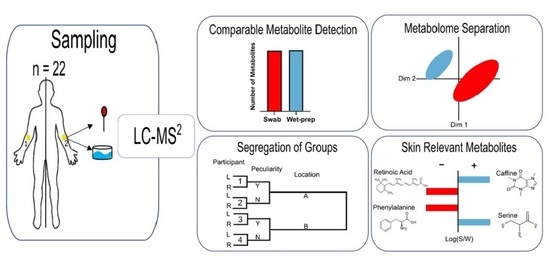
Enhanced Access to the Health-Related Skin Metabolome by Fast, Reproducible and Non-Invasive WET PREP Sampling
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Sample Collection and Processing
4.3. Data Analysis/Statistical Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dalgard, F.J.; Gieler, U.; Tomas-Aragones, L.; Lien, L.; Poot, F.; Jemec, G.B.E.; Misery, L.; Szabo, C.; Linder, D.; Sampogna, F.; et al. The Psychological Burden of Skin Diseases: A Cross-Sectional Multicenter Study among Dermatological Out-Patients in 13 European Countries. J. Investig. Dermatol. 2015, 135, 984–991. [Google Scholar] [CrossRef] [Green Version]
- Randhawa, M.; Southall, M.; Samaras, S.T. Metabolomic Analysis of Sun Exposed Skin. Mol. Biosyst. 2013, 9, 2045–2050. [Google Scholar] [CrossRef]
- Kuehne, A.; Hildebrand, J.; Soehle, J.; Wenck, H.; Terstegen, L.; Gallinat, S.; Knott, A.; Winnefeld, M.; Zamboni, N. An Integrative Metabolomics and Transcriptomics Study to Identify Metabolic Alterations in Aged Skin of Humans in Vivo. BMC Genom. 2017, 18, 169. [Google Scholar] [CrossRef] [Green Version]
- Ono, E.; Murota, H.; Mori, Y.; Yoshioka, Y.; Nomura, Y.; Munetsugu, T.; Yokozeki, H.; Katayama, I. Sweat Glucose and GLUT2 Expression in Atopic Dermatitis: Implication for Clinical Manifestation and Treatment. PLoS ONE 2018, 13, e0195960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouslimani, A.; da Silva, R.; Kosciolek, T.; Janssen, S.; Callewaert, C.; Amir, A.; Dorrestein, K.; Melnik, A.V.; Zaramela, L.S.; Kim, J.-N.; et al. The Impact of Skin Care Products on Skin Chemistry and Microbiome Dynamics. BMC Biol. 2019, 17, 1–20. [Google Scholar] [CrossRef]
- Barton, W.; O’Sullivan, O.; Cotter, P.D. Metabolic Phenotyping of the Human Microbiome. F1000 Res. 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Yan, D.; Afifi, L.; Jeon, C.; Trivedi, M.; Chang, H.W.; Lee, K.; Liao, W. The Metabolomics of Psoriatic Disease. Psoriasis Auckl. NZ 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ashrafi, M.; Xu, Y.; Muhamadali, H.; White, I.; Wilkinson, M.; Hollywood, K.; Baguneid, M.; Goodacre, R.; Bayat, A. A Microbiome and Metabolomic Signature of Phases of Cutaneous Healing Identified by Profiling Sequential Acute Wounds of Human Skin: An Exploratory Study. PLoS ONE 2020, 15, e0229545. [Google Scholar] [CrossRef]
- Melnik, B.C. Linking Diet to Acne Metabolomics, Inflammation, and Comedogenesis: An Update. Available online: https://www.dovepress.com/linking-diet-to-acne-metabolomics-inflammation-and-comedogenesis-an-up-peer-reviewed-article-CCID (accessed on 26 February 2021).
- Dutkiewicz, E.P.; Hsieh, K.-T.; Wang, Y.-S.; Chiu, H.-Y.; Urban, P.L. Hydrogel Micropatch and Mass Spectrometry-Assisted Screening for Psoriasis-Related Skin Metabolites. Clin. Chem. 2016, 62, 1120–1128. [Google Scholar] [CrossRef]
- Dutkiewicz, E.P.; Lin, J.-D.; Tseng, T.-W.; Wang, Y.-S.; Urban, P.L. Hydrogel Micropatches for Sampling and Profiling Skin Metabolites. Anal. Chem. 2014, 86, 2337–2344. [Google Scholar] [CrossRef]
- Dutkiewicz, E.P.; Chiu, H.-Y.; Urban, P.L. Probing Skin for Metabolites and Topical Drugs with Hydrogel Micropatches. Anal. Chem. 2017, 89, 2664–2670. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, T.; Klose, C.; Gerl, M.J.; Wójcik-Maciejewicz, A.; Herzog, R.; Simons, K.; Reich, A.; Surma, M.A. Large-Scale Human Skin Lipidomics by Quantitative, High-Throughput Shotgun Mass Spectrometry. Sci. Rep. 2017, 7, 43761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooton, K.; Han, W.; Li, L. Comprehensive and Quantitative Profiling of the Human Sweat Submetabolome Using High-Performance Chemical Isotope Labeling LC–MS. Available online: https://pubs.acs.org/doi/pdf/10.1021/acs.analchem.6b01930 (accessed on 8 July 2020).
