A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis
Abstract
:1. Introduction
2. Results
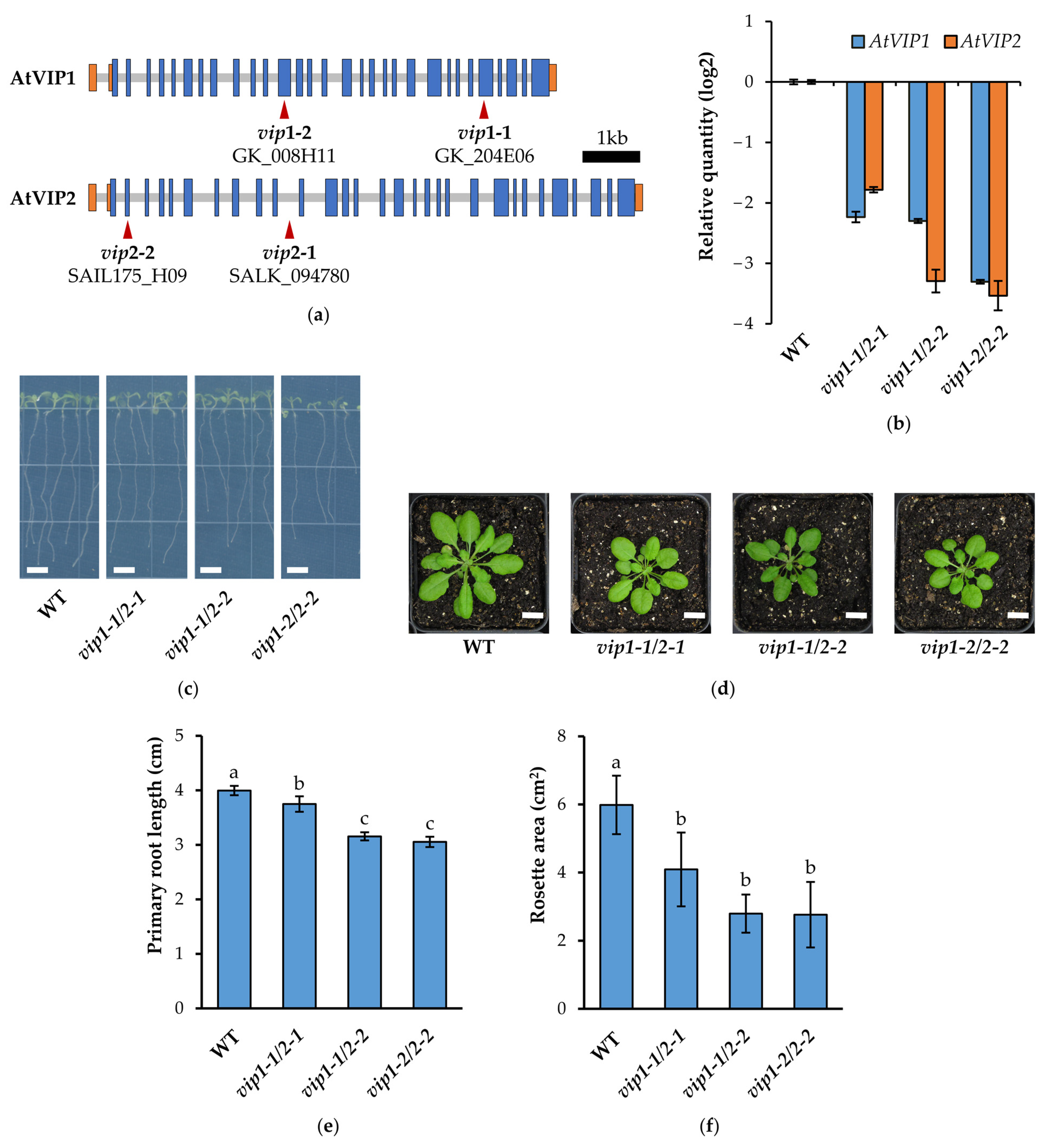
2.1. vip Double Mutants Are Characterized by a Reduced Growth Phenotype and Transcriptional Changes
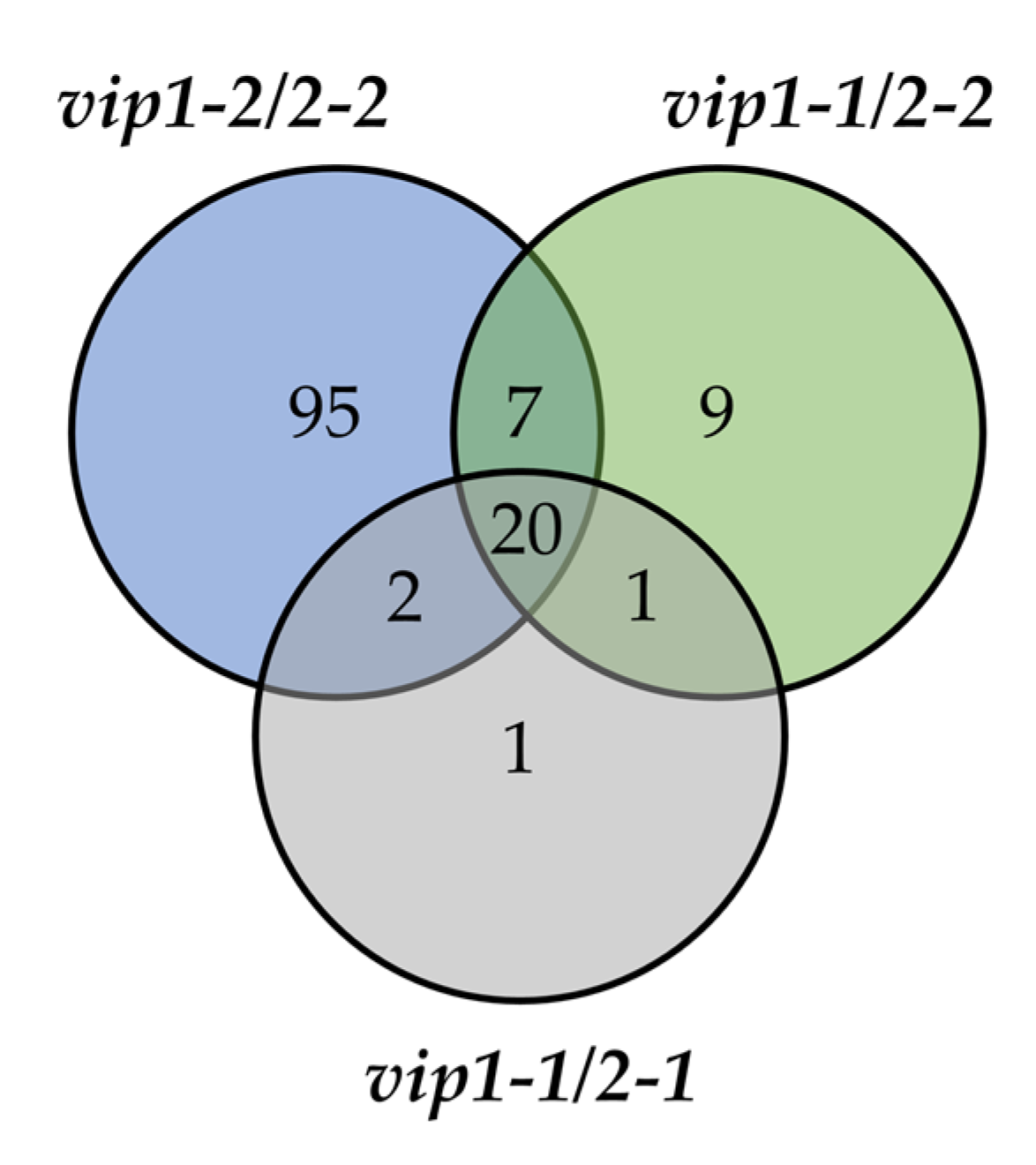
2.2. Systemic Phosphate Starvation Response Genes Are Downregulated in vip Double Mutants
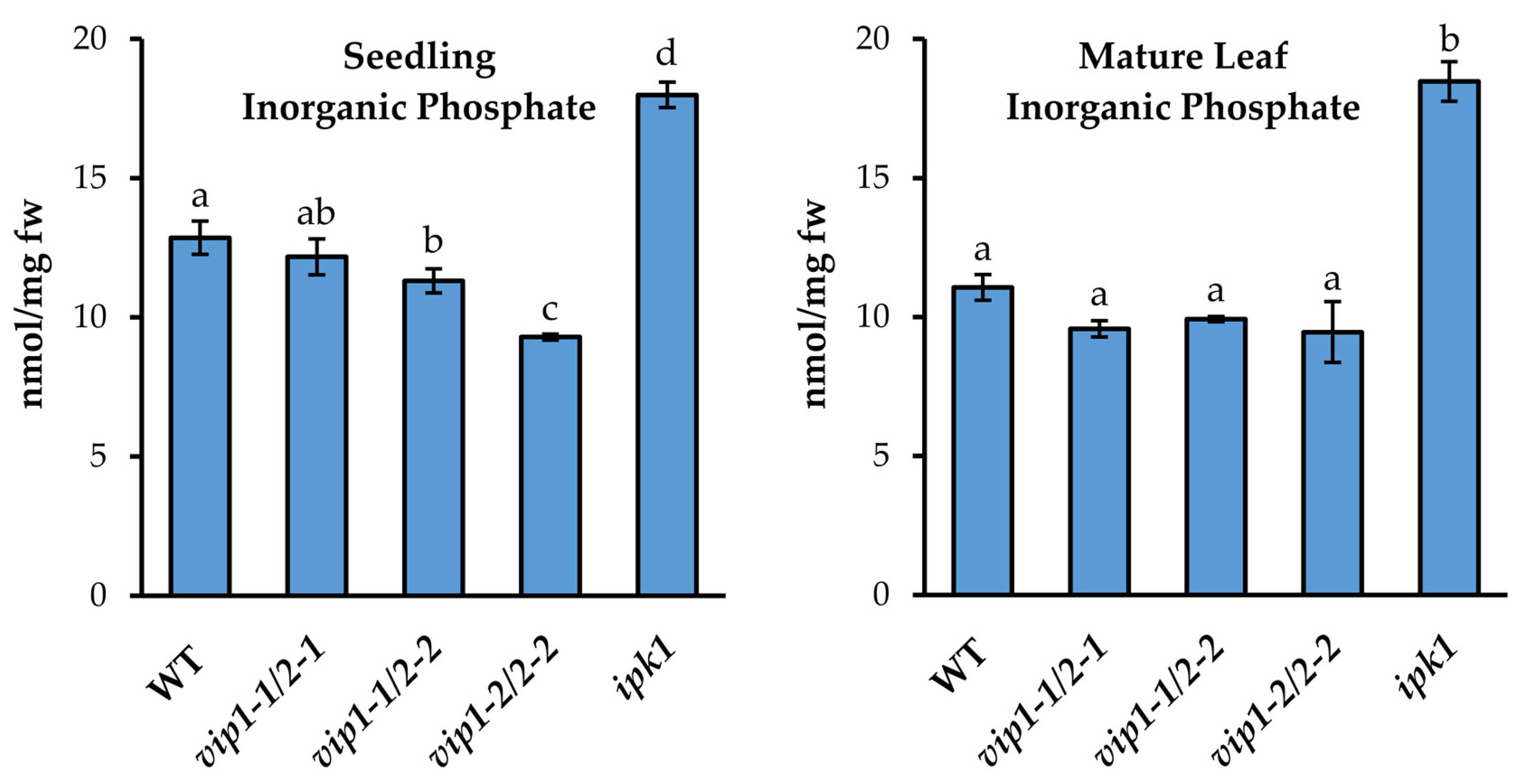
2.3. Inorganic Phosphate Levels Are Altered in vip Double Mutants
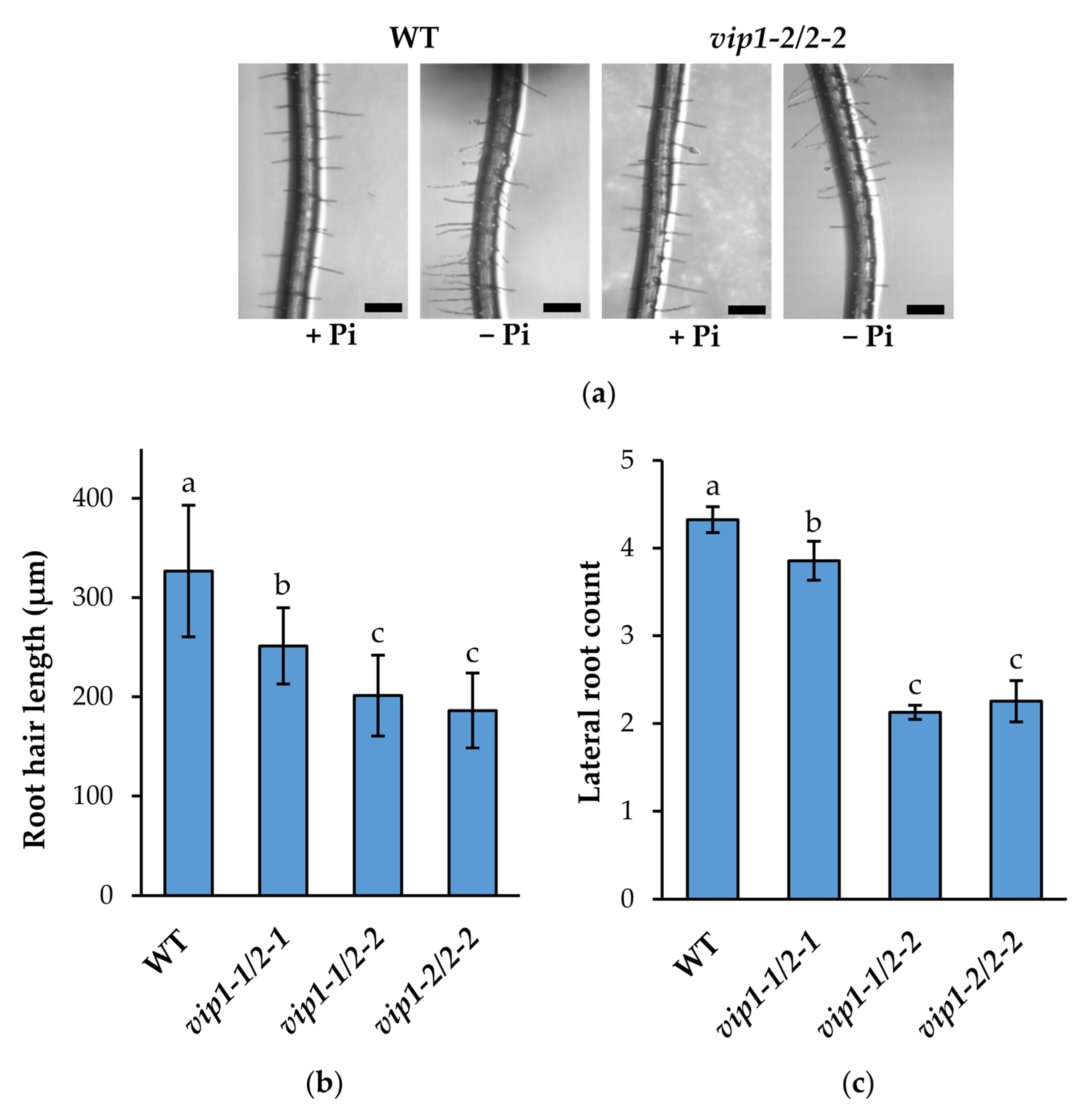
2.4. Root System Architecture Is Significantly Altered in vip Double Mutants’ Response to Low Phosphate
2.5. Anthocyanin Accumulation Is Reduced in vip Double Mutants in Response to Low Phosphate
2.6. Phosphate Starvation Response (PSR) Genes Are Inducible in vip Double Mutants
2.7. vip1-2/vip2-2 Mutants Are Deficient in InsP8
2.8. Altered Lipid Remodeling Responses in PP-InsP-Deficient Mutants
3. Discussion
3.1. Comparison of Mutants with Disrupted Inositol Pyrophosphate Biosynthesis
3.2. Transcriptional Responses of vip Mutants
3.3. Metabolite Changes in vip Mutants
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Basal Morphometric Analysis
4.3. Low Phosphate Treatment
4.4. RNA Isolation and NextGen RNA Sequencing
4.5. Inorganic Phosphate Assay
4.6. Anthocyanin Measurements
4.7. Statistical Analysis of Morphometric and Spectrophotometric Data
4.8. RNA Isolation, cDNA Synthesis, and qRT-PCR Analysis
4.9. Plant Inositol Polyphosphate Analysis
4.10. Lipidomics Methods
4.11. Lipidomics Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herrera-Estrella, L.; López-Arredondo, D. Phosphorus: The underrated element for feeding the world. Trends Plant Sci. 2016, 21, 461–463. [Google Scholar] [CrossRef]
