihara, S.; Sakai, Y.; Suzuki, H.; Ooga, T.; Sawaki, E.; Muramatsu, K.; Nakamura, A.; Yamashita, A.; Kitada, Y.; et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef]
- Madeo, F.; Eisenberg, T.; Pietrocola, F.; Kroemer, G. Spermidine in health and disease. Science 2018, 359, 6374. [Google Scholar] [CrossRef]
- Carriche, G.M.; Almeida, L.; Stüve, P.; Velasquez, L.; Dhillon-LaBrooy, A.; Roy, U.; Lindenberg, M.; Strowig, T.; Plaza-Sirvent, C.; Schmitz, I.; et al. Regulating T-cell differentiation through the polyamine spermidine. J. Allergy Clin. Immunol. 2021, 147, 335–348.e11. [Google Scholar] [CrossRef]
- Morón, B.; Spalinger, M.; Kasper, S.; Atrott, K.; Frey-Wagner, I.; Fried, M.; McCole, D.F.; Rogler, G.; Scharl, M. Activation of Protein Tyrosine Phosphatase Non-Receptor Type 2 by Spermidine Exerts Anti-Inflammatory Effects in Human THP-1 Monocytes and in a Mouse Model of Acute Colitis. PLoS ONE 2013, 8, e73703. [Google Scholar] [CrossRef]
- Madeo, F.; Bauer, M.A.; Carmona-Gutierrez, D.; Kroemer, G. Spermidine: A physiological autophagy inducer acting as an anti-aging vitamin in humans? Autophagy 2019, 15, 165–168. [Google Scholar] [CrossRef]
- Wirth, M.; Schwarz, C.; Benson, G.; Horn, N.; Buchert, R.; Lange, C.; Köbe, T.; Hetzer, S.; Maglione, M.; Michael, E.; et al. Effects of spermidine supplementation on cognition and biomarkers in older adults with subjective cognitive decline (SmartAge)—Study protocol for a randomized controlled trial. Alzheimer’s Res. Ther. 2019, 11, 36. [Google Scholar] [CrossRef]
- Wolosker, H.; Dumin, E.; Balan, L.; Foltyn, V.N. D-amino acids in the brain: D-serine in neurotransmission and neurodegeneration. FEBS J. 2008, 275, 3514–3526. [Google Scholar] [CrossRef]
- Cava, F.; Lam, H.; De Pedro, M.A.; Waldor, M.K. Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cell. Mol. Life Sci. 2011, 68, 817–831. [Google Scholar] [CrossRef]
- Sasabe, J.; Miyoshi, Y.; Rakoff-Nahoum, S.; Zhang, T.; Mita, M.; Davis, B.M.; Hamase, K.; Waldor, M.K. Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat. Microbiol. 2016, 1, 1–7. [Google Scholar] [CrossRef]
- Sasabe, J.; Suzuki, M. Emerging role of D-Amino acid metabolism in the innate defense. Front. Microbiol. 2018, 9, 933. [Google Scholar] [CrossRef]
- Yunes, R.A.; Poluektova, E.U.; Dyachkova, M.S.; Klimina, K.M.; Kovtun, A.S.; Averina, O.V.; Orlova, V.S.; Danilenko, V.N. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 2016, 42, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, S.; Oda, S.; Tsuboi, Y.; Hyeon, G.K.; Oshida, M.; Kumagai, H.; Suzuki, H. γ-glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 2008, 283, 19981–19990. [Google Scholar] [CrossRef] [PubMed]
- Barrett, E.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Ooga, T.; Kibe, R.; Aiba, Y.; Koga, Y.; Benno, Y. Colonic absorption of low-molecular-weight metabolites influenced by the intestinal microbiome: A pilot study. PLoS ONE 2017, 12, e0169207. [Google Scholar] [CrossRef]
- Ren, W.; Yin, J.; Xiao, H.; Chen, S.; Liu, G.; Tan, B.; Li, N.; Peng, Y.; Li, T.; Zeng, B.; et al. Intestinal microbiota-derived GABA mediates interleukin-17 expression during enterotoxigenic Escherichia coli infection. Front. Immunol. 2017, 7, 685. [Google Scholar] [CrossRef]
