The Proteomic Signature of Intestinal Acute Rejection in the Mouse
Abstract
:1. Introduction
2. Results
2.1. Intestinal Graft Histology
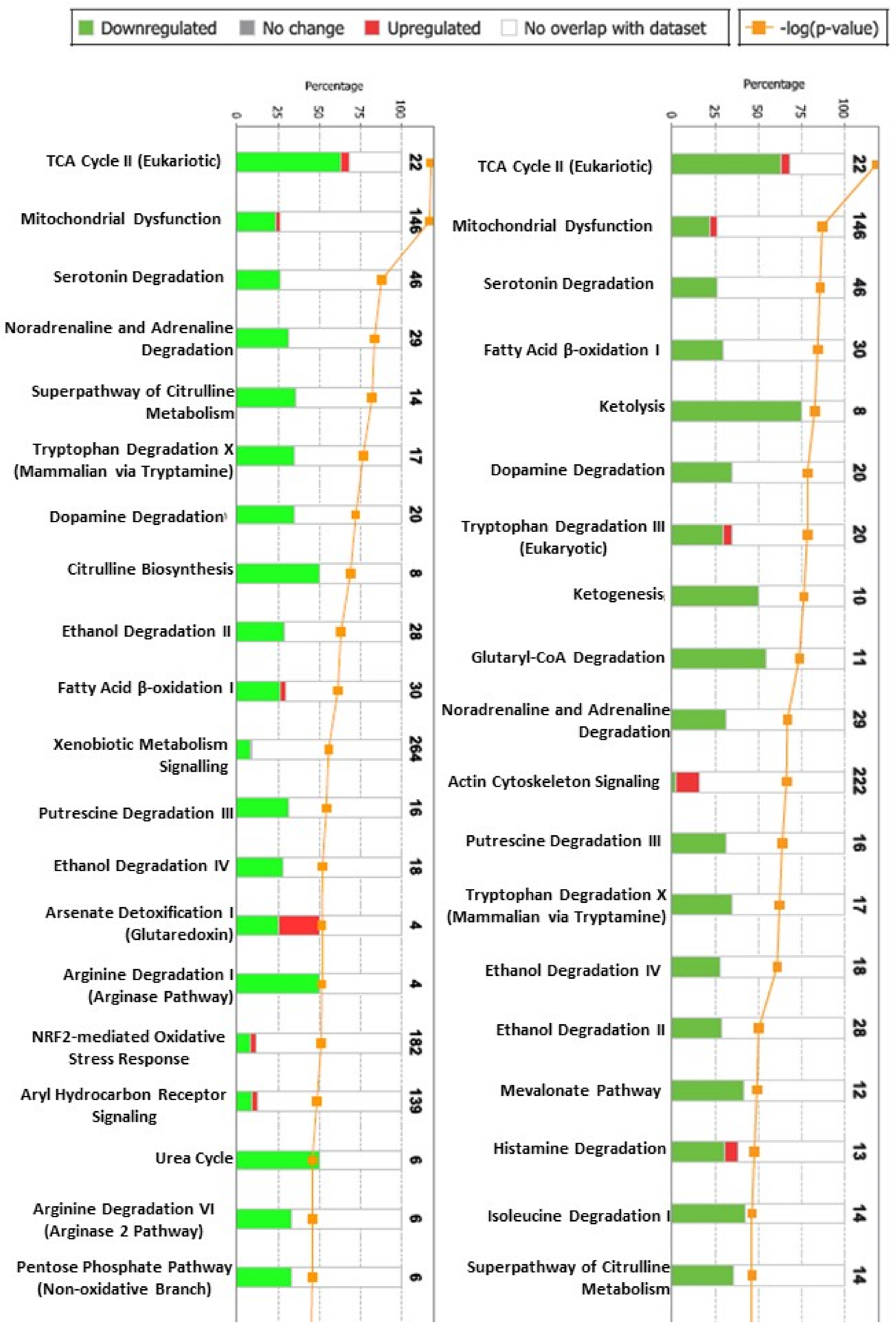
2.2. The Proteomic Analysis
2.3. Confirmatory Analysis: Western Blot Analysis and Immunohistochemistry
3. Discussion
4. Materials and Methods
4.1. Animals and Study Design
4.2. Surgery
4.3. Histology
4.4. Immunofluorescence
4.5. Proteomic Analysis
4.5.1. Sample Preparation
4.5.2. Strong Cation Exchange Chromatography (SCX) of TMT Labeled Peptides
4.5.3. LC-MS/MS Analysis on LTQ-Orbitrap Velos
4.5.4. Database Search and TMT Quantification
4.5.5. Bioinformatics Analysis of the Differentially Expressed Proteins
4.6. Western Blot Analyses of Intestinal Mucosa
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Venick, R.S. Grant monitoring after intestinal transplantation. Curr. Opin. Organ Transplant. 2021, 26, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Raghu, V.K.; Beaumont, J.L.; Everly, M.J.; Venick, R.S.; Lacaille, F.; Mazariegos, G.V. Pediatric intestinal transplantation: Analysis of the intestinal transplant registry. Pediatr. Transplant. 2019, 23, e13580. [Google Scholar] [CrossRef] [PubMed]
- Varkey, J. Graft assessment for acute rejection after intestinal transplantation: Current status and future perspective. Scand. J. Gastroenterol. 2021, 56, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Mercer, D.F. Hot topics in postsmall bowel transplantation: Noninvasive graft monitoring including stool calprotectin and plasma citrulline. Curr. Opin. Organ Transplant. 2011, 16, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Gondolesi, G.; Ghirardo, S.; Raymond, K.; Hoppenhauer, L.; Surillo, D.; Rumbo, C.; Fishbein, T.; Sansaricq, C.; Sauter, B. The value of plasma citrulline to predict mucosal injury in intestinal allografts. Am. J. Transplant. 2006, 6, 2786–2790. [Google Scholar] [CrossRef]
- Fujisaki, S.; Park, Y.J.; Kimizuka, Y.; Inoue, M.; Tomita, R.; Fukuzawa, M.; Matsumoto, K. Expression of mucosal addressin cell adhe-sion molecule-1 (MAdCAM-1) during small-bowel graft rejection in rats. Scand. J. Gastroenterol. 2003, 38, 437–442. [Google Scholar] [CrossRef]
- Asaoka, T.; Island, E.R.; Tryphonopoulos, P.; Selvaggi, G.; Moon, J.; Tekin, A.; Amador, A.; Levi, D.M.; Garcia, J.; Smith, L.; et al. Characteristic immune, apoptosis and inflammatory gene profiles associated with intestinal acute cellular rejection in formalin-fixed paraffin-embedded mucosal biopsies. Transpl. Int. 2011, 24, 697–707. [Google Scholar] [CrossRef]
- Joshi, M.; Dindelegan, G.; Olausson, M.; Oltean, M. Natural killer group 2 member D cell recruitment driven by major histo-compatibility complex class I chain-related antigens A and B: A possible mechanism during acute intestinal allograft rejection in the mouse. Transplant. Proc. 2010, 42, 4467–4469. [Google Scholar] [CrossRef]