- Pretty, J.; Bharucha, Z.P. Sustainable intensification in agricultural systems. Ann. Bot. 2014, 114, 1571–1596. [Google Scholar] [CrossRef] [PubMed]
- Péret, B.; Clément, M.; Nussaume, L.; Desnos, T. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 2011, 16, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Bouain, N.; Doumas, P.; Rouached, H. The Root System Response to Phosphate Deficiency in Arabidopsis. Curr. Genom. 2016, 17, 308–314. [Google Scholar] [CrossRef] [Green Version]
- Péret, B.; Desnos, T.; Jost, R.; Kanno, S.; Berkowitz, O.; Nussaume, L. Root architecture responses: In search of phosphate. Plant Physiol. 2014, 166, 1713–1723. [Google Scholar] [CrossRef] [Green Version]
- Chiou, T.-J.; Lin, S.-I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [Green Version]
- Chien, P.S.; Chiang, C.P.; Leong, S.J.; Chiou, T.J. Sensing and signaling of phosphate starvation: From local to long distance. Plant Cell Physiol. 2018, 59, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.P.; White, P.J. Sugar signaling in root responses to low phosphorus availability. Plant Physiol. 2011, 156, 1033–1040. [Google Scholar] [CrossRef] [Green Version]
- Bari, R.; Pant, B.D.; Stitt, M.; Golm, S.P. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [Green Version]
- Rubio, V.; Bustos, R.; Irigoyen, M.L.; Cardona-López, X.; Rojas-Triana, M.; Paz-Ares, J. Plant hormones and nutrient signaling. Plant Mol. Biol. 2009, 69, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y. Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 2013, 52, 43–50. [Google Scholar] [CrossRef]
- Cruz-Ramírez, A.; Oropeza-Aburto, A.; Razo-Hernández, F.; Ramírez-Chávez, E.; Herrera-Estrella, L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2006, 103, 6765–6770. [Google Scholar] [CrossRef] [Green Version]