- Braun, H.S.; Sponder, G.; Pieper, R.; Aschenbach, J.R.; Deiner, C. GABA selectively increases mucin-1 expression in isolated pig jejunum. Genes Nutr. 2015, 10, 1–8. [Google Scholar] [CrossRef]
- Bajic, S.S.; Djokic, J.; Dinic, M.; Veljovic, K.; Golic, N.; Mihajlovic, S.; Tolinacki, M. GABA-producing natural dairy isolate from artisanal zlatar cheese attenuates gut inflammation and strengthens gut epithelial barrier in vitro. Front. Microbiol. 2019, 10, 527. [Google Scholar] [CrossRef]
- Thompson, J.A.; Oliveira, R.A.; Xavier, K.B. Chemical conversations in the gut microbiota. Gut Microbes 2016, 7, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Bhatt, V.S. Quorum sensing mechanisms in gram positive bacteria. In Implication of Quorum Sensing System in Biofilm Formation and Virulence; Springer: Singapore, 2019; pp. 297–311. ISBN 9789811324291. [Google Scholar]
- Xavier, K.B.; Bassler, B.L. Interference with AI-2-mediated bacterial cell-cell communication. Nature 2005, 437, 750–753. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.A.; Oliveira, R.A.; Djukovic, A.; Ubeda, C.; Xavier, K.B. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015, 10, 1861–1871. [Google Scholar] [CrossRef]
- Li, Q.; Ren, Y.; Fu, X. Inter-kingdom signaling between gut microbiota and their host. Cell. Mol. Life Sci. 2019, 76, 2383–2389. [Google Scholar] [CrossRef] [PubMed]
- Pundir, P.; Liu, R.; Vasavda, C.; Serhan, N.; Limjunyawong, N.; Yee, R.; Zhan, Y.; Dong, X.; Wu, X.; Zhang, Y.; et al. A Connective Tissue Mast-Cell-Specific Receptor Detects Bacterial Quorum-Sensing Molecules and Mediates Antibacterial Immunity. Cell Host Microbe 2019, 26, 114–122.e8. [Google Scholar] [CrossRef]
- Li, Q.; Peng, W.; Wu, J.; Wang, X.; Ren, Y.; Li, H.; Peng, Y.; Tang, X.; Fu, X. Autoinducer-2 of gut microbiota, a potential novel marker for human colorectal cancer, is associated with the activation of TNFSF9 signaling in macrophages. Oncoimmunology 2019, 8, e1626192. [Google Scholar] [CrossRef]
- Goldstein, D.S.; Eisenhofer, G.; Kopin, I.J. Sources and significance of plasma levels of catechols and their metabolites in humans. J. Pharmacol. Exp. Ther. 2003, 305, 800–811. [Google Scholar] [CrossRef]
- Flierl, M.A.; Rittirsch, D.; Nadeau, B.A.; Chen, A.J.; Sarma, J.V.; Zetoune, F.S.; McGuire, S.R.; List, R.P.; Day, D.E.; Hoesel, L.M.; et al. Phagocyte-derived catecholamines enhance acute inflammatory injury. Nature 2007, 449, 721–725. [Google Scholar] [CrossRef]
- Costa, M.; Brookes, S.J.H.; Hennig, G.W. Anatomy and physiology of the enteric nervous system. Gut 2000, 47, iv15–iv19. [Google Scholar] [CrossRef]
- Eisenhofer, G.; Aneman, Å.; Friberg, P.; Hooper, D.; Fåndriks, L.; Lonroth, H.; Hunyady, B.; Mezey, E. Substantial production of Dopamine in the human gastrointestinal tract. J. Clin. Endocrinol. Metab. 1997, 82, 3864–3871. [Google Scholar] [CrossRef] [PubMed]
- Furness, J.B. The Enteric Nervous System. Available online: https://www.wiley.com/en-us/The+Enteric+Nervous+System-p-9781405133760 (accessed on 14 May 2021).
