Mycobacterium avium subsp. paratuberculosis Proteome Changes Profoundly in Milk
Abstract
1. Introduction
2. Results
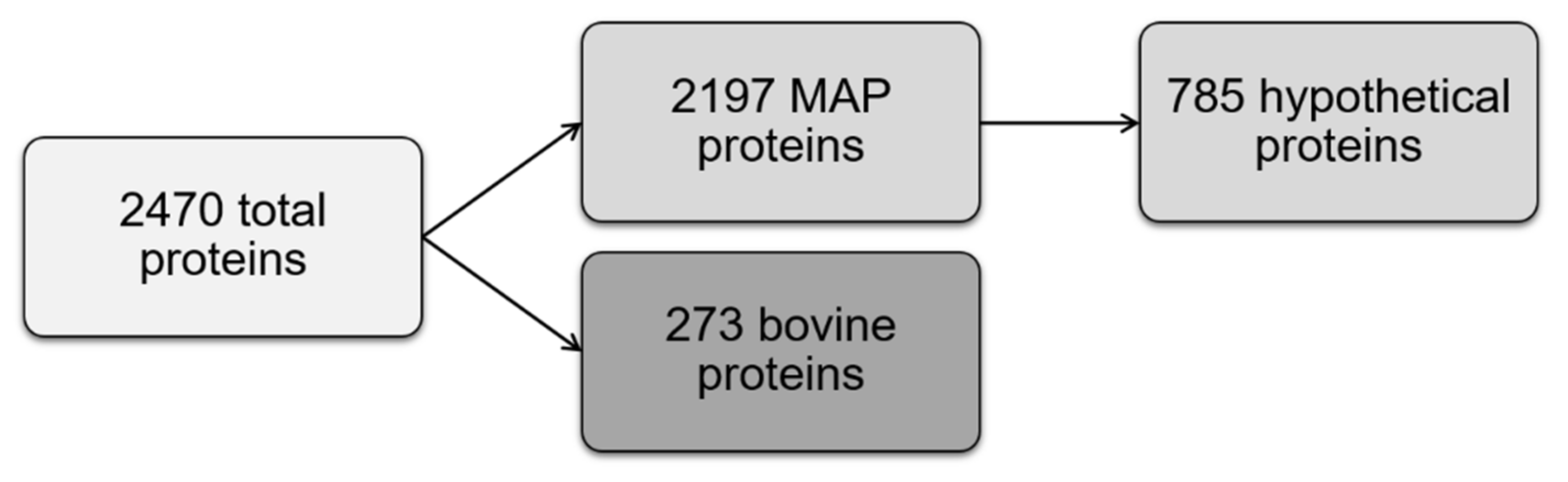
2.1. Identification of MAP Proteome with Label-Free Mass Spectrometry
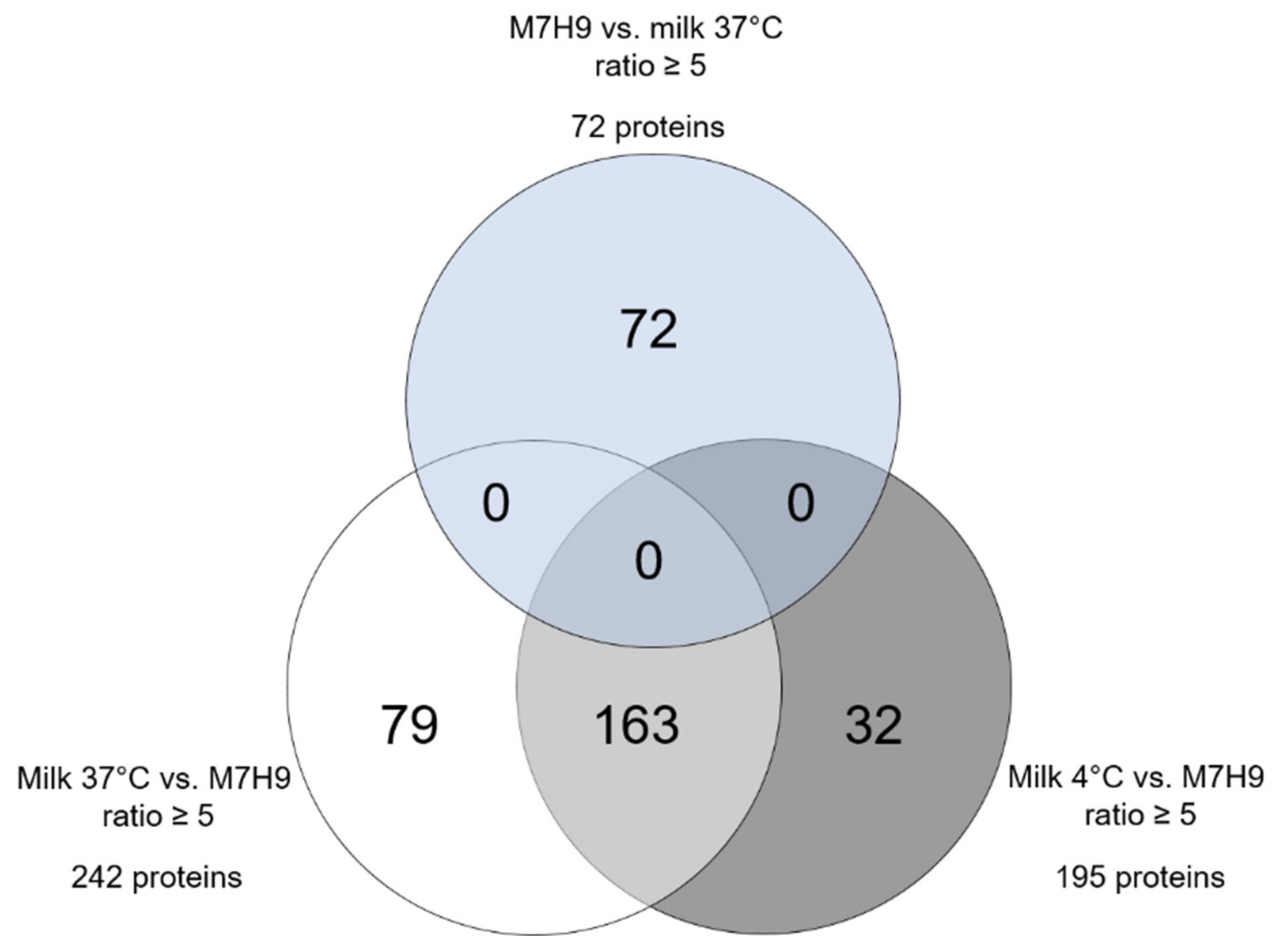
2.2. Highly Differential MAP Proteomes after Incubation in Standard Medium and Milk
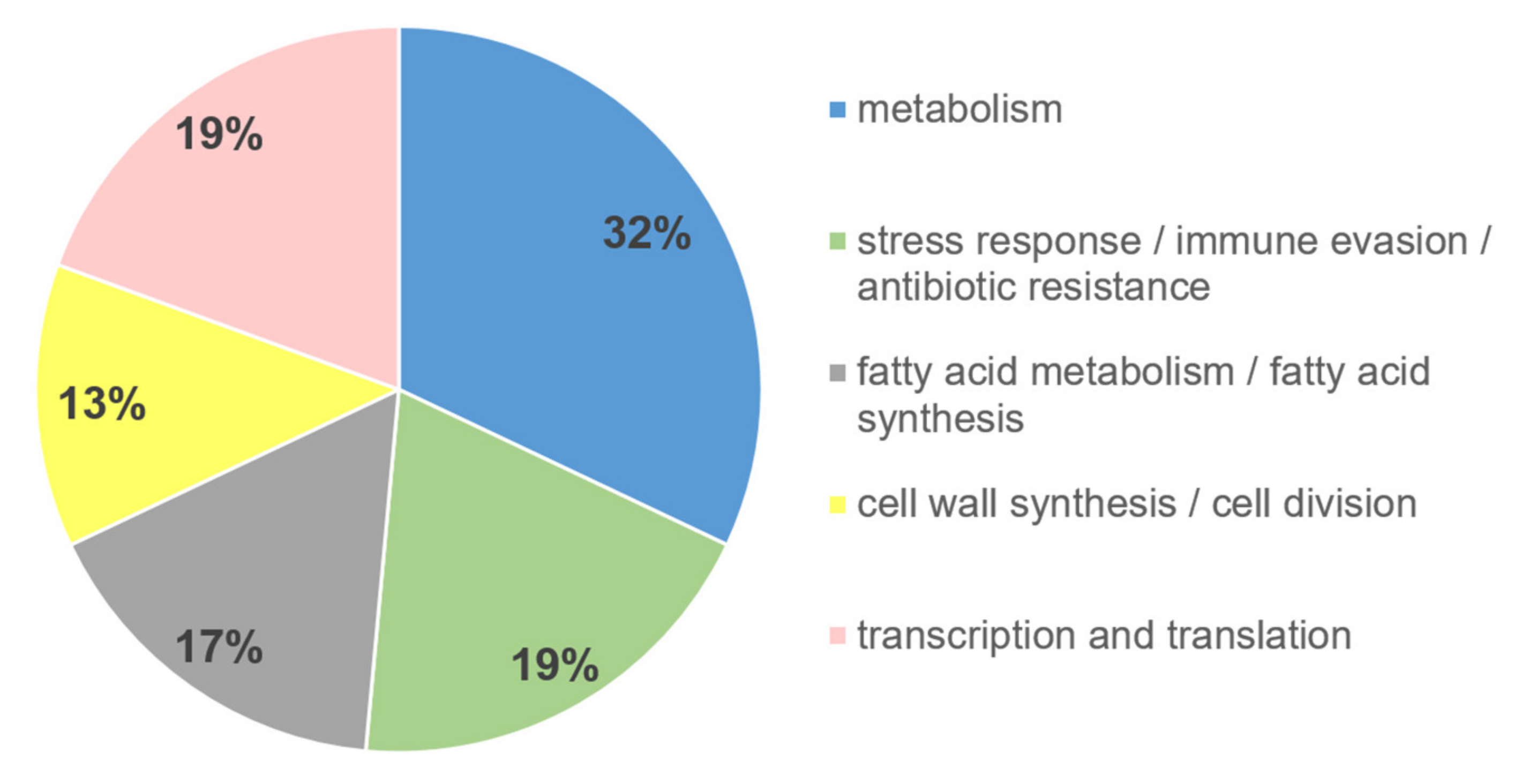
2.3. Functional Affiliation of Differential MAP Proteins
2.4. Stably Expressed Proteins as Possible Targets for Detection Methods
3. Discussion
4. Materials and Methods
4.1. Bacteria
4.2. Preparation of Milk
4.3. Incubation of MAP in Liquids
4.4. Harvesting and Refiltration of MAP
4.5. Lysis of MAP Samples
4.6. Sample Preparation for LC–MS/MS Mass Spectrometry
4.7. Mass Spectrometry
4.8. Protein Identification and Label-Free Quantification
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Twort, F.W.; Ingram, G.L.Y. Further Experiments with the Mycobacterium Enteritidis Chronicæ Pseudo-Tuberculosæ Bovis, Johne, and with Vaccines Prepared from this Micro-Organism. Vet. J. 1900 1913, 69, 4–15. [Google Scholar] [CrossRef]
- Twort, F.W.; Ingram, G.L.Y. A method for isolating and cultivating the mycobacterium enteritidis chronicæ pseudotuberculosæ bovis, Jöhne, and some experiments on the preparation of a diagnostic vaccine for pseudo-tuberculous enteritis of bovines. Proc. R. Soc. B 1912, 84, 517–542. [Google Scholar] [CrossRef]
- Eslami, M.; Shafiei, M.; Ghasemian, A.; Valizadeh, S.; Al-Marzoqi, A.H.; Shokouhi Mostafavi, S.K.; Nojoomi, F.; Mirforughi, S.A. Mycobacterium avium paratuberculosis and Mycobacterium avium complex and related subspecies as causative agents of zoonotic and occupational diseases. J. Cell. Physiol. 2019, 234, 12415–12421. [Google Scholar] [CrossRef]
- Sechi, L.A.; Edow, C.T. Mycobacterium avium ss. paratuberculosis Zoonosis—The Hundred Year War—Beyond Crohn’s Disease. Front. Immunol. 2015, 6, 96. [Google Scholar] [CrossRef]
- Van der Sloot, K.W.J.; Voskuil, M.D.; Blokzijl, T.; Dinkla, A.; Ravesloot, L.; Visschedijk, M.C.; van Dullemen, H.M.; Festen, E.A.M.; Alizadeh, B.Z.; van Leer-Buter, C.; et al. Isotype specific antibody responses to Mycobacterium Avium subspecies Paratuberculosis antigens are associated with the use of biological therapy in Inflammatory Bowel Disease. J. Crohns Colitis 2020. [Google Scholar] [CrossRef]
- Aitken, J.M.; Phan, K.; Bodman, S.E.; Sharma, S.; Watt, A.; George, P.M.; Agrawal, G.; Tie, A.B.M. A Mycobacterium species for Crohn’s disease? Pathology 2021. [Google Scholar] [CrossRef]
- Kuenstner, J.T.; Naser, S.; Chamberlin, W.; Borody, T.; Graham, D.Y.; McNees, A.; Hermon-Taylor, J.; Hermon-Taylor, A.; Dow, C.T.; Thayer, W.; et al. The Consensus from the Mycobacterium avium ssp. paratuberculosis (MAP) Conference 2017. Front. Public Health 2017, 5, 208. [Google Scholar] [CrossRef] [PubMed]
- Mullan, W.M.A. Are we closer to understanding why viable cells of Mycobacterium avium subsp. paratuberculosis are still being reported in pasteurised milk? Int. J. Dairy Technol. 2019, 72, 332–344. [Google Scholar] [CrossRef]