- Dörmann, P.; Benning, C. Galactolipids rule in seed plants. Trends Plant Sci. 2002, 7, 112–118. [Google Scholar] [CrossRef]
- Kalisch, B.; Dörmann, P.; Hölzl, G. Lipids in Plant and Algae Development; Springer: Cham, Switzerland, 2016; Volume 86. [Google Scholar] [CrossRef]
- Moellering, E.R.; Benning, C. Galactoglycerolipid metabolism under stress: A time for remodeling. Trends Plant Sci. 2011, 16, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Essigmann, B.; Güler, S.; Narang, R.A.; Linke, D.; Benning, C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1 a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 1950–1955. [Google Scholar] [CrossRef] [Green Version]
- Andersson, M.X.; Stridh, M.H.; Larsson, K.E.; Liljenberg, C.; Sandelius, A.S. Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 2003, 537, 128–132. [Google Scholar] [CrossRef] [Green Version]
- Shimojima, M. Biosynthesis and functions of the plant sulfolipid. Prog. Lipid Res. 2011, 50, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Burgos, A.; Pant, P.; Cuadros-Inostroza, A.; Willmitzer, L.; Scheible, W.R. The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 2015, 66, 1907–1918. [Google Scholar] [CrossRef] [Green Version]
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [Green Version]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [Green Version]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; De Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodríguez, J.; et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef] [Green Version]