- Sandrini, S.; Aldriwesh, M.; Alruways, M.; Freestone, P. Microbial endocrinology: Host-bacteria communication within the gut microbiome. J. Endocrinol. 2015, 225, R21–R34. [Google Scholar] [CrossRef]
- Patel, P.; Nankova, B.B.; LaGamma, E.F. Butyrate, a gut-derived environmental signal, regulates tyrosine hydroxylase gene expression via a novel promoter element. Dev. Brain Res. 2005, 160, 53–62. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Klimova, S.Y.; Cherdyntseva, T.A.; Netrusov, A.I. Hormones and hormone-like substances of microorganisms: A review. Appl. Biochem. Microbiol. 2006, 42, 229–235. [Google Scholar] [CrossRef]
- Asano, Y.; Hiramoto, T.; Nishino, R.; Aiba, Y.; Kimura, T.; Yoshihara, K.; Koga, Y.; Sudo, N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol.-Gastrointest. Liver Physiol. 2012, 303, 1288–1295. [Google Scholar] [CrossRef]
- Eisenhofer, G.; Kopin, I.J.; Goldstein, D.S. Catecholamine metabolism: A contemporary view with implications for physiology and medicine. Pharmacol. Rev. 2004, 56, 331–349. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M.; Ernst, S. Alpha and beta adrenergic receptor involvement in catecholamine-induced growth of gram-negative bacteria. Biochem. Biophys. Res. Commun. 1993, 190, 447–452. [Google Scholar] [CrossRef]
- Lyte, M.; Ernst, S. Catecholamine induced growth of gram negative bacteria. Life Sci. 1992, 50, 203–212. [Google Scholar] [CrossRef]
- Sperandio, V.; Torres, A.G.; Jarvis, B.; Nataro, J.P.; Kaper, J.B. Bacteria-host communication: The language of hormones. Proc. Natl. Acad. Sci. USA 2003, 100, 8951–8956. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.B.; Hughes, D.T.; Zhu, C.; Boedeker, E.C.; Sperandio, V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 10420–10425. [Google Scholar] [CrossRef]
- Bearson, B.L.; Bearson, S.M.D. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 2008, 44, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.G.; Weinshenker, D.; Sperandio, V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect. Immun. 2010, 78, 914–926. [Google Scholar] [CrossRef]
- Gart, E.V.; Suchodolski, J.S.; Welsh, T.H.; Alaniz, R.C.; Randel, R.D.; Lawhon, S.D. Salmonella typhimurium and multidirectional communication in the gut. Front. Microbiol. 2016, 7, 1827. [Google Scholar] [CrossRef] [PubMed]
- Madden, K.S.; Sanders, V.M.; Felten, D.L. Catecholamine influences and sympathetic neural modulation of immune responsiveness. Annu. Rev. Pharmacol. Toxicol. 1995, 35, 417–448. [Google Scholar] [CrossRef] [PubMed]
- Sanders, V.M.; Straub, R.H. Norepinephrine, the β-adrenergic receptor, and immunity. Brain. Behav. Immun. 2002, 16, 290–332. [Google Scholar] [CrossRef] [PubMed]
- Ottaway, C.A.; Husband, A.J. The influence of neuroendocrine pathways on lymphocyte migration. Immunol. Today 1994, 15, 511–517. [Google Scholar] [CrossRef]
- Elenkov, I.J.; Wilder, R.L.; Chrousos, G.P.; Vizi, E.S. The sympathetic nerve—An integrative interface between two supersystems: The brain and the immune system. Pharmacol. Rev. 2000, 52, 595–638. [Google Scholar]
- Flierl, M.A.; Rittirsch, D.; Huber-Lang, M.; Vidya Sarma, J.; Award, P. Catecholamines—Crafty weapons in the inflammatory arsenal of immune/inflammatory cells or opening Pandora’s box§? Mol. Med. 2008, 14, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.W.; Meyers, R.T.; Brennan, K.M.; Rumble, J.M.; Narasimhachari, N.; Perozzi, E.F.; Ryan, J.J.; Stewart, J.K.; Fischer-Stenger, K. Catecholamines in a macrophage cell line. J. Neuroimmunol. 2003, 135, 47–55. [Google Scholar] [CrossRef]
- Lyte, M.; Bailey, M.T. Neuroendocrine-bacterial interactions in a neurotoxin-induced model of trauma. J. Surg. Res. 1997, 70, 195–201. [Google Scholar] [CrossRef]
- Corrigan, R.M.; Gründling, A. Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 2013, 11, 513–524. [Google Scholar] [CrossRef]
- Woodward, J.J.; Lavarone, A.T.; Portnoy, D.A. C-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 2010, 328, 1703–1705. [Google Scholar] [CrossRef] [PubMed]