- Mueller, A.; Platz, K.-P.; Heckert, C.; Häusler, M.; Guckelberger, O.; Schuppan, D.; Lobeck, H.; Neuhaus, P. Extracellular matrix: An early target of preservation/reperfusion injury and acute rejection after small bowel transplantation. Transplant. Proc. 1998, 30, 2569–2571. [Google Scholar] [CrossRef]
- Oltean, M.; Dindelegan, G.; Kurlberg, G.; Nilsson, O.; Karlsson-Parra, A.; Olausson, M. Intragraft heat shock protein-60 expression after small bowel transplantation in the mouse. Transplant. Proc. 2004, 36, 350–352. [Google Scholar] [CrossRef]
- D’Errico, A.; Corti, B.; Pinna, A.; Altimari, A.; Gruppioni, E.; Gabusi, E.; Fiorentino, M.; Bagni, A.; Grigioni, W. Granzyme B and perforin as predictive markers for acute rejection in human intestinal transplantation. Transplant. Proc. 2003, 35, 3061–3065. [Google Scholar] [CrossRef]
- Girlanda, R.; Cheema, A.K.; Kaur, P.; Kwon, Y.; Li, A.; Guerra, J.; Matsumoto, C.S.; Zasloff, M.; Fishbein, T.M. Metabolomics of human intestinal transplant rejection. Am. J. Transplant. 2012, 12, S18–S26. [Google Scholar] [CrossRef]
- Kumar, A.R.; Li, X.; Leblanc, J.F.; Farmer, D.G.; Elashoff, D.; Braun, J.; Ziring, D. Proteomic Analysis Reveals Innate Immune Activity in Intestinal Transplant Dysfunction. Transplantation 2011, 92, 112–119. [Google Scholar] [CrossRef]
- Thomson, A.W.; Bonham, C.A.; Zeevi, A. Mode of Action of Tacrolimus (FK506): Molecular and Cellular Mechanisms. Ther. Drug Monit. 1995, 17, 584–591. [Google Scholar] [CrossRef]
- Yandza, T.; Gerhardt, M.F.; Saint-Paul, M.-C.; Braud, V.; Gugenheim, J.; Hébuterne, X. Significance of Serum Bile Acids in Small Bowel Allograft Rejection in Pigs. Transplantation 2009, 87, 24–28. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, L.; Quan, D.; Garcia, B.; Ozcay, N.; Duff, J.; Stiller, C.; Lazarovits, A.; Grant, D.; Zhong, R. Pattern of liver, kidney, heart, and intestine allograft rejection in different mouse strain combinations. Transplantation 1996, 62, 1267–1272. [Google Scholar] [CrossRef]
- Stojanovic, T.; Scheele, L.; Wagner, A.H.; Middel, P.; Bedke, J.; Lautenschläger, I.; Leister, I.; Panzner, S.; Hecker, M.; Lautenschl, I. STAT-1 decoy oligonucleotide improves microcirculation and reduces acute rejection in allogeneic rat small bowel transplants. Gene Ther. 2007, 14, 883–890. [Google Scholar] [CrossRef] [Green Version]
- Yarur, A.J.; Quintero, M.A.; Jain, A.; Czul, F.; Barkin, J.S.; Abreu, M.T. Serum Amyloid A as a Surrogate Marker for Mucosal and Histologic Inflammation in Patients with Crohn’s Disease. Inflamm. Bowel Dis. 2017, 23, 158–164. [Google Scholar] [CrossRef]