- Wild, R.; Gerasimaite, R.; Jung, J.Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jacob Jessen, H.; Poirier, Y.; Hothorn, M.; et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, C.; Saiardi, A. Eukaryotic phosphate homeostasis: The inositol pyrophosphate perspective. Trends Biochem. Sci. 2017, 42, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Ried, M.K.; Hothorn, M.; Poirier, Y. Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr. Opin. Biotechnol. 2018, 49, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Raboy, V. The ABCs of low-phytate crops. Nat. Biotechnol. 2007, 25, 874–875. [Google Scholar] [CrossRef] [PubMed]
- Adepoju, O.; Williams, S.P.; Craige, B.; Cridland, C.A.; Sharpe, A.K.; Brown, A.M.; Land, E.; Perera, I.Y.; Mena, D.; Sobrado, P.; et al. Inositol trisphosphate kinase and diphosphoinositol pentakisphosphate kinase enzymes constitute the inositol pyrophosphate synthesis pathway in plants. arXiv 2019, arXiv:10.1101/724914, 724914. [Google Scholar]
- Laha, D.; Parvin, N.; Hofer, A.; Giehl, R.F.H.; Fernandez-Rebollo, N.; Von Wirén, N.; Saiardi, A.; Jessen, H.J.; Schaaf, G. Arabidopsis ITPK1 and ITPK2 Have an Evolutionarily Conserved Phytic Acid Kinase Activity. ACS Chem. Biol. 2019, 14, 2127–2133. [Google Scholar] [CrossRef] [Green Version]
- Desai, M.; Rangarajan, P.; Donahue, J.L.; Williams, S.P.; Land, E.S.; Mandal, M.K.; Phillippy, B.Q.; Perera, I.Y.; Raboy, V.; Gillaspy, G.E. Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J. 2014, 80, 642–653. [Google Scholar] [CrossRef]
- Laha, D.; Johnen, P.; Azevedo, C.; Dynowski, M.; Weiß, M.; Capolicchio, S.; Mao, H.; Iven, T.; Steenbergen, M.; Freyer, M.; et al. VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in arabidopsis. Plant Cell 2015, 27, 1082–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Lau, K.; Puschmann, R.; Harmel, R.K.; Zhang, Y.; Pries, V.; Gaugler, P.; Broger, L.; Dutta, A.K.; Jessen, H.J.; et al. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. Elife 2019, 8, 1–25. [Google Scholar] [CrossRef]