- Rani, S.; Beaver, A.; Schukken, Y.H.; Pradhan, A.K. Modeling the effects of infection status and hygiene practices on Mycobacterium avium subspecies paratuberculosis contamination in bulk tank milk. Food Control 2019, 104, 367–376. [Google Scholar] [CrossRef]
- Sweeney, R.W.; Whitlock, R.H.; Rosenberger, A.E. Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J. Clin. Microbiol. 1992, 30, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Nigsch, A.; Robbe-Austerman, S.; Stuber, T.P.; Pavinski Bitar, P.D.; Gröhn, Y.T.; Schukken, Y.H. Who infects whom?—Reconstructing infection chains of Mycobacterium avium ssp. paratuberculosis in an endemically infected dairy herd by use of genomic data. PLoS ONE 2021, 16, e0246983. [Google Scholar] [CrossRef]
- Botsaris, G.; Swift, B.M.; Slana, I.; Liapi, M.; Christodoulou, M.; Hatzitofi, M.; Christodoulou, V.; Rees, C.E. Detection of viable Mycobacterium avium subspecies paratuberculosis in powdered infant formula by phage-PCR and confirmed by culture. Int. J. Food Microbiol. 2016, 216, 91–94. [Google Scholar] [CrossRef]
- Grant, I.R. Bacteriophage-Based Methods for Detection of Viable Mycobacterium avium subsp. paratuberculosis and Their Potential for Diagnosis of Johne’s Disease. Front. Vet. Sci. 2021, 8, 632498. [Google Scholar] [CrossRef] [PubMed]
- Hanifian, S. Behavior of Mycobacterium avium paratuberculosis in Lighvan cheese tracked by propidium monoazide qPCR and culture. LWT 2020, 133, 109886. [Google Scholar] [CrossRef]
- Chiodini, R.J.; Hermon-Taylor, J. The Thermal Resistance of Mycobacterium Paratuberculosis in Raw Milk under Conditions Simulating Pasteurization. J. Vet. Diagn. Investig. 1993, 5, 629–631. [Google Scholar] [CrossRef]
- Gerrard, Z.E.; Swift, B.M.C.; Botsaris, G.; Davidson, R.S.; Hutchings, M.R.; Huxley, J.N.; Rees, C.E.D. Survival of Mycobacterium avium subspecies paratuberculosis in retail pasteurised milk. Food Microbiol. 2018, 74, 57–63. [Google Scholar] [CrossRef]
- Grant, I.R.; Ball, H.J.; Rowe, M.T. Effect of higher pasteurization temperatures, and longer holding times at 72 °C, on the inactivation ofMycobacterium paratuberculosisin milk. Lett. Appl. Microbiol. 1999, 28, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Paolicchi, F.; Cirone, K.; Morsella, C.; Gioffré, A. First isolation of Mycobacterium avium subsp Paratuberculosis from commercial pasteurized milk in Argentina. Braz. J. Microbiol. 2012, 43, 1034–1037. [Google Scholar] [CrossRef] [PubMed]
- Fechner, K.; Dreymann, N.; Schimkowiak, S.; Czerny, C.-P.; Teitzel, J. Efficacy of dairy on-farm high-temperature, short-time pasteurization of milk on the viability of Mycobacterium avium ssp. paratuberculosis. J. Dairy Sci. 2019, 102, 11280–11290. [Google Scholar] [CrossRef]
- Robertson, R.E.; Cerf, O.; Condron, R.J.; Donaghy, J.A.; Heggum, C.; Jordan, K. Review of the controversy over whether or not Mycobacterium avium subsp. paratuberculosis poses a food safety risk with pasteurised dairy products. Int. Dairy J. 2017, 73, 10–18. [Google Scholar] [CrossRef]