- Poynton, R.A.; Hampton, M.B. Peroxiredoxins as biomarkers of oxidative stress. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 906–912. [Google Scholar] [CrossRef]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive Oxygen Species in Inflammation and Tissue Injury. Antioxid. Redox Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Zhou, L.; Jiang, J.; Qin, Y.; Xie, H.; Feng, X.; Gao, F.; Zheng, S. Proteomic Analysis of Differentially Expressed Proteins in Rat Liver Allografts Developed Acute Rejection. Eur. Surg. Res. 2010, 44, 43–51. [Google Scholar] [CrossRef]
- Kienzl, K.; Sarg, B.; Golderer, G.; Obrist, P.; Werner, E.R.; Werner-Felmayer, G.; Lindner, H.; Maglione, M.; Schneeberger, S.; Margreiter, R.; et al. Proteomic Profiling of Acute Cardiac Allograft Rejection. Transplantation 2009, 88, 553–560. [Google Scholar] [CrossRef]
- Ogita, K.; Hopkinson, K.; Nakao, M.; Wood, R.F.M.; Pockley, A.G. Stress responses in graft and native intestine after rat heterotopic small bowel transplantation. Transplantation 2000, 69, 2273–2277. [Google Scholar] [CrossRef]
- Mehta, N.; Carroll, M.; Sykes, D.; Tan, Z.; Bergsland, J.; Canty, J.; Bhayana, J.; Hoover, E.; Salerno, T. Heat Shock Protein 70 Expression in Native and Heterotopically Transplanted Rat Hearts. J. Surg. Res. 1997, 70, 151–155. [Google Scholar] [CrossRef]
- Tamaki, K.; Otaka, M.; Takada, M.; Yamamoto, S.; Odashima, M.; Itoh, H.; Watanabe, S. Evidence for Enhanced Cytoprotective Function of HSP90-Overexpressing Small Intestinal Epithelial Cells. Dig. Dis. Sci. 2011, 56, 1954–1961. [Google Scholar] [CrossRef]
- Duquesnoy, R.J.; Liu, K.; Fu, X.-F.; Murase, N.; Ye, Q.; Demetris, A.J. Evidence for heat shock protein immunity in a rat cardiac allograft model of chronic rejection. Transplantation 1999, 67, 156–164. [Google Scholar] [CrossRef]
- Qian, B.F.; El-Salhy, M.; Danielsson, A.; Shalaby, A.; Axelsson, H. Changes in intestinal endocrine cells in the mouse after unilateral cervical vagotomy. Histol. Histopathol. 1999, 14, 453–460. [Google Scholar]
- Acosta, S.; Dizeyi, N.; Pierzynowski, S.; Alm, P.; Abrahamsson, P.A. Neuroendocrine cells and nerves in the prostate of the guinea pig: Effects of peripheral denervation and castration. Prostate 2001, 46, 191–199. [Google Scholar] [CrossRef]