- Stevenson-Paulik, J.; Bastidas, R.J.; Chiou, S.-T.T.; Frye, R.A.; York, J.D. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 2005, 102, 12612–12617. [Google Scholar] [CrossRef] [Green Version]
- Kuo, H.F.; Chang, T.Y.; Chiang, S.F.; Wang, W.D.; Charng, Y.Y.; Chiou, T.J. Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J. 2014, 80, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.F.; Hsu, Y.Y.; Lin, W.C.; Chen, K.Y.; Munnik, T.; Brearley, C.A.; Chiou, T.J. Arabidopsis inositol phosphate kinases IPK1 and ITPK1 constitute a metabolic pathway in maintaining phosphate homeostasis. Plant J. 2018, 95, 613–630. [Google Scholar] [CrossRef] [Green Version]
- Shears, S.B.; Baughman, B.M.; Gu, C.; Nair, V.S.; Wang, H. The significance of the 1-kinase/1-phosphatase activities of the PPIP5K family. Adv. Biol. Regul. 2017, 63, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Nguyen, H.-N.; Hofer, A.; Jessen, H.J.; Dai, X.; Wang, H.; Shears, S.B. The Significance of the bifunctional kinase/phosphatase activities of diphosphoinositol pentakisphosphate kinases (PPIP5Ks) for coupling inositol pyrophosphate cell signaling to cellular phosphate homeostasis. J. Biol. Chem. 2017, 292, 4544–4555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Ma, G.; Sui, L.; Wei, M.; Satheesh, V.; Zhang, R.; Ge, S.; Li, J.; Zhang, T.E.; Wittwer, C.; et al. Inositol pyrophosphate insp8 acts as an intracellular phosphate signal in Arabidopsis. Mol. Plant 2019, 12, 1463–1473. [Google Scholar] [CrossRef]
- Ried, M.K.; Wild, R.; Zhu, J.; Pipercevic, J.; Sturm, K.; Broger, L.; Harmel, R.K.; Abriata, L.A.; Hothorn, L.A.; Fiedler, D.; et al. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 2021, 12, 384. [Google Scholar] [CrossRef] [PubMed]