- Grant, I.R.; Williams, A.G.; Rowe, M.T.; Muir, D.D. Efficacy of Various Pasteurization Time-Temperature Conditions in Combination with Homogenization on Inactivation of Mycobacterium avium subsp. paratuberculosis in Milk. Appl. Environ. Microbiol. 2005, 71, 2853–2861. [Google Scholar] [CrossRef] [PubMed]
- Lamont, E.A.; Bannantine, J.P.; Armién, A.; Ariyakumar, D.S.; Sreevatsan, S. Identification and Characterization of a Spore-Like Morphotype in Chronically Starved Mycobacterium avium Subsp. Paratuberculosis Cultures. PLoS ONE 2012, 7, e30648. [Google Scholar] [CrossRef]
- Guo, M.S.; Gross, C.A. Stress-Induced Remodeling of the Bacterial Proteome. Curr. Biol. 2014, 24, R424–R434. [Google Scholar] [CrossRef]
- Lippolis, J.D.; Bayles, D.O.; Reinhardt, T.A. Proteomic Changes in Escherichia coli When Grown in Fresh Milk versus Laboratory Media. J. Proteome Res. 2009, 8, 149–158. [Google Scholar] [CrossRef]
- Um, J.; Manguy, J.; Anes, J.; Jacquier, J.-C.; Hurley, D.; Dillon, E.T.; Wynne, K.; Fanning, S.; O’Sullivan, M.; Shields, D.C. Enriching antimicrobial peptides from milk hydrolysates using pectin/alginate food-gels. Food Chem. 2021, 352, 129220. [Google Scholar] [CrossRef]
- Derzelle, S.; Bolotin, A.; Mistou, M.-Y.; Rul, F. Proteome Analysis of Streptococcus thermophilus Grown in Milk Reveals Pyruvate Formate-Lyase as the Major Upregulated Protein. Appl. Environ. Microbiol. 2005, 71, 8597–8605. [Google Scholar] [CrossRef] [PubMed]
- Gitton, C.; Meyrand, M.; Wang, J.; Caron, C.; Trubuil, A.; Guillot, A.; Mistou, M.-Y. Proteomic Signature of Lactococcus lactis NCDO763 Cultivated in Milk. Appl. Environ. Microbiol. 2005, 71, 7152–7163. [Google Scholar] [CrossRef][Green Version]
- Chiodini, R.J.; Van Kruiningen, H.J.; Thayer, W.R.; Coutu, J.A. Spheroplastic phase of mycobacteria isolated from patients with Crohn’s disease. J. Clin. Microbiol. 1986, 24, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.; Shafran, I.; Romero, C.; Piromalli, C.; Biggerstaff, J.; Naser, N.; Chamberlin, W.; Naser, S.A. Use of short-term culture for identification of Mycobacterium avium subsp. paratuberculosis in tissue from Crohn’s disease patients. Clin. Microbiol. Infect. 2000, 6, 303–307. [Google Scholar] [CrossRef][Green Version]
- Sechi, L.A.; Scanu, A.M.; Molicotti, P.; Cannas, S.; Mura, M.; Dettori, G.; Fadda, G.; Zanetti, S. Detection and isolation of Mycobacterium avium subspecies paratuberculosis from intestinal mucosal biopsies of patients with and without Crohn’s disease in Sardinia. Am. J. Gastroenterol. 2005, 100, 1529–1536. [Google Scholar] [CrossRef]
- Kleinwort, K.J.H.; Hauck, S.M.; Degroote, R.L.; Scholz, A.M.; Hölzel, C.; Maertlbauer, E.P.; Deeg, C. Peripheral blood bovine lymphocytes and MAP show distinctly different proteome changes and immune pathways in host-pathogen interaction. PeerJ 2019, 7, e8130. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, L.E.; Petrofsky, M.; Sommer, S.; Barletta, R.G. Peyer’s Patch-Deficient Mice Demonstrate That Mycobacterium avium subsp. paratuberculosis Translocates across the Mucosal Barrier via both M Cells and Enterocytes but Has Inefficient Dissemination. Infect. Immun. 2010, 78, 3570–3577. [Google Scholar] [CrossRef]
