Unlike Glycerophosphocholine or Choline Chloride, Dietary Phosphatidylcholine Does Not Increase Plasma Trimethylamine-N-Oxide Levels in Sprague-Dawley Rats
Abstract
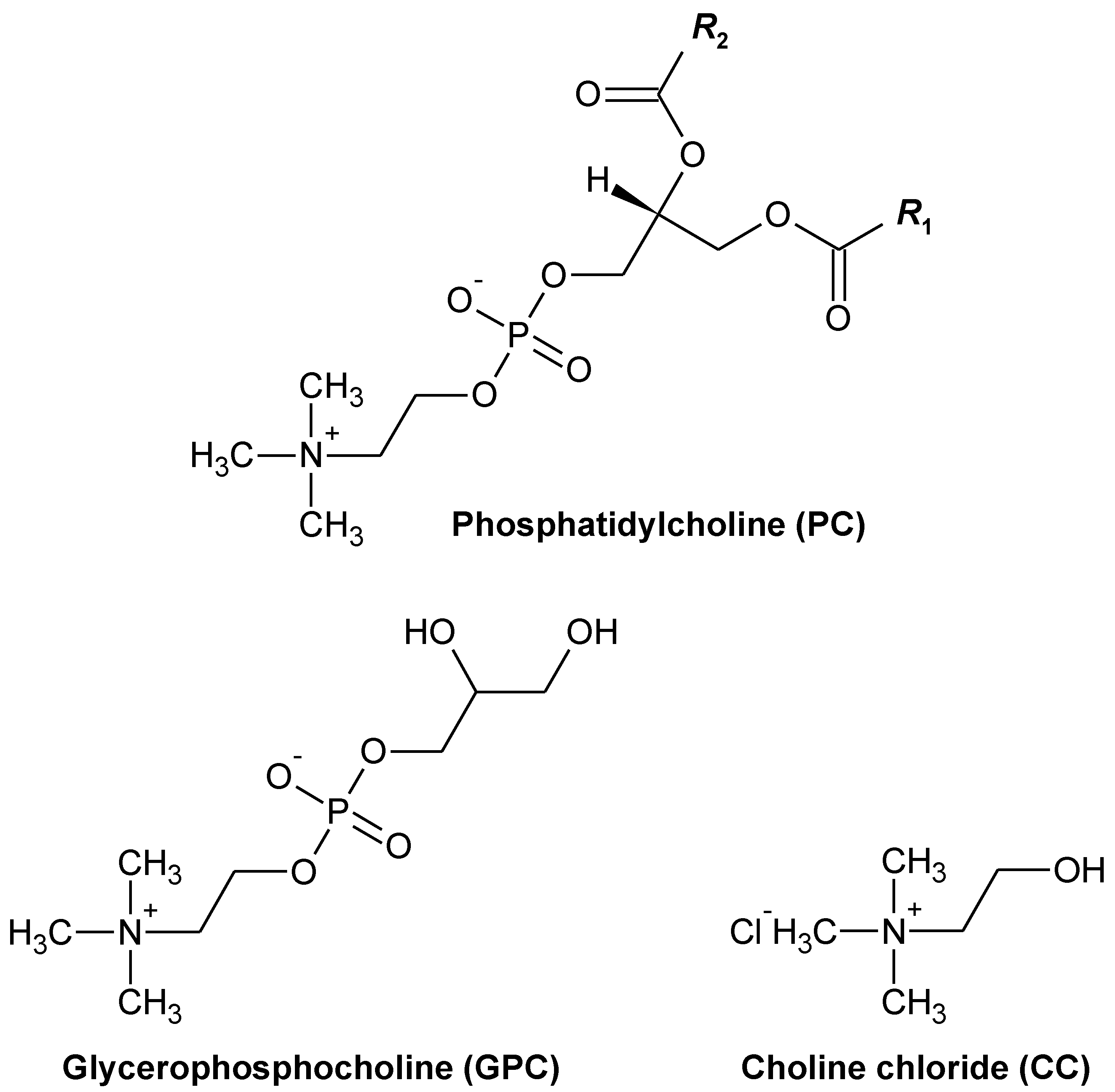
:1. Introduction
2. Results and Discussion
2.1. Effects of Dietary PC, GPC, and CC on the Growth and Metabolic Parameters in SD Rats
2.2. Effects of Dietary PC, GPC, and CC on Plasma TMAO Levels and mRNA-rElated TMAO Generation in Livers of SD Rats
2.3. Effects of Dietary PC, GPC, and CC on Gut Microbiota in SD Rats: Identification of Gut Microbes Associated with Changes in Plasma TMAO Levels
3. Materials and Methods
3.1. Animals and Diets
3.2. Analysis of Plasma Biochemical Parameters
3.3. Analysis of Hepatic Phospholipids Contents
3.4. Analysis of Plasma TMAO Levels
3.5. Analysis of Hepatic Messenger Ribonucleic Acid (mRNA) Levels
3.6. Analysis of Gut Microbiota in Rats Fed Each Choline Compound
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shirouchi, B.; Yamanaka, R.; Tanaka, S.; Kawatou, F.; Hayashi, T.; Takeyama, A.; Nakao, A.; Goromaru, R.; Iwamoto, M.; Sato, M. Quantities of Phospholipid Molecular Classes in Japanese Meals and Prediction of Their Sources by Multiple Regression Analysis. J. Nutr. Sci. Vitaminol. 2018, 64, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Cohn, J.S.; Wat, E.; Kamili, A.; Tandy, S. Dietary Phospholipids, Hepatic Lipid Metabolism and Cardiovascular Disease. Curr. Opin. Lipidol. 2008, 19, 257–262. [Google Scholar] [CrossRef]
- Küllenberg, D.; Taylor, L.A.; Schneider, M.; Massing, U. Health Effects of Dietary Phospholipids. Lipids Health Dis. 2012, 11, 3. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.T.; Xu, J.; Wang, Y.M.; Xue, C.H. Health Benefits of Dietary Marine DHA/EPA-Enriched Glycerophospholipids. Prog. Lipid Res. 2019, 75, 100997. [Google Scholar] [CrossRef]
- Zeisel, S.H.; da Costa, K.A. Choline: An Essential Nutrient for Public Health. Nutr. Rev. 2009, 67, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut Flora Metabolism of Phosphatidylcholine Promotes Cardiovascular Disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Waiz, M.; Mikov, M.; Mitchell, S.C.; Smith, R.L. The Exogenous Origin of Trimethylamine in the Mouse. Metabolism 1992, 41, 135–136. [Google Scholar] [CrossRef]
- Lang, D.H.; Yeung, C.K.; Peter, R.M.; Ibarra, C.; Gasser, R.; Itagaki, K.; Philpot, R.M.; Rettie, A.E. Isoform Specificity of Trimethylamine N-Oxygenation by Human Flavin-Containing Monooxygenase (FMO) and P450 Enzymes: Selective Catalysis by FMO3. Biochem. Pharmacol. 1998, 56, 1005–1012. [Google Scholar] [CrossRef]
- Yu, Z.L.; Zhang, L.Y.; Jiang, X.M.; Xue, C.H.; Chi, N.; Zhang, T.T.; Wang, Y.M. Effects of Dietary Choline, Betaine, and L-Carnitine on the Generation of Trimethylamine-N-Oxide in Healthy Mice. J. Food Sci. 2020, 85, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.; Ulrich, C.M.; Neuhouser, M.L.; Malysheva, O.; Bailey, L.B.; Xiao, L.; Brown, E.C.; Cushing-Haugen, K.L.; Zheng, Y.; Cheng, T.Y.; et al. Plasma Choline Metabolites and Colorectal Cancer Risk in the Women’s Health Initiative Observational Study. Cancer