Regional Brain Analysis of Modified Amino Acids and Dipeptides during the Sleep/Wake Cycle
Abstract
:1. Introduction
2. Results
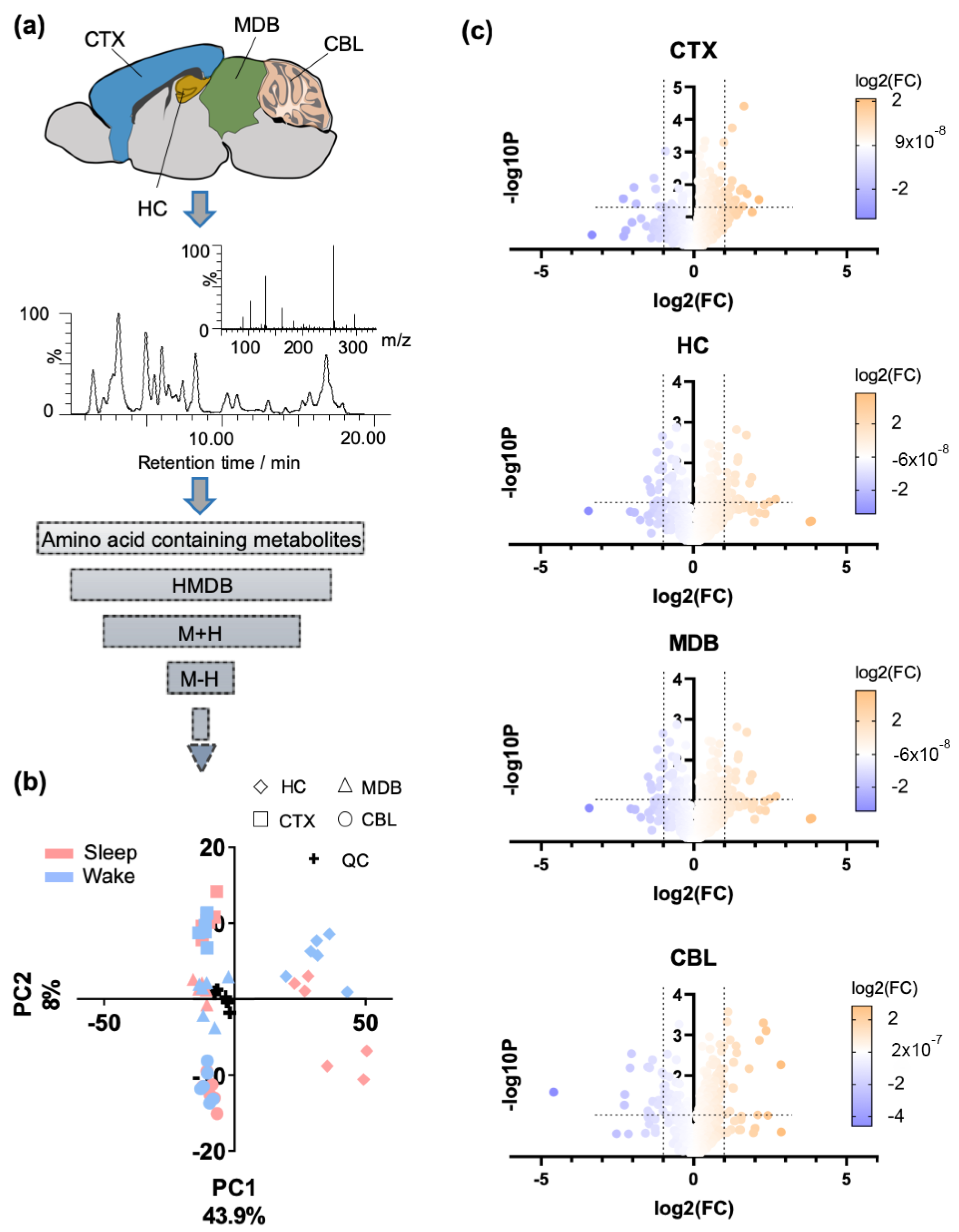
2.1. Regional Brain Metabolomics of Amino Acid Analogues
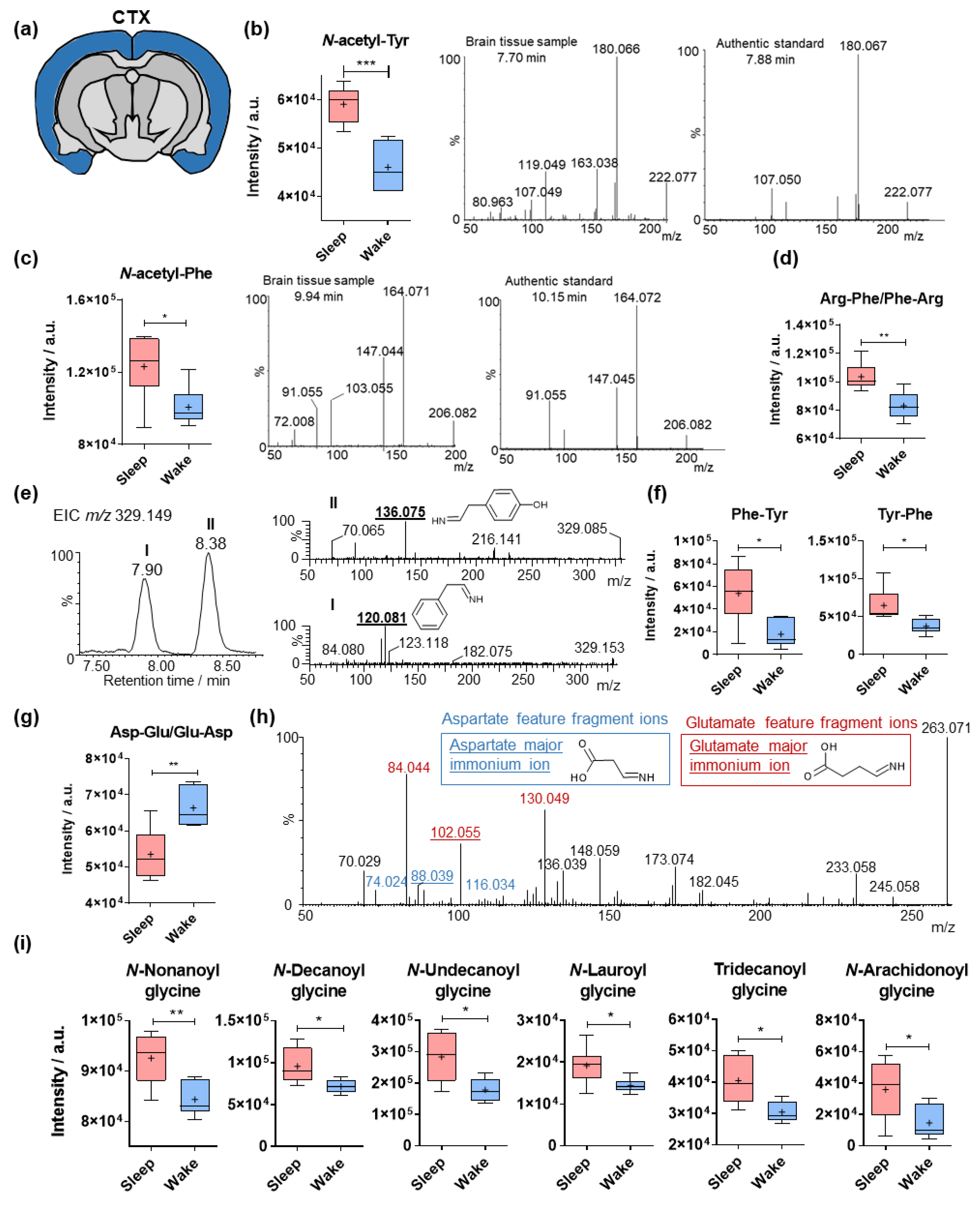
2.2. Sleep-Specific Effects on Acylated Amino Acids and Dipeptides in the Cerebral Cortex
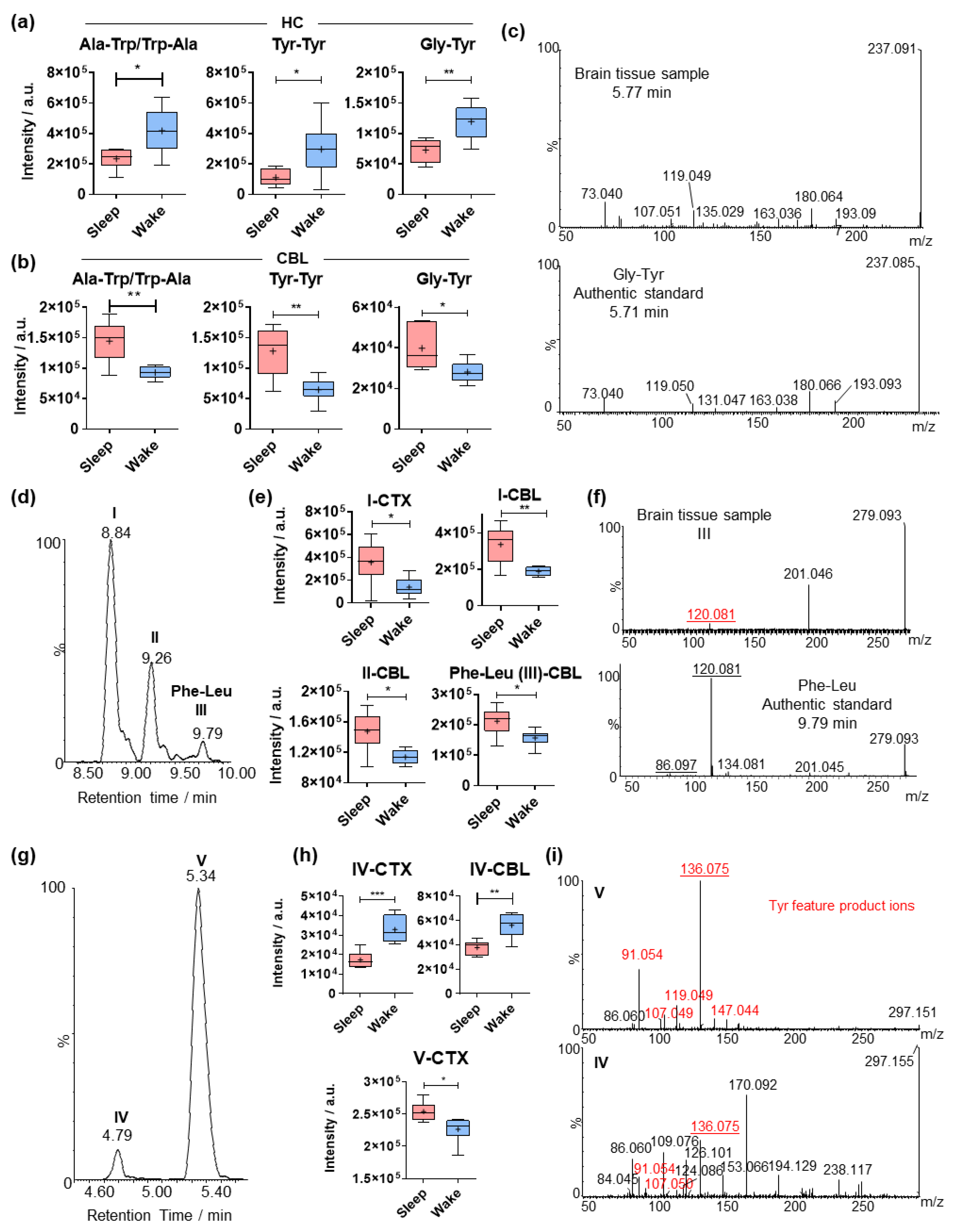
2.3. Circadian Effects on Different Amino Acid Analogues in the Hippocampus, Midbrain and Cerebellum
2.4. Comparative Analysis of Dipeptides between the Investigated Brain Regions
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animal Experiments
4.3. Tissue Processing and Sample Preparation
4.4. UPLC Mass Spectrometry
4.5. Identification of Metabolites
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joiner, W.J. Unraveling the Evolutionary Determinants of Sleep. Curr. Biol. 2016, 26, R1073–R1087. [Google Scholar] [CrossRef] [Green Version]
- DiNuzzo, M.; Nedergaard, M. Brain energetics during the sleep-wake cycle. Curr. Opin. Neurobiol. 2017, 47, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Aalling, N.N.; Nedergaard, M.; DiNuzzo, M. Cerebral Metabolic Changes During Sleep. Curr. Neurol. Neurosci. Rep. 2018, 18, 57. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Kang, H.; Xu, Q.; Chen, M.J.; Liao, Y.; Thiyagarajan, M.; O’Donnell, J.; Christensen, D.J.; Nicholson, C.; Iliff, J.J.; et al. Sleep drives metabolite clearance from the adult brain. Science 2013, 342, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Malik, D.M.; Paschos, G.K.; Sehgal, A.; Weljie, A.M. Circadian and Sleep Metabolomics Across Species. J. Mol. Biol. 2020, 432, 3578–3610. [Google Scholar] [CrossRef] [PubMed]
- Dalangin, R.; Kim, A.; Campbell, R.E. The Role of Amino Acids in Neurotransmission and Fluorescent Tools for Their Detection. Int. J. Mol. Sci. 2020, 21, 6197. [Google Scholar] [CrossRef] [PubMed]