- Alonso-Hearn, M.; Eckstein, T.M.; Sommer, S.; Bermudez, L.E. A Mycobacterium avium subsp. paratuberculosis LuxR regulates cell envelope and virulence. Innate Immun. 2010, 16, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Johansen, M.D.; Herrmann, J.-L.; Kremer, L. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 2020, 18, 392–407. [Google Scholar] [CrossRef]
- Richter, E.; Wessling, J.; Lügering, N.; Domschke, W.; Rüsch-Gerdes, S. Mycobacterium aviumsubsp.paratuberculosisInfection in a Patient with HIV, Germany. Emerg. Infect. Dis. 2002, 8, 729–731. [Google Scholar] [CrossRef]
- Zhu, Z.; Guo, W. Recent developments on rapid detection of main constituents in milk: A review. Crit. Rev. Food Sci. Nutr. 2021, 61, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-H.; Xu, D.; Liu, J.-Q.; Zhao, X.-H. Enhanced degradation of five organophosphorus pesticides in skimmed milk by lactic acid bacteria and its potential relationship with phosphatase production. Food Chem. 2014, 164, 173–178. [Google Scholar] [CrossRef]
- Ijaq, J.; Malik, G.; Kumar, A.; Das, P.S.; Meena, N.; Bethi, N.; Sundararajan, V.S.; Suravajhala, P. A model to predict the function of hypothetical proteins through a nine-point classification scoring schema. BMC Bioinform. 2019, 20, 14. [Google Scholar] [CrossRef]
- Kumar, K.; Prakash, A.; Tasleem, M.; Islam, A.; Ahmad, F.; Hassan, M.I. Functional annotation of putative hypothetical proteins from Candida dubliniensis. Gene 2014, 543, 93–100. [Google Scholar] [CrossRef]
- Desler, C.; Suravajhala, P.; Sanderhoff, M.; Rasmussen, M.; Rasmussen, L.J. In Silico screening for functional candidates amongst hypothetical proteins. BMC Bioinform. 2009, 10, 289. [Google Scholar] [CrossRef]
- Yang, Z.; Zeng, X.; Tsui, S.K.-W. Investigating function roles of hypothetical proteins encoded by the Mycobacterium tuberculosis H37Rv genome. BMC Genom. 2019, 20, 394. [Google Scholar] [CrossRef] [PubMed]
- Aguttu, C.; Okech, B.A.; Mukisa, A.; Lubega, G.W. Screening and characterization of hypothetical proteins of Plasmodium falciparum as novel vaccine candidates in the fight against malaria using reverse vaccinology. J. Genet. Eng. Biotechnol. 2021, 19, 103. [Google Scholar] [CrossRef]
- Sette, A.; Rappuoli, R. Reverse Vaccinology: Developing Vaccines in the Era of Genomics. Immunity 2010, 33, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.E.; Gawronski, J.D.; De Jesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLOS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef]
- Guillet, M.; Van Der Kemp, P.A.; Boiteux, S. dUTPase activity is critical to maintain genetic stability in Saccharomyces cerevisiae. Nucleic Acids Res. 2006, 34, 2056–2066. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Q.; Ye, F.; Huang, C.Y.; Chen, W.-M.; Huang, W.-Q. Alda-1, an ALDH2 activator, protects against hepatic ischemia/reperfusion injury in rats via inhibition of oxidative stress. Free Radic. Res. 2018, 52, 629–638. [Google Scholar] [CrossRef]