- Crombez, H.; Motte, H.; Beeckman, T. Tackling plant phosphate starvation by the roots. Dev. Cell 2019, 48, 599–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-Specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zhang, X.; Li, L.; Sun, Z.; Li, J.; Chen, X.; Hong, G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 2021, 230, 205–217. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhou, W.; El Sheery, N.I.; Peters, C.; Li, M.; Wang, X.; Huang, J. Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 2011, 66, 781–795. [Google Scholar] [CrossRef]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Raghothama, K.G.; Smith, A.P. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef] [Green Version]
- Acevedo-Hernández, G.; Oropeza-Aburto, A.; Herrera-Estrella, L. A specific variant of the PHR1 binding site is highly enriched in the Arabidopsis phosphate-responsive phospholipase DZ2 coexpression network. Plant Signal. Behav. 2012, 7, 914–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickman, R.; Van Verk, M.C.; Van Dijken, A.J.H.; Mendes, M.P.; Vroegop-Vos, I.A.; Caarls, L.; Steenbergen, M.; Van der Nagel, I.; Wesselink, G.J.; Jironkin, A.; et al. Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell 2017, 29, 2086–2105. [Google Scholar] [CrossRef] [Green Version]
- Zander, M.; Lewsey, M.G.; Clark, N.M.; Yin, L.; Bartlett, A.; Saldierna Guzmán, J.P.; Hann, E.; Langford, A.E.; Jow, B.; Wise, A.; et al. Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 2020, 6, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z. Molecular basis for jasmonate and ethylene signal interactions in Arabidopsis. J. Exp. Bot. 2014, 65, 5743–5748. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T. Chloroplast NDH: A different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 1015–1022. [Google Scholar] [CrossRef]