- Vallianatou, T.; Lin, W.; Bèchet, N.B.; Correia, M.S.; Shanbhag, N.C.; Lundgaard, I.; Globisch, D. Differential regulation of oxidative stress, microbiota-derived, and energy metabolites in the mouse brain during sleep. J. Cereb. Blood Flow Metab. 2021, 41, 3324–3338. [Google Scholar] [CrossRef]
- Ozawa, H.; Hirayama, A.; Shoji, F.; Maruyama, M.; Suzuki, K.; Yamanaka-Okumura, H.; Tatano, H.; Morine, Y.; Soga, T.; Shimada, M.; et al. Comprehensive Dipeptide Analysis Revealed Cancer-Specific Profile in the Liver of Patients with Hepatocellular Carcinoma and Hepatitis. Metabolites 2020, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, H.; Hirayama, A.; Ishikawa, T.; Kudo, R.; Maruyama, M.; Shoji, F.; Doke, T.; Ishimoto, T.; Maruyama, S.; Soga, T.; et al. Comprehensive Dipeptide Profiling and Quantitation by Capillary Electrophoresis and Liquid Chromatography Coupled with Tandem Mass Spectrometry. Anal. Chem. 2020, 92, 9799–9806. [Google Scholar] [CrossRef]
- Tang, Y.; Li, R.; Lin, G.; Li, L. PEP search in MyCompoundID: Detection and identification of dipeptides and tripeptides using dimethyl labeling and hydrophilic interaction liquid chromatography tandem mass spectrometry. Anal. Chem. 2014, 86, 3568–3574. [Google Scholar] [CrossRef]
- Fonteh, A.N.; Harrington, R.J.; Tsai, A.; Liao, P.; Harrington, M.G. Free amino acid and dipeptide changes in the body fluids from Alzheimer’s disease subjects. Amino Acids 2007, 32, 213–224. [Google Scholar] [CrossRef]
- Mizushige, T.; Kanegawa, N.; Yamada, A.; Ota, A.; Kanamoto, R.; Ohinata, K. Aromatic amino acid-leucine dipeptides exhibit anxiolytic-like activity in young mice. Neurosci. Lett. 2013, 543, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Ano, Y.; Kita, M.; Kitaoka, S.; Furuyashiki, T. Leucine-Histidine Dipeptide Attenuates Microglial Activation and Emotional Disturbances Induced by Brain Inflammation and Repeated Social Defeat Stress. Nutrients 2019, 11, 2161. [Google Scholar] [CrossRef] [Green Version]