- Perez-Miller, S.; Younus, H.; Vanam, R.; Chen, C.-H.; Mochly-Rosen, D.; Hurley, T.D. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat. Struct. Mol. Biol. 2010, 17, 159–164. [Google Scholar] [CrossRef]
- Chen, C.-H.; Ferreira, J.C.; Gross, E.R.; Mochly-Rosen, D. Targeting Aldehyde Dehydrogenase 2: New Therapeutic Opportunities. Physiol. Rev. 2014, 94, 1–34. [Google Scholar] [CrossRef]
- Fu, S.-H.; Zhang, H.-F.; Yang, Z.B.; Li, T.-B.; Liu, B.; Lou, Z.; Ma, Q.-L.; Luo, X.-J.; Peng, J. Alda-1 reduces cerebral ischemia/reperfusion injury in rat through clearance of reactive aldehydes. Naunyn-Schmiedebergs Arch. Pharmacol. 2014, 387, 87–94. [Google Scholar] [CrossRef]
- Lu, Q.; Mundy, M.; Chambers, E.; Lange, T.; Newton, J.; Borgas, D.; Yao, H.; Choudhary, G.; Basak, R.; Oldham, M.; et al. Alda-1 Protects Against Acrolein-Induced Acute Lung Injury and Endothelial Barrier Dysfunction. Am. J. Respir. Cell Mol. Biol. 2017, 57, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; He, G.; Wang, J.; Wang, Y.; Chen, W. Pretreatment with the ALDH2 agonist Alda-1 reduces intestinal injury induced by ischaemia and reperfusion in mice. Clin. Sci. 2017, 131, 1123–1136. [Google Scholar] [CrossRef]
- Patil, S.S.; Hernández-Cuervo, H.; Fukumoto, J.; Narala, V.R.; Saji, S.; Borra, M.; Alleyn, M.; Lin, M.; Soundararajan, R.; Lockey, R.; et al. Alda-1 attenuates hyperoxia-induced mitochondrial dysfunction in lung vascular endothelial cells. Aging 2019, 11, 3909–3918. [Google Scholar] [CrossRef] [PubMed]
- Hua, T.; Yang, M.; Zhou, Y.; Chen, L.; Wu, H.; Liu, R. Alda-1 Prevents Pulmonary Epithelial Barrier Dysfunction following Severe Hemorrhagic Shock through Clearance of Reactive Aldehydes. BioMed Res. Int. 2019, 2019, 2476252. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Chen, M.; Che, R.; Ding, W.; Yu, F.; Zhou, Y.; Cui, W.; Xiaoxu, X.; God’Spower, B.-O.; et al. Comparative proteomic analysis reveals drug resistance of Staphylococcus xylosus ATCC700404 under tylosin stress. BMC Vet. Res. 2019, 15, 224. [Google Scholar] [CrossRef]
- Briozzo, P.; Evrin, C.; Meyer, P.; Assairi, L.; Joly, N.; Bārzu, O.; Gilles, A.-M. Structure of Escherichia coli UMP Kinase Differs from That ofOther Nucleoside Monophosphate Kinases and Sheds New Light on EnzymeRegulation. J. Biol. Chem. 2005, 280, 25533–25540. [Google Scholar] [CrossRef]
- Kim, Y.R.; Lee, S.E.; Kim, C.M.; Kim, S.Y.; Shin, E.K.; Shin, D.H.; Chung, S.S.; Choy, H.E.; Progulske-Fox, A.; Hillman, J.D.; et al. Characterization and Pathogenic Significance of Vibrio vulnificus Antigens Preferentially Expressed in Septicemic Patients. Infect. Immun. 2003, 71, 5461–5471. [Google Scholar] [CrossRef]
- Lee, S.E.; Kim, S.Y.; Kim, C.M.; Kim, M.-K.; Kim, Y.R.; Jeong, K.; Ryu, H.-J.; Lee, Y.S.; Chung, S.S.; Choy, H.E.; et al. The pyrH Gene of Vibrio vulnificus Is an Essential In Vivo Survival Factor. Infect. Immun. 2007, 75, 2795–2801. [Google Scholar] [CrossRef] [PubMed]
- Brzostek, A.; Płociński, P.; Minias, A.; Ciszewska, A.; Gąsior, F.; Pawełczyk, J.; Dziadek, B.; Słomka, M.; Dziadek, J. Dissecting the RecA-(In)dependent Response to Mitomycin C in Mycobacterium tuberculosis Using Transcriptional Profiling and Proteomics Analyses. Cells 2021, 10, 1168. [Google Scholar] [CrossRef]