Res. 2014, 74, 7442–7452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.H.; Wang, Z.; Kennedy, D.J.; Wu, Y.; Buffa, J.A.; Agatisa-Boyle, B.; Li, X.S.; Levison, B.S.; Hazen, S.L. Gut Microbiota-Dependent Trimethylamine N-Oxide (TMAO) Pathway Contributes to Both Development of Renal Insufficiency and Mortality Risk in Chronic Kidney Disease. Circ. Res. 2015, 116, 448–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernstrom, M.H. Lecithin, Choline, and Cholinergic Transmission. Nutr. Pharmacol. 1981, 4, 5–29. [Google Scholar]
- Smolders, L.; de Wit, N.J.W.; Balvers, M.G.J.; Obeid, R.; Vissers, M.M.M.; Esser, D. Natural Choline From Egg Yolk Phospholipids Is More Efficiently Absorbed Compared With Choline Bitartrate; Outcomes of a Randomized Trial in Healthy Adults. Nutrients 2019, 11, 2758. [Google Scholar] [CrossRef] [Green Version]
- Craciun, S.; Balskus, E.P. Microbial Conversion of Choline to Trimethylamine Requires a Glycyl Radical Enzyme. Proc. Natl. Acad. Sci. USA. 2012, 109, 21307–21312. [Google Scholar] [CrossRef] [Green Version]
- Bodea, S.; Funk, M.A.; Balskus, E.P.; Drennan, C.L. Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase. Cell Chem. Biol. 2016, 23, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.E.; Aardema, N.D.J.; Bunnell, M.L.; Larson, D.P.; Aguilar, S.S.; Bergeson, J.R.; Malysheva, O.V.; Caudill, M.A.; Lefevre, M. Effect of Choline Forms and Gut Microbiota Composition on Trimethylamine- N-Oxide Response in Healthy Men. Nutrients 2020, 12, 2220. [Google Scholar] [CrossRef]
- Tetko, I.V.; Tanchuk, V.Y. Application of Associative Neural Networks for Prediction of Lipophilicity in ALOGPS 2.1 Program. J. Chem. Inf. Comput. Sci. 2002, 42, 1136–1145. [Google Scholar] [CrossRef]
- Azzouz, L.L.; Sharma, S. Physiology, Large Intestine. Physiol. Large Intest. 2018. [Google Scholar]
- Novick, R.M.; Mitzey, A.M.; Brownfield, M.S.; Elfarra, A.A. Differential Localization of Flavin-Containing Monooxygenase (FMO) Isoforms 1, 3, and 4 in Rat Liver and Kidney and Evidence for Expression of FMO4 in Mouse, Rat, and Human Liver and Kidney Microsomes. J. Pharmacol. Exp. Ther. 2009, 329, 1148–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Lin, S.; Vanhoutte, P.M.; Woo, C.W.; Xu, A. Akkermansia muciniphila Protects Against Atherosclerosis by Preventing Metabolic Endotoxemia-Induced Inflammation in Apoe−/− Mice. Circulation 2016, 133, 2434–2446. [Google Scholar] [CrossRef] [Green Version]
- Dao, M.C.; Everard, A.; Aron-Wisnewsky, J.; Sokolovska, N.; Prifti, E.; Verger, E.O.; Kayser, B.D.; Levenez, F.; Chilloux, J.; Hoyles, L.; et al. Akkermansia muciniphila and Improved Metabolic Health During a Dietary Intervention in Obesity: Relationship With Gut Microbiome Richness and Ecology. Gut 2016, 65, 426–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yassour, M.; Lim, M.Y.; Yun, H.S.; Tickle, T.L.; Sung, J.; Song, Y.M.; Lee, K.; Franzosa, E.A.; Morgan, X.C.; Gevers, D.; et al. Sub-Clinical Detection of Gut Microbial Biomarkers of Obesity and type 2 Diabetes. Genome Med. 2016, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut Microbiota Dysbiosis Contributes to the Development of Hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation With Akkermansia muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Falony, G.; Vieira-Silva, S.; Raes, J. Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine. Annu. Rev. Microbiol. 2015, 69, 305–321. [Google Scholar] [CrossRef]