- McFarland, A.P.; Luo, S.; Ahmed-Qadri, F.; Zuck, M.; Thayer, E.F.; Goo, Y.A.; Hybiske, K.; Tong, L.; Woodward, J.J. Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-κB Activation and Shapes a Proinflammatory Antibacterial State. Immunity 2017, 46, 433–445. [Google Scholar] [CrossRef]
- Whiteley, A.T.; Eaglesham, J.B.; de Oliveira Mann, C.C.; Morehouse, B.R.; Lowey, B.; Nieminen, E.A.; Danilchanka, O.; King, D.S.; Lee, A.S.Y.; Mekalanos, J.J.; et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 2019, 567, 194–199. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Ahn, S.H.; Inagaki, T.; Choi, M.; Ito, S.; Guo, G.L.; Kliewer, S.A.; Gonzalez, F.J. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 2007, 48, 2664–2672. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile acids activated receptors regulate innate immunity. Front. Immunol. 2018, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, S.; Mencarelli, A.; Chini, M.G.; Distrutti, E.; Renga, B.; Bifulco, G.; Baldelli, F.; Donini, A.; Fiorucci, S. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS ONE 2011, 6, e25637. [Google Scholar] [CrossRef]
- Mencarelli, A.; Renga, B.; Migliorati, M.; Cipriani, S.; Distrutti, E.; Santucci, L.; Fiorucci, S. The Bile Acid Sensor Farnesoid X Receptor Is a Modulator of Liver Immunity in a Rodent Model of Acute Hepatitis. J. Immunol. 2009, 183, 6657–6666. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, R.M.; Van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.L.; Renooij, W.; Murzilli, S.; Klomp, L.W.J.; Siersema, P.D.; Schipper, M.E.I.; Danese, S.; et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- Yang, B.H.; Hagemann, S.; Mamareli, P.; Lauer, U.; Hoffmann, U.; Beckstette, M.; Föhse, L.; Prinz, I.; Pezoldt, J.; Suerbaum, S.; et al. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016, 9, 444–457. [Google Scholar] [CrossRef]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef] [PubMed]
- MacEyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef]
- Abdel Hadi, L.; Di Vito, C.; Riboni, L. Fostering Inflammatory Bowel Disease: Sphingolipid Strategies to Join Forces. Mediat. Inflamm. 2016, 2016. [Google Scholar] [CrossRef]
- Stoffel, W.; Dittmar, K.; Wilmes, R. Sphingolipid Metabolism in Bacteroideaceae. Hoppe Seylers Z. Physiol. Chem. 1975, 356, 715–726. [Google Scholar] [CrossRef]
- LaBach, J.P.; White, D.C. Identification of ceramide phosphorylethanolamine and ceramide phosphorylglycerol in the lipids of an anaerobic bacterium. J. Lipid Res. 1969, 10, 528–534. [Google Scholar] [CrossRef]
- Ana, D.; Na, C.; Bielawski, J.; Hannun, Y.A.; Kasper, D.L. Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc. Natl. Acad. Sci. USA 2011, 108, 4666–4671. [Google Scholar] [CrossRef]
- Hickey, C.A.; Kuhn, K.A.; Donermeyer, D.L.; Porter, N.T.; Jin, C.; Cameron, E.A.; Jung, H.; Kaiko, G.E.; Wegorzewska, M.; Malvin, N.P.; et al. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe 2015, 17, 672–680. [Google Scholar] [CrossRef]
- Kinjo, Y.; Wu, D.; Kim, G.; Xing, G.W.; Poles, M.A.; Ho, D.D.; Tsuji, M.; Kawahara, K.; Wong, C.H.; Kronenberg, M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 2005, 434, 520–525. [Google Scholar] [CrossRef]
- An, D.; Oh, S.F.; Olszak, T.; Neves, J.F.; Avci, F.Y.; Erturk-Hasdemir, D.; Lu, X.; Zeissig, S.; Blumberg, R.S.; Kasper, D.L. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014, 156, 123–133. [Google Scholar] [CrossRef]
- Olszak, T.; An, D.; Zeissig, S.; Vera, M.P.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.M.; Ke, X.; Hitchcock, D.; Jeanfavre, S.; Avila-Pacheco, J.; Nakata, T.; Arthur, T.D.; Fornelos, N.; Heim, C.; Franzosa, E.A.; et al. Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe 2019, 25, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, R.F.; Holtje, J.V.; Wicken, A.J.; Tomasz, A.; Daneo-Moore, L.; Shockman, G.D. Inhibition of bacterial wall lysins by lipoteichoic acids and related compounds. Biochem. Biophys. Res. Commun. 1975, 67, 1128–1135. [Google Scholar] [CrossRef]