- Knyazev, D.G.; Kuttner, R.; Bondar, A.-N.; Zimmerman, M.; Siligan, C.; Pohl, P. Voltage Sensing in Bacterial Protein Translocation. Biomolecules 2020, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Abdellrazeq, G.S.; Elnaggar, M.M.; Bannantine, J.P.; Park, K.T.; Souza, C.D.; Backer, B.; Hulubei, V.; Fry, L.M.; Khaliel, S.A.; Torky, H.A.; et al. A Mycobacterium avium subsp. paratuberculosis relA deletion mutant and a 35 kDa major membrane protein elicit development of cytotoxic T lymphocytes with ability to kill intracellular bacteria. Vet. Res. 2018, 49, 53. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-W.; Schmoller, S.K.; Shin, S.J.; Talaat, A.M. Defining the Stressome of Mycobacterium avium subsp. paratuberculosis In Vitro and in Naturally Infected Cows. J. Bacteriol. 2007, 189, 7877–7886. [Google Scholar] [CrossRef]
- Khanam, T.; Rai, N.; Ramachandran, R. Mycobacterium tuberculosis class II apurinic/apyrimidinic-endonuclease/3′-5′ exonuclease III exhibits DNA regulated modes of interaction with the sliding DNA β-clamp. Mol. Microbiol. 2015, 98, 46–68. [Google Scholar] [CrossRef] [PubMed]
- Gaudion, A.; Dawson, L.; Davis, E.; Smollett, K. Characterisation of the Mycobacterium tuberculosis alternative sigma factor SigG: Its operon and regulon. Tuberculosis 2013, 93, 482–491. [Google Scholar] [CrossRef][Green Version]
- Dunphy, K.Y.; Senaratne, R.H.; Masuzawa, M.; Kendall, L.V.; Riley, L.W. Attenuation ofMycobacterium tuberculosisFunctionally Disrupted in a Fatty Acyl–Coenzyme A Synthetase GenefadD5. J. Infect. Dis. 2010, 201, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Domenech, P.; Reed, M.B.; Barry, C.E., 3rd. Contribution of the Mycobacterium tuberculosis MmpL Protein Family to Virulence and Drug Resistance. Infect. Immun. 2005, 73, 3492–3501. [Google Scholar] [CrossRef]
- Danelishvili, L.; Chinison, J.J.J.; Pham, T.; Gupta, R.; Bermudez, L.E. The Voltage-Dependent Anion Channels (VDAC) of Mycobacterium avium phagosome are associated with bacterial survival and lipid export in macrophages. Sci. Rep. 2017, 7, 7007. [Google Scholar] [CrossRef]
- Wang, R.; Ehrt, S. Rv0954 Is a Member of the Mycobacterial Cell Division Complex. Front. Microbiol. 2021, 12, 626461. [Google Scholar] [CrossRef]
- Melly, G.C.; Stokas, H.; Davidson, P.M.; Roma, J.S.; Rhodes, H.L.; Purdy, G.E. Identification of residues important for M. tuberculosis MmpL11 function reveals that function is modulated by phosphorylation in the C-terminal domain. Mol. Microbiol. 2021, 115, 208–221. [Google Scholar] [CrossRef]
- McDowell, M.A.; Heimes, M.; Sinning, I. Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat. Struct. Mol. Biol. 2021, 28, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Hobmaier, B.F.; Lutterberg, K.; Kleinwort, K.J.H.; Mayer, R.; Hirmer, S.; Amann, B.; Hölzel, C.; Märtlbauer, E.P.; Deeg, C.A. Characterization of plant lectins for their ability to isolate Mycobacterium avium subsp. paratuberculosis from milk. Food Microbiol. 2019, 82, 231–239. [Google Scholar] [CrossRef]