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MetPA: A Web-Based Metabolomics Tool for Pathway Analysis and Visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [Green Version]
- Wolf, K.; Cyrys, J.; Harciníková, T.; Gu, J.; Kusch, T.; Hampel, R.; Schneider, A.; Peters, A. Land Use Regression Modeling of Ultrafine Particles, Ozone, Nitrogen Oxides and Markers of Particulate Matter Pollution in Augsburg, Germany. Sci. Total Environ. 2017, 579, 1531–1540. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, A.V.; Scott, I.R.; Harding, C.R.; Bowser, P.A. Stratum Corneum Moisturization at the Molecular Level. J. Investig. Dermatol. 1994, 103, 731–740. [Google Scholar] [CrossRef] [Green Version]
- Gökmen, S.S.; Aygit, A.C.; Ayhan, M.S.; Yorulmaz, F.; Gülen, S. Significance of Arginase and Ornithine in Malignant Tumors of the Human Skin. J. Lab. Clin. Med. 2001, 137, 340–344. [Google Scholar] [CrossRef]
- Burke, R.C.; Lee, T.H.; Buettner-Janusch, V. Free Amino Acids and Water Soluble Peptides in Stratum Corneum and Skin Surface Film in Human Beings. Yale J. Biol. Med. 1966, 38, 355–373. [Google Scholar]
- Proksch, E. PH in Nature, Humans and Skin. J. Dermatol. 2018, 45, 1044–1052. [Google Scholar] [CrossRef]
- Jang, H.; Matsuda, A.; Jung, K.; Karasawa, K.; Matsuda, K.; Oida, K.; Ishizaka, S.; Ahn, G.; Amagai, Y.; Moon, C.; et al. Skin PH Is the Master Switch of Kallikrein 5-Mediated Skin Barrier Destruction in a Murine Atopic Dermatitis Model. J. Investig. Dermatol. 2016, 136, 127–135. [Google Scholar] [CrossRef]
- Eijsink, V.G.H.; Axelsson, L.; Diep, D.B.; Håvarstein, L.S.; Holo, H.; Nes, I.F. Production of Class II Bacteriocins by Lactic Acid Bacteria; an Example of Biological Warfare and Communication. Antonie Van Leeuwenhoek 2002, 81, 639–654. [Google Scholar] [CrossRef]
- Traisaeng, S.; Herr, D.R.; Kao, H.-J.; Chuang, T.-H.; Huang, C.-M. A Derivative of Butyric Acid, the Fermentation Metabolite of Staphylococcus Epidermidis, Inhibits the Growth of a Staphylococcus aureus Strain Isolated from Atopic Dermatitis Patients. Toxins 2019, 11, 311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaro-Ortiz, A.; Yan, B.; D’Orazio, J.A. Ultraviolet Radiation, Aging and the Skin: Prevention of Damage by Topical CAMP Manipulation. Molecules 2014, 19, 6202–6219. [Google Scholar] [CrossRef] [PubMed]
- Maeno, K. Direct Quantification of Natural Moisturizing Factors in Stratum Corneum Using Direct Analysis in Real Time Mass Spectrometry with Inkjet-Printing Technique. Sci. Rep. 2019, 9, 17789. [Google Scholar] [CrossRef]
- Bouslimani, A.; Porto, C.; Rath, C.M.; Wang, M.; Guo, Y.; Gonzalez, A.; Berg-Lyon, D.; Ackermann, G.; Christensen, G.J.M.; Nakatsuji, T.; et al. Molecular Cartography of the Human Skin Surface in 3D. Proc. Natl. Acad. Sci. USA 2015, 112, E2120–E2129. [Google Scholar] [CrossRef] [Green Version]
- Sillner, N.; Walker, A.; Harrieder, E.-M.; Schmitt-Kopplin, P.; Witting, M. Development and Application of a HILIC UHPLC-MS Method for Polar Fecal Metabolome Profiling. J. Chromatogr. B 2019, 1109, 142–148. [Google Scholar] [CrossRef]
- Suhre, K.; Schmitt-Kopplin, P. MassTRIX: Mass Translator into Pathways. Nucleic Acids Res. 2008, 36, W481–W484. [Google Scholar] [CrossRef] [Green Version]
- RStudio Team. RStudio: Integrated Development for R. RStudio Inc. 2020. Available online: http://www.rstudio.com/ (accessed on 11 August 2020).