- David, A.I.; Selvaggi, G.; Ruiz, P.; Gaynor, J.J.; Tryphonopoulos, P.; Kleiner, G.I.; Moon, J.I.; Nishida, S.; Pappas, P.A.; Conanan, L.; et al. Blood Citrulline Level Is an Exclusionary Marker for Significant Acute Rejection After Intestinal Transplantation. Transplantation 2007, 84, 1077–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dindelegan, G.; Liden, H.; Kurlberg, G.; Oltean, M.; Nilsson, O.; Åneman, A.; Lycke, N.; Olausson, M. Laser-Doppler flowmetry is reliable for early diagnosis of small-bowel acute rejection in the mouse. Microsurgery 2003, 23, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Casselbrant, A.; Söfteland, J.M.; Hellström, M.; Malinauskas, M.; Oltean, M. Luminal polyethylene glycol alleviates intestinal preservation injury irrespective of molecular size. J. Pharmacol. Exp. Ther. 2018, 366, 29–36. [Google Scholar] [CrossRef] [Green Version]



| Accession | Symbol | Description | Fold Change | Molecular Function | Biological Process |
|---|---|---|---|---|---|
| Upregulated tissue expression | |||||
| P28776 | Ido1 | Indoleamine 2,3-dioxygenase 1 | 4.72 | Dioxygenase, Oxidoreductase | Inflammatory response |
| Q01514 | Gbp1 | Interferon-induced guanylate-binding protein 1 | 4.44 | Hydrolase | Inflammatory response |
| P52624 | Upp1 | Uridine phosphorylase 1 | 4.21 | Glycosyltransferase, Transferase | Inflammatory response |
| Q91WP6 | Ser3na | Serine protease inhibitor | 3.19 | Protease inhibitor | Inflammatory response |
| P27005 | S100a8 | Protein S100-A8 | 3.17 | Antimicrobial | Cell death and survival |
| P42225 | Stat1 | Signal transducer and activator of transcription 1 | 3.04 | Activator, DNA-binding | Cell death and survival |
| P05367 | Saa2 | Serum amyloid A-2 protein | 2.91 | Cytokine | Inflammatory response |
| Q8VCM7 | Fgg | Fibrinogen gamma chain | 2.44 | Binding protein | Hemostasis |
| O35744 | Chi3l1 | Chitinase-3-like protein 3 | 2.38 | Antimicrobial | Inflammatory response |
| Q99KQ4 | Nampt | Nicotinamide phosphoribosyltransferase | 2.12 | Cytokine, Glycosyltransferase | Cell death and survival |
| E9Q555 | Rnf213 | E3 ubiquitin-protein ligase RNF213 | 1.82 | Hydrolase, Transferase | Angiogenesis |
| P01899 | H2-d1 | H-2 class I histocompatibility antigen, D-B alpha chain | 1.69 | Binding protein | Immunology |
| Q9R233 | Tapbp | Tapasin | 1.59 | Binding protein | Immunology |
| Q9JIK5 | Ddx21 | Nucleolar RNA helicase 2 | 1.56 | Binding protein | Immunology |
| P17918 | Pcna | Proliferating cell nuclear antigen | 1.55 | DNA-Binding | Cell death and survival |
| P31001 | Des | Desmin | 1.52 | Muscle protein | Cell structure |
| P26041 | Msn | Moesin | 1.49 | Signal protein | Inflammatory response |
| Q60590 | Orm1 | Alpha-1-acid glycoprotein 1 | 1.46 | Transport protein | Inflammatory response |
| P25206 | Mcm3 | DNA replication licensing factor MCM3 | 1.4 | DNA-binding, Helicase, Hydrolase | Cell death and survival |
| P09405 | Ncl | Nucleolin | 1.36 | Binding protein | Angiogenesis |
| P16858 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | 1.35 | Oxidoreductase, Transferase | Inflammatory response |