- Couso, I.; Evans, B.S.; Li, J.; Liu, Y.; Ma, F.; Diamond, S.; Allen, D.K.; Umen, J.G. Synergism between inositol polyphosphates and TOR kinase signaling in nutrient sensing, growth control, and lipid metabolism in chlamydomonas. Plant Cell 2016, 28, 2026–2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinboelting, N.; Huep, G.; Kloetgen, A.; Viehoever, P.; Weisshaar, B. GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012, 40, D1211–D1215. [Google Scholar] [CrossRef] [Green Version]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-Wide insertional mutagenesis Arabidopsis thaliana. Science 2003, 301, 5633. [Google Scholar] [CrossRef] [Green Version]
- Sessions, A.; Burke, E.; Presting, G.; Aux, G.; McElver, J.; Patton, D.; Dietrich, B.; Ho, P.; Bacwaden, J.; Ko, C.; et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell 2002, 14, 2985–2994. [Google Scholar] [CrossRef]
- Ames, B.N. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966, 8, 115–118. [Google Scholar] [CrossRef]
- He, X.; Li, Y.; Lawson, D.; Xie, D.Y. Metabolic engineering of anthocyanins in dark tobacco varieties. Physiol. Plant. 2017, 159, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Saiardi, A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 2006, 1, 2416–2422. [Google Scholar] [CrossRef] [PubMed]
- Shiva, S.; Enninful, R.; Roth, M.R.; Tamura, P.; Jagadish, K.; Welti, R. An efficient modified method for plant leaf lipid extraction results in improved recovery of phosphatidic acid. Plant Methods 2018, 14, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- The LIPID MAPS®. Lipidomics Gateway. Available online: https://www.lipidmaps.org/ (accessed on 1 May 2021).