- Battista, N.; Bari, M.; Bisogno, T. N-Acyl Amino Acids: Metabolism, Molecular Targets, and Role in Biological Processes. Biomolecules 2019, 9, 822. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Dohgu, S.; Komabayashi, G.; Kiyohara, H.; Takata, F.; Kataoka, Y.; Nirasawa, T.; Maebuchi, M.; Matsui, T. Brain-transportable dipeptides across the blood-brain barrier in mice. Sci. Rep. 2019, 9, 5769. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Kiyohara, H.; Yoshino, A.; Nakano, A.; Takata, F.; Dohgu, S.; Kataoka, Y.; Matsui, T. Brain-transportable soy dipeptide, Tyr-Pro, attenuates amyloid β peptide. NPJ Sci. Food 2020, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Moriyasu, K.; Ichinose, T.; Nakahata, A.; Tanaka, M.; Matsui, T.; Furuya, S. The Dipeptides Ile-Tyr and Ser-Tyr Exert Distinct Effects on Catecholamine Metabolism in the Mouse Brainstem. Int. J. Pept. 2016, 2016, 6020786. [Google Scholar] [CrossRef] [Green Version]
- Ano, Y.; Ayabe, T.; Ohya, R.; Kondo, K.; Kitaoka, S.; Furuyashiki, T. Tryptophan-Tyrosine Dipeptide, the Core Sequence of β-Lactolin, Improves Memory by Modulating the Dopamine System. Nutrients 2019, 11, 348. [Google Scholar] [CrossRef] [Green Version]
- Snyder, S.H. Brain peptides as neurotransmitters. Science 1980, 209, 976–983. [Google Scholar] [CrossRef] [Green Version]
- Conway, L.P.; Rendo, V.; Correia, M.S.P.; Bergdahl, I.A.; Sjoblom, T.; Globisch, D. Unexpected Acetylation of Endogenous Aliphatic Amines by Arylamine N-Acetyltransferase NAT2. Angew. Chem. Int. Ed. 2020, 59, 14342–14346. [Google Scholar] [CrossRef]
- Globisch, D.; Moreno, A.Y.; Hixon, M.S.; Nunes, A.A.K.; Denery, J.R.; Specht, S.; Hoerauf, A.; Janda, K.D. Onchocerca volvulus-neurotransmitter tyramine is a biomarker for river blindness. Proc. Natl. Acad. Sci. USA 2013, 110, 4218–4223. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.T.; Terrell, S.M.; Li, V.L.; Wei, W.; Fischer, C.R.; Long, J.Z. Cooperative enzymatic control of N-acyl amino acids by PM20D1 and FAAH. Elife 2020, 9, e55211. [Google Scholar] [CrossRef]