- Slana, I.; Paolicchi, F.; Janštová, B.; Navrátilová, P.; Pavlík, I. Detection methods for Mycobacterium avium subsp paratuberculosis in milk and milk products: A review. Veterinární Med. 2008, 53, 283–306. [Google Scholar] [CrossRef]
- Sweeney, R.W.; Collins, M.T.; Koets, A.P.; McGuirk, S.M.; Roussel, A.J. Paratuberculosis (Johne’s Disease) in Cattle and Other Susceptible Species. J. Vet. Intern. Med. 2012, 26, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Soumya, M.P.; Pillai, R.M.; Antony, P.X.; Mukhopadhyay, H.K.; Rao, V.N. Comparison of faecal culture and IS900 PCR assay for the detection of Mycobacterium avium subsp. paratuberculosis in bovine faecal samples. Vet. Res. Commun. 2009, 33, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-T.; Park, H.-E.; Jung, Y.-H.; Yoo, H.S. An ISMap 02—Like insertion sequence in Mycobacterium spp. interferes with specific detection of Mycobacterium avium subsp. paratuberculosis. Vet. Microbiol. 2018, 216, 1–6. [Google Scholar] [CrossRef]
- Grosche, A.; Hauser, A.; Lepper, M.F.; Mayo, R.; von Toerne, C.; Merl-Pham, J.; Hauck, S.M. The Proteome of Native Adult Müller Glial Cells from Murine Retina. Mol. Cell. Proteom. 2016, 15, 462–480. [Google Scholar] [CrossRef] [PubMed]
- Hauck, S.M.; Hofmaier, F.; Dietter, J.; Swadzba, M.E.; Blindert, M.; Amann, B.; Behler, J.; Kremmer, E.; Ueffing, M.; Deeg, C.A. Label-free LC-MSMS analysis of vitreous from autoimmune uveitis reveals a significant decrease in secreted Wnt signalling inhibitors DKK3 and SFRP2. J. Proteom. 2012, 75, 4545–4554. [Google Scholar] [CrossRef] [PubMed]
- Hauck, S.M.; Lepper, M.F.; Hertl, M.; Sekundo, W.; Deeg, C.A. Proteome Dynamics in Biobanked Horse Peripheral Blood Derived Lymphocytes (PBL) with Induced Autoimmune Uveitis. Proteomics 2017, 17, 1700013. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
| Protein ID | Accession | Peptide Count | Ratio | Functional Affiliation | ||
|---|---|---|---|---|---|---|
| Milk 4 °C/M7H9 | Milk 37 °C/M7H9 | Milk 37 °C/Milk 4 °C | ||||
| Dut | AAS05131 | 1 | infinite | infinite | 0.5 | metabolism |
| AldA_1 | AAS02919 | 1 | 75.9 | 23.4 | 0.3 | |
| PyrH | AAS05263 | 2 | 21.2 | 8.8 | 0.4 | |
| DnaE1 | AAS03574 | 2 | 939.7 | 1365.0 | 1.5 | transcription and translation |
| SigG | AAS06171 | 1 | 23.7 | 42.9 | 1.8 | |
| XthA | AAS06466 | 1 | 17.1 | 16.6 | 1.0 | |
| CrgA | AAS02330 | 4 | 14.1 | 18.5 | 1.3 | cell wall synthesis/cell division |
| MmpL11 | AAS06187 | 3 | 12.2 | 13.5 | 1.1 | |
| YidC | AAS06897 | 4 | 6.7 | 7.4 | 1.1 | |
| MmpL4_1 | AAS02393 | 1 | infinite | infinite | 1.6 | fatty acid metabolism/fatty acid synthesis |
| FadD5 | AAS06151 | 2 | 122.9 | 103.1 | 0.8 | |
| LipN | AAS05554 | 2 | 6.0 | 5.9 | 1.0 | |
| RecA | AAS05165 | 1 | infinite | infinite | 0.7 | stress response/immune evasion/antibiotic resistance |
| RelA | AAS03364 | 3 | 313.2 | 8371.5 | 26.7 | |
| SecE | AAS06660 | 3 | 26.5 | 25.4 | 1.0 | |
| Protein ID | Accession | Peptide Count | Ratio | |||
|---|---|---|---|---|---|---|
| Milk 4 °C/M7H9 | Milk 37 °C/M7H9 | Milk 4 °C/Milk 37 °C | ||||