- Fischer, W. Bacterial Phosphoglycolipids and Lipoteichoic Acids. In Glycolipids, Phosphoglycolipids, and Sulfoglycolipids; Springer: New York, NY, USA, 1990; pp. 123–234. [Google Scholar]
- Brown, S.; Santa Maria, J.P.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [PubMed]
- Vasselon, T.; Detmers, P.A. Toll receptors: A central element in innate immune responses. Infect. Immun. 2002, 70, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Draing, C.; Pfitzenmaier, M.; Zummo, S.; Mancuso, G.; Geyer, A.; Hartung, T.; Von Aulock, S. Comparison of lipoteichoic acid from different serotypes of Streptococcus pneumoniae. J. Biol. Chem. 2006, 281, 33849–33859. [Google Scholar] [CrossRef]
- Schröder, N.W.J.; Morath, S.; Alexander, C.; Hamann, L.; Hartung, T.; Zähringer, U.; Göbel, U.B.; Weber, J.R.; Schumann, R.R. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J. Biol. Chem. 2003, 278, 15587–15594. [Google Scholar] [CrossRef]
- Trianiafilou, M.; Manukyan, M.; Mackie, A.; Morath, S.; Hartung, T.; Heine, H.; Triantafilou, K. Lipoteichoic acid and Toll-like receptor 2 internalization and targeting to the Golgi are lipid raft-dependent. J. Biol. Chem. 2004, 279, 40882–40889. [Google Scholar] [CrossRef]
- Michelsen, K.S.; Aicher, A.; Mohaupt, M.; Hartung, T.; Dimmeler, S.; Kirschning, C.J.; Schumann, R.R. The role of toll-like receptors (TLRs) in bacteria-induced maturation of murine dendritic cells (DCs): Peptidoglycan and lipoteichoic acid are inducers of DC maturation and require TLR2. J. Biol. Chem. 2001, 276, 25680–25686. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Baik, J.E.; Kang, S.S.; Yun, C.H.; Seo, D.G.; Han, S.H. Lipoteichoic acid of Streptococcus mutans interacts with Toll-like receptor 2 through the lipid moiety for induction of inflammatory mediators in murine macrophages. Mol. Immunol. 2014, 57, 284–291. [Google Scholar] [CrossRef]
- Dunne, D.W.; Resnick, D.; Greenberg, J.; Krieger, M.; Joiner, K.A. The type I macrophage scavenger receptor binds to Gram-positive bacteria and recognizes lipoteichoic acid. Proc. Natl. Acad. Sci. USA 1994, 91, 1863–1867. [Google Scholar] [CrossRef] [PubMed]
- Ellingsen, E.; Morath, S.; Flo, T.; Schromm, A.; Hartung, T.; Thiemermann, C.; Espevik, T.; Golenbock, D.; Foster, D.; Solberg, R.; et al. Induction of Cytokine Production in Human T Cells and Monocytes by Highly Purified Lipoteichoic Acid: Involvement of Toll-Like Receptors and CD14. Available online: https://www.medscimonit.com/download/index/idArt/420854 (accessed on 15 May 2021).
- von Aulock, S.; Morath, S.; Hareng, L.; Knapp, S.; van Kessel, K.P.M.; van Strijp, J.A.G.; Hartung, T. Lipoteichoic acid from Staphylococcus aureus is a potent stimulus for neutrophil recruitment. Immunobiology 2003, 208, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.E.; Dahle, M.K.; McDonald, M.; Foster, S.J.; Aasen, A.O.; Thiemermann, C. Peptidoglycan and lipoteichoic acid in gram-positive bacterial sepsis: Receptors, signal transduction, biological effects, and synergism. Shock 2003, 20, 402–414. [Google Scholar] [CrossRef]
- Hara, H.; Seregin, S.S.; Yang, D.; Fukase, K.; Chamaillard, M.; Alnemri, E.S.; Inohara, N.; Chen, G.Y.; Núñez, G. The NLRP6 Inflammasome Recognizes Lipoteichoic Acid and Regulates Gram-Positive Pathogen Infection. Cell 2018, 175, 1651–1664.e14. [Google Scholar] [CrossRef]
- Delgado, S.; Sánchez, B.; Margolles, A.; Ruas-Madiedo, P.; Ruiz, L. Molecules produced by probiotics and intestinal microorganisms with immunomodulatory activity. Nutrients 2020, 12, 391. [Google Scholar] [CrossRef]
- Riehl, T.E.; Alvarado, D.; Ee, X.; Zuckerman, A.; Foster, L.; Kapoor, V.; Thotala, D.; Ciorba, M.A.; Stenson, W.F. Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut 2019, 68, 1003–1013. [Google Scholar] [CrossRef]