- Long, J.Z.; Svensson, K.J.; Bateman, L.A.; Lin, H.; Kamenecka, T.; Lokurkar, I.A.; Lou, J.; Rao, R.R.; Chang, M.R.; Jedrychowski, M.P.; et al. The Secreted Enzyme PM20D1 Regulates Lipidated Amino Acid Uncouplers of Mitochondria. Cell 2016, 166, 424–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.; O’Dell, D.K.; Yu, Y.W.; Monn, M.F.; Hughes, H.V.; Burstein, S.; Walker, J.M. Identification of endogenous acyl amino acids based on a targeted lipidomics approach. J. Lipid Res. 2010, 51, 112–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.; Yu, Y.W.; Monn, M.F.; Hughes, H.V.; O’Dell, D.K.; Walker, J.M. Targeted lipidomics approach for endogenous N-acyl amino acids in rat brain tissue. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009, 877, 2890–2894. [Google Scholar] [CrossRef]
- Bradshaw, H.B.; Rimmerman, N.; Hu, S.S.; Burstein, S.; Walker, J.M. Novel endogenous N-acyl glycines identification and characterization. Vitam. Horm. 2009, 81, 191–205. [Google Scholar] [CrossRef]
- Vallianatou, T.; Shariatgorji, R.; Nilsson, A.; Karlgren, M.; Hulme, H.; Fridjonsdottir, E.; Svenningsson, P.; Andrén, P.E. Integration of Mass Spectrometry Imaging and Machine Learning Visualizes Region-Specific Age-Induced and Drug-Target Metabolic Perturbations in the Brain. ACS Chem. Neurosci. 2021, 12, 1811–1823. [Google Scholar] [CrossRef]
- Bonfanti, L.; Peretto, P.; De Marchis, S.; Fasolo, A. Carnosine-related dipeptides in the mammalian brain. Prog. Neurobiol. 1999, 59, 333–353. [Google Scholar] [CrossRef]