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35, 606–612. [Google Scholar] [CrossRef] [Green Version]
- Castro-Perez, J.M.; Kamphorst, J.; Degroot, J.; Lafeber, F.; Goshawk, J.; Yu, K.; Shockcor, J.P.; Vreeken, R.J.; Hankemeier, T. Comprehensive LC-MSE lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J. Proteome Res. 2010, 9, 2377–2389. [Google Scholar] [CrossRef]
- Vu, H.S.; Tamura, P.; Galeva, N.A.; Chaturvedi, R.; Roth, M.R.; Williams, T.D.; Wang, X.; Shah, J.; Welti, R. Direct infusion mass spectrometry of oxylipin-containing Arabidopsis membrane lipids reveals varied patterns in different stress responses. Plant Physiol. 2012, 158, 324–339. [Google Scholar] [CrossRef] [PubMed]



| Locus ID | Gene Description (Symbol) | −1 kb P1BS |
|---|---|---|
| AT1G17710 | Phosphoethanolamine/Phosphocholine Phosphatase 1 (PEPC1) | 2 |
| AT1G23110 | Fold Protein | 1 |
| AT1G58280 | Phosphoglycerate Mutase Family Protein | 1 |
| AT1G73010 | Phosphate Starvation-induced Gene 2 (PS2) | 2 |
| AT2G11810 | Monogalactosyldiacylglycerol Synthase Type C (MGDC) | 0 |
| AT2G46880 | Purple Acid Phosphatase 14 (PAP14) | 2 |
| AT3G01345 | Expressed Protein | 0 |
| AT3G02040 | Glycerophosphodiester Phosphodiesterase 1 (GDPD1) | 4 |
| AT3G05630 | Phospholipase D Zeta 2 (PLDZ2) | 4 |
| AT3G09922 | Induced By Phosphate Starvation1 (IPS1) | 2 |
| AT3G17790 | Purple Acid Phosphatase 17 (PAP17) | 0 |
| AT3G30122 | Pseudogene | 1 |
| AT3G43110 | Transmembrane Protein | 1 |
| AT4G33030 | Sulfoquinovosyldiacylglycerol 1 (SQD1) | 0 |
| AT5G03545 | Induced By Pi Starvation 2 (ATIPS2, AT4) | 2 |
| AT5G15070 | Arabidopsis Homolog Of Yeast Vip1 2 (ATVIP2) | 0 |
| AT5G20150 | Spx Domain Gene 1 (SPX1) | 1 |
| AT5G20410 | Monogalactosyldiacylglycerol Synthase 2 (MGD2) | 2 |
| AT5G20790 | Transmembrane Protein | 0 |
| AT5G53048 | Natural Antisense Transcripts | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Land, E.S.; Cridland, C.A.; Craige, B.; Dye, A.; Hildreth, S.B.; Helm, R.F.; Gillaspy, G.E.; Perera, I.Y. A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis. Metabolites 2021, 11, 601. https://doi.org/10.3390/metabo11090601
Land ES, Cridland CA, Craige B, Dye A, Hildreth SB, Helm RF, Gillaspy GE, Perera IY. A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis. Metabolites. 2021; 11(9):601. https://doi.org/10.3390/metabo11090601
Chicago/Turabian StyleLand, Eric S., Caitlin A. Cridland, Branch Craige, Anna Dye, Sherry B. Hildreth, Rich F. Helm, Glenda E. Gillaspy, and Imara Y. Perera. 2021. "A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis" Metabolites 11, no. 9: 601. https://doi.org/10.3390/metabo11090601
APA StyleLand, E. S., Cridland, C. A., Craige, B., Dye, A., Hildreth, S. B., Helm, R. F., Gillaspy, G. E., & Perera, I. Y. (2021). A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis. Metabolites, 11(9), 601. https://doi.org/10.3390/metabo11090601