- Peña, J.A.; Versalovic, J. Lactobacillus rhamnosus GG decreases TNF-α production in lipopolysaccharide-activated murine macrophages by a contact-independent mechanism. Cell. Microbiol. 2003, 5, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ahmadi, S.; Nagpal, R.; Jain, S.; Mishra, S.P.; Kavanagh, K.; Zhu, X.; Wang, Z.; McClain, D.A.; Kritchevsky, S.B.; et al. Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3-5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: From C. elegans to mice. GeroScience 2020, 42, 333–352. [Google Scholar] [CrossRef]
- Grangette, C.; Nutten, S.; Palumbo, E.; Morath, S.; Hermann, C.; Dewulf, J.; Pot, B.; Hartung, T.; Hols, P.; Mercenier, A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl. Acad. Sci. USA 2005, 102, 10321–10326. [Google Scholar] [CrossRef] [PubMed]
- Melmed, G.; Thomas, L.S.; Lee, N.; Tesfay, S.Y.; Lukasek, K.; Michelsen, K.S.; Zhou, Y.; Hu, B.; Arditi, M.; Abreu, M.T. Human Intestinal Epithelial Cells Are Broadly Unresponsive to Toll-Like Receptor 2-Dependent Bacterial Ligands: Implications for Host-Microbial Interactions in the Gut. J. Immunol. 2003, 170, 1406–1415. [Google Scholar] [CrossRef]
- Ott, S.J.; Musfeldt, M.; Wenderoth, D.F.; Hampe, J.; Brant, O.; Fölsch, U.R.; Timmis, K.N.; Schreiber, S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Sokol, H.; Seksik, P.; Rigottier-Gois, L.; Lay, C.; Lepage, P.; Podglajen, I.; Marteau, P.; Doré, J. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 2006, 12, 106–111. [Google Scholar] [CrossRef]
- Roy, U.; Gálvez, E.J.C.; Iljazovic, A.; Lesker, T.R.; Błażejewski, A.J.; Pils, M.C.; Heise, U.; Huber, S.; Flavell, R.A.; Strowig, T. Distinct Microbial Communities Trigger Colitis Development upon Intestinal Barrier Damage via Innate or Adaptive Immune Cells. Cell Rep. 2017, 21, 994–1008. [Google Scholar] [CrossRef]
- Schirmer, M.; Franzosa, E.A.; Lloyd-Price, J.; McIver, L.J.; Schwager, R.; Poon, T.W.; Ananthakrishnan, A.N.; Andrews, E.; Barron, G.; Lake, K.; et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 2018, 3, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Patwa, L.G.; Fan, T.J.; Tchaptchet, S.; Liu, Y.; Lussier, Y.A.; Sartor, R.B.; Hansen, J.J. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology 2011, 141, 1842–1851. [Google Scholar] [CrossRef]
- Ilott, N.E.; Bollrath, J.; Danne, C.; Schiering, C.; Shale, M.; Adelmann, K.; Krausgruber, T.; Heger, A.; Sims, D.; Powrie, F. Defining the microbial transcriptional response to colitis through integrated host and microbiome profiling. ISME J. 2016, 10, 2389–2404. [Google Scholar] [CrossRef]
- Becattini, S.; Sorbara, M.T.; Kim, S.G.; Littmann, E.L.; Dong, Q.; Walsh, G.; Wright, R.; Amoretti, L.; Fontana, E.; Hohl, T.M.; et al. Rapid transcriptional and metabolic adaptation of intestinal microbes to host immune activation. Cell Host Microbe 2021, 29, 378–393.e5. [Google Scholar] [CrossRef]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef]
- Rubino, S.J.; Geddes, K.; Girardin, S.E. Innate IL-17 and IL-22 responses to enteric bacterial pathogens. Trends Immunol. 2012, 33, 112–118. [Google Scholar] [CrossRef]
- Bouskra, D.; Brézillon, C.; Bérard, M.; Werts, C.; Varona, R.; Boneca, I.G.; Eberl, G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008, 456, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Salzman, N.H.; Bevins, C.L. Dysbiosis-A consequence of Paneth cell dysfunction. Semin. Immunol. 2013, 25, 334–341. [Google Scholar] [CrossRef]
- Ehmann, D.; Wendler, J.; Koeninger, L.; Larsen, I.S.; Klag, T.; Berger, J.; Marette, A.; Schaller, M.; Stange, E.F.; Malek, N.P.; et al. Paneth cell α-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 3746–3751. [Google Scholar] [CrossRef]
- Bantug, G.R.; Galluzzi, L.; Kroemer, G.; Hess, C. The spectrum of T cell metabolism in health and disease. Nat. Rev. Immunol. 2018, 18, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Deaglio, S.; Dwyer, K.M.; Gao, W.; Friedman, D.; Usheva, A.; Erat, A.; Chen, J.F.; Enjyoji, K.; Linden, J.; Oukka, M.; et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 2007, 204, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.A.; Hall, J.A.; Sun, C.M.; Cai, Q.; Ghyselinck, N.; Chambon, P.; Belkaid, Y.; Mathis, D.; Benoist, C. Retinoic Acid Enhances Foxp3 Induction Indirectly by Relieving Inhibition from CD4+CD44hi Cells. Immunity 2008, 29, 758–770. [Google Scholar] [CrossRef]