- Swann, J.R.; Spitzer, S.O.; Diaz Heijtz, R. Developmental Signatures of Microbiota-Derived Metabolites in the Mouse Brain. Metabolites 2020, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Epstein, A.A.; Kurczy, M.E.; Benton, P.H.; Uritboonthai, W.; Fox, H.S.; Boska, M.D.; Gendelman, H.E.; Siuzdak, G. Brain region mapping using global metabolomics. Chem. Biol. 2014, 21, 1575–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.T.; Tosun, M.; Jeong, H.H.; Karakas, C.; Semerci, F.; Liu, Z.; Maletić-Savatić, M. Metabolomics of mammalian brain reveals regional differences. BMC Syst. Biol. 2018, 12, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, J.; Ji, J.; Rabow, Z.; Shen, T.; Folz, J.; Brydges, C.R.; Fan, S.; Lu, X.; Mehta, S.; Showalter, M.R.; et al. A metabolome atlas of the aging mouse brain. Nat. Commun. 2021, 12, 6021. [Google Scholar] [CrossRef]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Chan, W.; Ang, I.L.; Wei, R.; Lam, M.M.T.; Lei, K.M.K.; Poon, T.C.W. Revisiting Fragmentation Reactions of Protonated α-Amino Acids by High-Resolution Electrospray Ionization Tandem Mass Spectrometry with Collision-Induced Dissociation. Sci. Rep. 2019, 9, 6453. [Google Scholar] [CrossRef]
- Ballet, C.; Correia, M.S.P.; Conway, L.P.; Locher, T.L.; Lehmann, L.C.; Garg, N.; Vujasinovic, M.; Deindl, S.; Lohr, J.M.; Globisch, D. New enzymatic and mass spectrometric methodology for the selective investigation of gut microbiota-derived metabolites. Chem. Sci. 2018, 9, 6233–6239. [Google Scholar] [CrossRef] [Green Version]
- Correia, M.S.P.; Lin, W.; Aria, A.J.; Jain, A.; Globisch, D. Rapid Preparation of a Large Sulfated Metabolite Library for Structure Validation in Human Samples. Metabolites 2020, 10, 415. [Google Scholar] [CrossRef]
- Globisch, D.; Pearson, D.; Hienzsch, A.; Bruckl, T.; Wagner, M.; Thoma, I.; Thumbs, P.; Reiter, V.; Kneuttinger, A.C.; Muller, M.; et al. Systems-Based Analysis of Modified tRNA Bases. Angew. Chem. Int. Ed. 2011, 50, 9739–9742. [Google Scholar] [CrossRef]