- Dusa, A.V. Draw Venn Diagrams. R Package Version 19. 2020. Available online: https://CRAN.R-project.org/package=venn (accessed on 11 August 2020).
- Eulerr, L.J.; Area-Proportional Euler and Venn Diagrams with Ellipses. R Package Version 610. 2020. Available online: https://cran.r-project.org/package=eulerr (accessed on 3 May 2021).



| Category | Compound | Focus | RP | HILIC | Reference | ||
|---|---|---|---|---|---|---|---|
| Significant Different Detection between WET PREP and Swab | log2 Fold Change (Average WET PREP/Average Swab) | Significant Different Detection between WET PREP and Swab | log2 Fold Change (Average WET PREP/Average Swab) | ||||
| Amino Acid | Taurine | Age | n.d. | + | 0.04 | Kuehne et al., 2017 | |
| Serine | Psoriasis | + | 0.21 | + | 0.21 | Kim et al., 2009 | |
| Proline | Age | +++ | 0.11 | + | 0.03 | Kuehne et al., 2017 | |
| Threonine | Age | n.d. | ++ | 0.10 | Kuehne et al., 2017 | ||
| Aspartic acid | Dock8 deficiency | n.d. | n.d. | Jacob et al., 2019 * | |||
| Glutamine | Psoriasis | +++ | 1.19 | +++ | only WET PREP | Kim et al., 2009 | |
| Glutamic acid | Psoriasis | + | 0.61 | − | −0.14 | Dutkiewics et al., 2016 | |
| Histidine | Cancer | +++ | 0.32 | n.d. | Taylor et al., 2020 | ||
| Phenyl alanine | Psoriasis | + | −0.09 | +++ | −0.11 | Dutkiewics et al., 2016 | |
| Tyrosine | Age | + | 0.06 | + | −0.07 | Kuehne et al., 2017 | |
| Tryptophan | Age | + | −0.09 | − | −0.04 | Kuehne et al., 2017 | |
| Amino Acid Derivative | Hypotaurine | Dock8 deficiency | +++ | only WET PREP | n.d. | Jacob et al., 2019 * | |
| Pyroglutamic acid | Skin | - | 0.02 | − | 0.14 | Joo et al., 2012 | |
| Ornithine | Age | +++ | 0.72 | scattered detection | Kuehne et al., 2017 | ||
| Acid | Lactic acid | Psoriasis | − | −0.08 | n.d. | Dutkiewics et al., 2016 | |
| Retinoic acid | Age | +++ | −0.22 | n.d. | Kuehne et al., 2017 | ||
| Sugar | Fucose | Age | n.d. | +++ | −0.15 | Kuehne et al., 2017 | |
| Glucose | Age | scattered detection | + | 0.04 | Kuehne et al., 2017 | ||
| Nucleo(t/s)ides | Uracil | Age | − | 0.03 | + | −0.18 | Kuehne et al., 2017 |
| Guanosine | Atopic Eczema | scattered detection | scattered detection | Jacob et al., 2019 * | |||
| Aromatic | Cresol | Age | scattered detection | +++ | 1.11 | Kuehne et al., 2017 | |
| Caffeine | Atopic Eczema | scattered detection | + | 0.49 | Jacob et al., 2019 * | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afghani, J.; Huelpuesch, C.; Schmitt-Kopplin, P.; Traidl-Hoffmann, C.; Reiger, M.; Mueller, C. Enhanced Access to the Health-Related Skin Metabolome by Fast, Reproducible and Non-Invasive WET PREP Sampling. Metabolites 2021, 11, 415. https://doi.org/10.3390/metabo11070415
Afghani J, Huelpuesch C, Schmitt-Kopplin P, Traidl-Hoffmann C, Reiger M, Mueller C. Enhanced Access to the Health-Related Skin Metabolome by Fast, Reproducible and Non-Invasive WET PREP Sampling. Metabolites. 2021; 11(7):415. https://doi.org/10.3390/metabo11070415
Chicago/Turabian StyleAfghani, Jamie, Claudia Huelpuesch, Philippe Schmitt-Kopplin, Claudia Traidl-Hoffmann, Matthias Reiger, and Constanze Mueller. 2021. "Enhanced Access to the Health-Related Skin Metabolome by Fast, Reproducible and Non-Invasive WET PREP Sampling" Metabolites 11, no. 7: 415. https://doi.org/10.3390/metabo11070415
APA StyleAfghani, J., Huelpuesch, C., Schmitt-Kopplin, P., Traidl-Hoffmann, C., Reiger, M., & Mueller, C. (2021). Enhanced Access to the Health-Related Skin Metabolome by Fast, Reproducible and Non-Invasive WET PREP Sampling. Metabolites, 11(7), 415. https://doi.org/10.3390/metabo11070415