| 1 | RpoC | AAS06681 | 55 | 1.0 | 1.4 | 0.7 |
| 2 | Fas | AAS04649 | 41 | 1.4 | 0.7 | 2.0 |
| 3 | Kgd | AAS04853 | 41 | 0.8 | 1.1 | 0.7 |
| 4 | RpoB | AAS06680 | 41 | 0.6 | 0.8 | 0.8 |
| 5 | MetE | AAS04978 | 39 | 0.7 | 0.9 | 0.8 |
| 6 | AceAb | AAS03960 | 34 | 0.9 | 1.3 | 0.7 |
| 7 | NAD-glutamate dehydrogenase | ETB03859 | 33 | 1.5 | 1.3 | 1.1 |
| 8 | Catalase/hydroperoxidase HPI(I) | ETB03832 | 32 | 1.2 | 0.6 | 1.8 |
| 9 | Aconitate hydratase | ETB10875 | 30 | 1.3 | 1.3 | 1.0 |
| 10 | GyrA | AAS02323 | 29 | 0.9 | 1.0 | 0.9 |
| 11 | Glutamate synthase | ETB00293 | 29 | 1.8 | 1.5 | 1.2 |
| 12 | Rne | AAS04584 | 28 | 0.8 | 1.0 | 0.8 |
| 13 | MmpL3 | AAS06191 | 28 | 0.8 | 0.8 | 0.9 |
| 14 | 3-Hydroxyacyl-CoA dehydrogenase | ETB02135 | 28 | 1.2 | 1.1 | 1.2 |
| 15 | IleS | AAS03563 | 27 | 0.9 | 0.9 | 1.1 |
| 16 | MAP_3698c | AAS06248 | 27 | 1.0 | 1.2 | 0.8 |
| 17 | Long-chain fatty acid-CoA ligase | ETB02782 | 27 | 0.5 | 0.6 | 0.9 |
| 18 | CtpI | AAS06048 | 26 | 1.4 | 1.1 | 1.3 |
| 19 | PckA | AAS06196 | 26 | 1.0 | 0.5 | 1.9 |
| 20 | ClpC | AAS02778 | 25 | 0.6 | 0.9 | 0.7 |
| 21 | Transketolase | ETB00031 | 25 | 1.0 | 0.8 | 1.2 |
| 22 | DNA topoisomerase I | ETB01242 | 25 | 1.5 | 1.7 | 0.8 |
| 23 | ABC transporter ATP-binding protein | ETB03858 | 25 | 0.8 | 0.9 | 1.0 |
| 24 | PonA_1 | AAS02381 | 24 | 0.6 | 0.9 | 0.6 |
| 25 | PolA | AAS03639 | 24 | 0.7 | 0.8 | 0.8 |
| 26 | ValS | AAS04588 | 24 | 0.7 | 0.8 | 0.8 |
| 27 | Rho | AAS04781 | 24 | 1.0 | 1.2 | 0.8 |
| 28 | SahH | AAS05912 | 24 | 1.8 | 2.0 | 0.9 |
| 29 | Pyruvate dehydrogenase E1 | ETB03102 | 24 | 0.5 | 0.8 | 0.7 |
| 30 | ATP synthase subunit beta | ETB04389 | 24 | 1.4 | 1.1 | 1.3 |
| 31 | Pca | AAS02611 | 23 | 1.6 | 1.3 | 1.2 |
| 32 | GltA2 | AAS03146 | 23 | 0.9 | 0.7 | 1.2 |
| 33 | GlnE | AAS04282 | 22 | 1.6 | 1.7 | 0.9 |
| 34 | InfB | AAS05224 | 21 | 0.6 | 0.9 | 0.7 |
| 35 | MAP_3291c | AAS05841 | 21 | 0.7 | 1.1 | 0.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinwort, K.J.H.; Hobmaier, B.F.; Mayer, R.; Hölzel, C.; Degroote, R.L.; Märtlbauer, E.; Hauck, S.M.; Deeg, C.A. Mycobacterium avium subsp. paratuberculosis Proteome Changes Profoundly in Milk. Metabolites 2021, 11, 549. https://doi.org/10.3390/metabo11080549
Kleinwort KJH, Hobmaier BF, Mayer R, Hölzel C, Degroote RL, Märtlbauer E, Hauck SM, Deeg CA. Mycobacterium avium subsp. paratuberculosis Proteome Changes Profoundly in Milk. Metabolites. 2021; 11(8):549. https://doi.org/10.3390/metabo11080549
Chicago/Turabian StyleKleinwort, Kristina J. H., Bernhard F. Hobmaier, Ricarda Mayer, Christina Hölzel, Roxane L. Degroote, Erwin Märtlbauer, Stefanie M. Hauck, and Cornelia A. Deeg. 2021. "Mycobacterium avium subsp. paratuberculosis Proteome Changes Profoundly in Milk" Metabolites 11, no. 8: 549. https://doi.org/10.3390/metabo11080549
APA StyleKleinwort, K. J. H., Hobmaier, B. F., Mayer, R., Hölzel, C., Degroote, R. L., Märtlbauer, E., Hauck, S. M., & Deeg, C. A. (2021). Mycobacterium avium subsp. paratuberculosis Proteome Changes Profoundly in Milk. Metabolites, 11(8), 549. https://doi.org/10.3390/metabo11080549