- Goverse, G.; Labao-Almeida, C.; Ferreira, M.; Molenaar, R.; Wahlen, S.; Konijn, T.; Koning, J.; Veiga-Fernandes, H.; Mebius, R.E. Vitamin A Controls the Presence of RORγ + Innate Lymphoid Cells and Lymphoid Tissue in the Small Intestine. J. Immunol. 2016, 196, 5148–5155. [Google Scholar] [CrossRef]
- Mucida, D.; Park, Y.; Kim, G.; Turovskaya, O.; Scott, I.; Kronenberg, M.; Cheroutre, H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 2007, 317, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Jaensson-Gyllenbäck, E.; Kotarsky, K.; Zapata, F.; Persson, E.K.; Gundersen, T.E.; Blomhoff, R.; Agace, W.W. Bile retinoids imprint intestinal CD103+ dendritic cells with the ability to generate gut-tropic T cells. Mucosal Immunol. 2011, 4, 438–447. [Google Scholar] [CrossRef]
- Mielke, L.A.; Jones, S.A.; Raverdeau, M.; Higgs, R.; Stefanska, A.; Groom, J.R.; Misiak, A.; Dungan, L.S.; Sutton, C.E.; Streubel, G.; et al. Retinoic acid expression associates with enhanced IL-22 production by γδ T cells and innate lymphoid cells and attenuation of intestinal inflammation. J. Exp. Med. 2013, 210, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Osanai, M.; Nishikiori, N.; Murata, M.; Chiba, H.; Kojima, T.; Sawada, N. Cellular retinoic acid bioavailability determines epithelial integrity: Role of retinoic acid receptor α agonists in colitis. Mol. Pharmacol. 2007, 71, 250–258. [Google Scholar] [CrossRef]
- Kubota, H.; Chiba, H.; Takakuwa, Y.; Osanai, M.; Tobioka, H.; Kohama, G.I.; Mori, M.; Sawada, N. Retinoid X receptor α and retinoic acid receptor γ mediate expression of genes encoding tight-junction proteins and barrier function in F9 cells during visceral endodermal differentiation. Exp. Cell Res. 2001, 263, 163–172. [Google Scholar] [CrossRef]
- Grizotte-Lake, M.; Zhong, G.; Duncan, K.; Kirkwood, J.; Iyer, N.; Smolenski, I.; Isoherranen, N.; Vaishnava, S. Commensals Suppress Intestinal Epithelial Cell Retinoic Acid Synthesis to Regulate Interleukin-22 Activity and Prevent Microbial Dysbiosis. Immunity 2018, 49, 1103–1115.e6. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.S.; Valastyan, J.S.; Bassler, B.L. A Host-Produced Autoinducer-2 Mimic Activates Bacterial Quorum Sensing. Cell Host Microbe 2016, 19, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Perez de Souza, L.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 2021, 1–14. [Google Scholar] [CrossRef]
- Da Silva, R.R.; Dorrestein, P.C.; Quinn, R.A. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. USA 2015, 112, 12549–12550. [Google Scholar] [CrossRef]
- Jarmusch, A.K.; Wang, M.; Aceves, C.M.; Advani, R.S.; Aguirre, S.; Aksenov, A.A.; Aleti, G.; Aron, A.T.; Bauermeister, A.; Bolleddu, S.; et al. ReDU: A framework to find and reanalyze public mass spectrometry data. Nat. Methods 2020, 17, 901–904. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.; Petras, D.; Nothias, L.F.; Wang, M.; Aron, A.T.; Jagels, A.; Tsugawa, H.; Rainer, J.; Garcia-Aloy, M.; Dührkop, K.; et al. Ion identity molecular networking in the GNPS environment. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shahaf, N.; Rogachev, I.; Heinig, U.; Meir, S.; Malitsky, S.; Battat, M.; Wyner, H.; Zheng, S.; Wehrens, R.; Aharoni, A. The WEIZMASS spectral library for high-confidence metabolite identification. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Vinaixa, M.; Schymanski, E.L.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC-Trends Anal. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef]
- NIST20: Updates to the NIST Tandem and Electron Ionization Spectral Libraries|NIST. Available online: https://www.nist.gov/programs-projects/nist20-updates-nist-tandem-and-electron-ionization-spectral-libraries (accessed on 14 May 2021).