| P68033 | Actc1 | Actin, alpha cardiac muscle 1 | 1.35 | Muscle protein | Cell movement |
| Q6NZJ6 | Eif4g1 | Eukaryotic translation initiation factor 4 gamma 1 | 1.35 | Initiation factor, RNA-binding, Translational shunt | Cell death and survival |
| P52480 | Pkm | Pyruvate kinase isozymes M1/M2 | 1.35 | Allosteric enzyme, Kinase, Transferase | Cancer |
| Q9CPY7 | Lap3 | Cytosol aminopeptidase | 1.32 | Aminopeptidase, Hydrolase, Protease | Cell death and survival |
| Q9Z1Q5 | Clic1 | Chloride intracellular channel protein 1 | 1.32 | Ion channel | Cellular growth and proliferation |
| Q99JG3 | Anxa13 | Annexin A13 | 1.31 | Binding protein | Cell death and survival |
| P97372 | Psme2 | Proteasome activator complex subunit 2 | 1.29 | Immunoproteasome assembly | Cell death and survival |
| Q61029 | Tmpo | Lamina-associated polypeptide 2 | 1.27 | DNA-Binding | Cell structure |
| P11499 | Hsp90ab1 | Heat shock protein HSP 90-beta | 1.25 | Chaperon | Cell death and survival |
| P05784 | Krt18 | Keratin, type I cytoskeletal 18 | 1.24 | Structural protein | Cell structure |
| Q80 × 90 | Flnb | Filamin-B | 1.24 | Actin-binding, | Cell movement |
| P60710 | Actb | Actin, cytoplasmic 1 | 1.21 | Muscle protein | Cell movement |
| P97371 | Psme1 | Proteasome activator complex subunit 1 | 1.21 | Immunoproteasome assembly | Inflammatory response |
| P62137 | Ppp1ca | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | 1.2 | Hydrolase, phosphatase | Cellular growth and proliferation |
| P37040 | Por | NADPH--cytochrome P450 reductase | 1.2 | Oxidoreductase | Cellular function and maintenance |
| O08808 | Diaph1 | Protein diaphanous homolog 1 | 1.2 | Actin-binding | Cell structure |
| Downregulated tissue expression | |||||
| Q9CZ13 | Uqcrc1 | Cytochrome b-c1 complex subunit 1, mitochondrial | 0.79 | Electron transport, Respiratory chain, Transport | Cellular function and maintenance |
| Q9ERG0 | Lima1 | LIM domain and actin-binding protein 1 | 0.79 | Binding protein | Lipid metabolism |
| Q9D0F3 | Aldh1b1 | Protein ERGIC-53 | 0.79 | Oxidoreductase | Cancer |
| Q921H8 | Acaa1 | 3-ketoacyl-CoA thiolase A, peroxisomal | 0.79 | Acyltransferase, Transferase | Lipid metabolism |
| Q02819 | Nucb1 | Nucleobindin-1 | 0.78 | DNA-binding, Guanine-nucleotide releasing factor | Cellular growth and proliferation |
| P09103 | Pdia1 | Protein disulfide-isomerase | 0.78 | Isomerases | Cell death and survival |
| Q9CY27 | Tecr | Trans-2,3-enoyl-CoA reductase | 0.78 | Oxidoreductase | Lipid metabolism |
| Q9JII6 | Ak1a1 | Alcohol dehydrogenase | 0.77 | Dehydrogenase/reductase | Small molecule biochemistry |
| P28271 | Aco1 | Cytoplasmic aconitate hydratase | 0.77 | Lyase, RNA-binding | Cellular growth and proliferation |