- Lin, W.F.; Conway, L.P.; Block, A.; Sommi, G.; Vujasinovic, M.; Lohr, J.M.; Globisch, D. Sensitive mass spectrometric analysis of carbonyl metabolites in human urine and fecal samples using chemoselective modification. Analyst 2020, 145, 3822–3831. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Mostyn, S.N.; Carland, J.E.; Shimmon, S.; Ryan, R.M.; Rawling, T.; Vandenberg, R.J. Synthesis and Characterization of Novel Acyl-Glycine Inhibitors of GlyT2. ACS Chem. Neurosci. 2017, 8, 1949–1959. [Google Scholar] [CrossRef]
- Schumann-Gillett, A.; O’Mara, M.L. Lipid-Based Inhibitors Act Directly on GlyT2. ACS Chem. Neurosci. 2019, 10, 1668–1678. [Google Scholar] [CrossRef]
- Girardeau, G.; Lopes-Dos-Santos, V. Brain neural patterns and the memory function of sleep. Science 2021, 374, 560–564. [Google Scholar] [CrossRef]
- Ano, Y.; Yoshino, Y.; Kutsukake, T.; Ohya, R.; Fukuda, T.; Uchida, K.; Takashima, A.; Nakayama, H. Tryptophan-related dipeptides in fermented dairy products suppress microglial activation and prevent cognitive decline. Aging (Albany N. Y.) 2019, 11, 2949–2967. [Google Scholar] [CrossRef]
- Ano, Y.; Yoshino, Y.; Uchida, K.; Nakayama, H. Preventive Effects of Tryptophan-Methionine Dipeptide on Neural Inflammation and Alzheimer’s Pathology. Int. J. Mol. Sci. 2019, 20, 6206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, T.C.; Obiang, P.; Torres-Herraez, A.; Watilliaux, A.; Coulon, P.; Rochefort, C.; Rondi-Reig, L. Anatomical and physiological foundations of cerebello-hippocampal interaction. Elife 2019, 8, e41896. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Guo, S.; Tang, W.; Li, B. Characterizing the spatial distribution of dipeptides in rodent tissue using MALDI MS imaging with on-tissue derivatization. Chem. Commun. 2021, 57, 12460–12463. [Google Scholar] [CrossRef]
- Perazzo, J.; Castanho, M.A.; Sá Santos, S. Pharmacological Potential of the Endogenous Dipeptide Kyotorphin and Selected Derivatives. Front. Pharmacol. 2016, 7, 530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidenreich, E.; Pfeffer, T.; Kracke, T.; Mechtel, N.; Nawroth, P.; Hoffmann, G.F.; Schmitt, C.P.; Hell, R.; Poschet, G.; Peters, V. A Novel UPLC-MS/MS Method Identifies Organ-Specific Dipeptide Profiles. Int. J. Mol. Sci. 2021, 22, 9979. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. J. Cereb. Blood Flow Metab. 2020, 40, 1769–1777. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Conway, L.P.; Ballet, C.; Correia, M.S.P.; Olsson, F.K.S.; Vujasinovic, M.; Lohr, J.M.; Globisch, D. Chemoselective Probe Containing a Unique Bioorthogonal Cleavage Site for Investigation of Gut Microbiota Metabolism. Angew. Chem. Int. Ed. 2018, 57, 13805–13809. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, A.; Mitani, T.; Tanaka, S.; Fujii, H.; Maebuchi, M.; Amiya, Y.; Tanaka, M.; Matsui, T.; Nakamura, S.; Katayama, S. Soybean-Derived Glycine-Arginine Dipeptide Administration Promotes Neurotrophic Factor Expression in the Mouse Brain. J. Agric. Food Chem. 2018, 66, 7935–7941. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vallianatou, T.; Bèchet, N.B.; Correia, M.S.P.; Lundgaard, I.; Globisch, D. Regional Brain Analysis of Modified Amino Acids and Dipeptides during the Sleep/Wake Cycle. Metabolites 2022, 12, 21. https://doi.org/10.3390/metabo12010021
Vallianatou T, Bèchet NB, Correia MSP, Lundgaard I, Globisch D. Regional Brain Analysis of Modified Amino Acids and Dipeptides during the Sleep/Wake Cycle. Metabolites. 2022; 12(1):21. https://doi.org/10.3390/metabo12010021
Chicago/Turabian StyleVallianatou, Theodosia, Nicholas B. Bèchet, Mario S. P. Correia, Iben Lundgaard, and Daniel Globisch. 2022. "Regional Brain Analysis of Modified Amino Acids and Dipeptides during the Sleep/Wake Cycle" Metabolites 12, no. 1: 21. https://doi.org/10.3390/metabo12010021
APA StyleVallianatou, T., Bèchet, N. B., Correia, M. S. P., Lundgaard, I., & Globisch, D. (2022). Regional Brain Analysis of Modified Amino Acids and Dipeptides during the Sleep/Wake Cycle. Metabolites, 12(1), 21. https://doi.org/10.3390/metabo12010021