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Advanced Mass Spectral Database—mzCloud. Available online: https://www.mzcloud.org/ (accessed on 14 May 2021).
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Sawada, Y.; Nakabayashi, R.; Yamada, Y.; Suzuki, M.; Sato, M.; Sakata, A.; Akiyama, K.; Sakurai, T.; Matsuda, F.; Aoki, T.; et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry 2012, 82, 38–45. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Quinn, T.P.; Erb, I. Examining microbe–metabolite correlations by linear methods. Nat. Methods 2021, 18, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.T.; Aksenov, A.A.; Nothias, L.F.; Foulds, J.R.; Quinn, R.A.; Badri, M.H.; Swenson, T.L.; Van Goethem, M.W.; Northen, T.R.; Vazquez-Baeza, Y.; et al. Learning representations of microbe–metabolite interactions. Nat. Methods 2019, 16, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.T.; McDonald, D.; Aksenov, A.A.; Nothias, L.F.; Foulds, J.R.; Quinn, R.A.; Badri, M.H.; Swenson, T.L.; Van Goethem, M.W.; Northen, T.R.; et al. Reply to: Examining microbe–metabolite correlations by linear methods. Nat. Methods 2021, 18, 40–41. [Google Scholar] [CrossRef] [PubMed]
- Rosshart, S.P.; Herz, J.; Vassallo, B.G.; Hunter, A.; Wall, M.K.; Badger, J.H.; McCulloch, J.A.; Anastasakis, D.G.; Sarshad, A.A.; Leonardi, I.; et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 2019, 365. [Google Scholar] [CrossRef]
- Cassotta, M.; Forbes-Hernández, T.Y.; Iglesias, R.C.; Ruiz, R.; Zabaleta, M.E.; Giampieri, F.; Battino, M. Links between nutrition, infectious diseases, and microbiota: Emerging technologies and opportunities for human-focused research. Nutrients 2020, 12, 1827. [Google Scholar] [CrossRef]
- Venema, K.; Van Den Abbeele, P. Experimental models of the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 115–126. [Google Scholar] [CrossRef]
- Marzorati, M.; Van De Wiele, T. An Advanced in Vitro Technology Platform to Study the Mechanism of Action of Prebiotics and Probiotics in the Gastrointestinal Tract. J. Clin. Gastroenterol. 2016, 50, S124–S125. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caffaratti, C.; Plazy, C.; Mery, G.; Tidjani, A.-R.; Fiorini, F.; Thiroux, S.; Toussaint, B.; Hannani, D.; Le Gouellec, A. What We Know So Far about the Metabolite-Mediated Microbiota-Intestinal Immunity Dialogue and How to Hear the Sound of This Crosstalk. Metabolites 2021, 11, 406. https://doi.org/10.3390/metabo11060406
Caffaratti C, Plazy C, Mery G, Tidjani A-R, Fiorini F, Thiroux S, Toussaint B, Hannani D, Le Gouellec A. What We Know So Far about the Metabolite-Mediated Microbiota-Intestinal Immunity Dialogue and How to Hear the Sound of This Crosstalk. Metabolites. 2021; 11(6):406. https://doi.org/10.3390/metabo11060406
Chicago/Turabian StyleCaffaratti, Clément, Caroline Plazy, Geoffroy Mery, Abdoul-Razak Tidjani, Federica Fiorini, Sarah Thiroux, Bertrand Toussaint, Dalil Hannani, and Audrey Le Gouellec. 2021. "What We Know So Far about the Metabolite-Mediated Microbiota-Intestinal Immunity Dialogue and How to Hear the Sound of This Crosstalk" Metabolites 11, no. 6: 406. https://doi.org/10.3390/metabo11060406
APA StyleCaffaratti, C., Plazy, C., Mery, G., Tidjani, A.-R., Fiorini, F., Thiroux, S., Toussaint, B., Hannani, D., & Le Gouellec, A. (2021). What We Know So Far about the Metabolite-Mediated Microbiota-Intestinal Immunity Dialogue and How to Hear the Sound of This Crosstalk. Metabolites, 11(6), 406. https://doi.org/10.3390/metabo11060406