| Q9JLQ0 | Cd2ap | CD2-associated protein | 0.76 | Adapter protein | Cell cycle |
| Q99KI0 | Acon | Aconitate hydratase, mitochondrial | 0.76 | Lyase | Cellular growth and proliferation |
| P47738 | Aldh2 | Aldehyde dehydrogenase, mitochondrial | 0.75 | Oxidoreductase | Small molecule biochemistry |
| P19783 | Cox41 | Cytochrome c oxidase subunit 4 isoform 1 | 0.75 | Oxidoreductase | Cellular function and maintenance |
| P35700 | Prdx1 | Peroxiredoxin-1 | 0.74 | Peroxidase | Cellular function and maintenance |
| P45952 | Acadm | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 0.74 | Oxidoreductase | Lipid metabolism |
| P24270 | Cat | Catalase | 0.74 | Catalase | Cellular function and maintenance |
| Q60598 | Cttn | Src substrate cortactin | 0.72 | Unknown | Cell structure |
| Q80XN0 | Bdh1 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial | 0.71 | Dehydrogenase/reductase | Lipid metabolism |
| Q9EPB4 | Pycard | Apoptosis-associated speck-like protein containing a CARD | 0.71 | Unknown | Inflammatory response |
| Q9DBS5 | Klc4 | Kinesin light chain 4 | 0.71 | Motor protein | Cell movement |
| P99028 | Uqcrh | Cytochrome b-c1 complex subunit 6, mitochondrial | 0.71 | Oxidoreductase | Cellular function and maintenance |
| Q9D855 | Uqcrb | Cytochrome b-c1 complex subunit 7 | 0.71 | Electron transport, Respiratory chain, Transport | Cellular function and maintenance |
| Q5SYD0 | Myo1d | Myosin-Id | 0.71 | Motor protein | Cell structure |
| Q8K2B3 | Sdha | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 0.7 | Oxidoreductase | Cellular function and maintenance |
| Q9CQW5 | Lgals2 | Galectin-2 | 0.7 | Binding protein | Unknown |
| Q99K01 | Pdxdc1 | Pyridoxal-dependent decarboxylase domain-containing protein 1 | 0.69 | Decarboxylase, Lyase | Cell cycle |
| P10852 | Slc3a2 | 4F2 cell-surface antigen heavy chain | 0.68 | transport protein | Cellular function and maintenance |
| Q3UMR5 | Mcu | Coiled-coil domain-containing protein 109A | 0.68 | Calcium channel, Ion channel | Cellular function and maintenance |
| Q9Z2I8 | Suclg2 | Succinyl-CoA ligase subunit beta, mitochondrial | 0.68 | Ligase | Cellular function and maintenance |
| Q8VC30 | Tkfc | Bifunctional ATP-dependent dihydroxyacetone kinase | 0.66 | Multifunctional enzyme | Cellular function and maintenance |
| Q9R100 | Cdh17 | Cadherin-17 | 0.65 | Adhesion protein | Cell structure |
| P56391 | Cox6b1 | Cytochrome c oxidase subunit 6B1 | 0.65 | Oxidoreductase | Cellular function and maintenance |
| P14152 | Mdh1–2 | Malate dehydrogenase, cytoplasmic | 0.64 | Oxidoreductase | Cellular function and maintenance |
| Q8C196 | Cps1 | Carbamoyl-phosphate synthase, mitochondrial | 0.64 | Ligase | Cellular function and maintenance |
| P31786 | Acbp | Acyl-CoA-binding protein | 0.63 | Binding protein | Unknown |
| Q9DCN2 | Nb5r3 | NADH-cytochrome b5 reductase 3 | 0.61 | Oxidoreductase | Lipid metabolism |
| P57016 | Lad1 | Ladinin-1 | 0.59 | Anchoring filament | Cell structure |
| O09131 | Gsto1 | Glutathione S-transferase omega-1 | 0.58 | Oxidoreductase, Transferase | Oxidative stress |
| Q8K0C9 | Gmds | GDP-mannose 4,6 dehydratase | 0.57 | Lyase | Cellular function and maintenance |
| Q9CZS1 | Al1b1 | Aldehyde dehydrogenase X, mitochondrial | 0.57 | Oxidoreductase | Small molecule biochemistry |
| Q9CQ62 | Decr | 2,4-dienoyl-CoA reductase, mitochondrial | 0.54 | Oxidoreductase | Lipid metabolism |
| Q8R0Y6 | Fthfd | 10-formyltetrahydrofolate dehydrogenase | 0.54 | Oxidoreductase | Cellular function and maintenance |
| Q9D8W7 | Ocad2 | OCIA domain-containing protein 2 | 0.5 | Unknown | Cancer |
| Q9QWG7 | St1b1 | Sulfotransferase family cytosolic 1B member 1 | 0.49 | Sulfotransferase | Cellular function and maintenance |
| P29758 | Oat | Ornithine aminotransferase, mitochondrial | 0.47 | Aminotransferase, Transferase | Cellular function and maintenance |
| Q64133 | Aofa | Amine oxidase A | 0.46 | Oxidoreductase | Cellular function and maintenance |
| O88310 | Itl1a | Intelectin-1a | 0.37 | Antimicrobial | Inflammatory response |
| P26339 | Cmga | Chromogranin-A | 0.31 | Inhibitor protein | Immunology |
| P35230 | Reg3b | Regenerating islet-derived protein 3-beta | 0.28 | Antibacterial protein | Immunology |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oltean, M.; Bagge, J.; Dindelegan, G.; Kenny, D.; Molinaro, A.; Hellström, M.; Nilsson, O.; Sihlbom, C.; Casselbrant, A.; Davila, M.; et al. The Proteomic Signature of Intestinal Acute Rejection in the Mouse. Metabolites 2022, 12, 23. https://doi.org/10.3390/metabo12010023
Oltean M, Bagge J, Dindelegan G, Kenny D, Molinaro A, Hellström M, Nilsson O, Sihlbom C, Casselbrant A, Davila M, et al. The Proteomic Signature of Intestinal Acute Rejection in the Mouse. Metabolites. 2022; 12(1):23. https://doi.org/10.3390/metabo12010023
Chicago/Turabian StyleOltean, Mihai, Jasmine Bagge, George Dindelegan, Diarmuid Kenny, Antonio Molinaro, Mats Hellström, Ola Nilsson, Carina Sihlbom, Anna Casselbrant, Marcela Davila, and et al. 2022. "The Proteomic Signature of Intestinal Acute Rejection in the Mouse" Metabolites 12, no. 1: 23. https://doi.org/10.3390/metabo12010023
APA StyleOltean, M., Bagge, J., Dindelegan, G., Kenny, D., Molinaro, A., Hellström, M., Nilsson, O., Sihlbom, C., Casselbrant, A., Davila, M., & Olausson, M. (2022). The Proteomic Signature of Intestinal Acute Rejection in the Mouse. Metabolites, 12(1), 23. https://doi.org/10.3390/metabo12010023