- Lin, H.; Yanpeng, A.; Hao, F.; Wang, Y.; Tang, H. Correlations of Fecal Metabonomic and Microbiomic Changes Induced by High-Fat Diet in the Pre-Obesity State. Sci. Rep. 2016, 6, 21618. [Google Scholar] [CrossRef]
- American Institute of Nutrition. Report of the American Institute of Nurtition Ad hoc Committee on Standards for Nutritional Studies. J. Nutr. 1977, 107, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Rouser, G.; Siakotos, A.N.; Fleischer, S. Quantitative Analysis of Phospholipids by Thin-Layer Chromatography and Phosphorus Analysis of Spots. Lipids 1966, 1, 85–86. [Google Scholar] [CrossRef]
- Ufnal, M.; Jazwiec, R.; Dadlez, M.; Drapala, A.; Sikora, M.; Skrzypecki, J. Trimethylamine-N-Oxide: A Carnitine-Derived Metabolite That Prolongs the Hypertensive Effect of Angiotensin II in Rats. Can. J. Cardiol. 2014, 30, 1700–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar]
- Hisada, T.; Endoh, K.; Kuriki, K. Inter- and Intra-Individual Variations in Seasonal and Daily Stabilities of the Human Gut Microbiota in Japanese. Arch. Microbiol. 2015, 197, 919–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aronesty, E. Comparison of Sequencing Utility Programs. TOBIOIJ 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Gordon, A.; Hannon, G.J. FASTQ/A Short-Reads Preprocessing Tools (2 February 2020-Version 0.0.13). Available online: http://hannonlab.cshl.edu/fastx_toolkit/ (accessed on 18 March 2021).
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences Into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the Gut Microbiota Composition Between Obese and Non-Obese Individuals in a Japanese Population, as Analyzed by Terminal Restriction Fragment Length Polymorphism and Next-Generation Sequencing. B.M.C. Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-Resolution Sample Inference From Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S RRNA Gene Database and Workbench Compatible With ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akinwande, M.O.; Dikko, H.G.; Samson, A. Variance Inflation Factor: As a Condition for the Inclusion of Suppressor Variable(S) in Regression Analysis. Open J. Stat. 2015, 05, 754–767. [Google Scholar] [CrossRef] [Green Version]
- Cleveland Clinic. Blood Tests to Determine Risk of Coronary Artery Disease. Available online: https://my.clevelandclinic.org/health/diagnostics/16792-blood-tests-to-determine-risk-of-coronary-artery-disease (accessed on 13 December 2021).
- Kanda, Y. Investigation of the Freely Available Easy-to-Use Software “EZR” for Medical Statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MetaboAnalyst 5.0—User-Friendly, Streamlined Metabolomics Data Analysis. Available online: https://www.metaboanalyst.ca/home.xhtml (accessed on 13 December 2021).







| Control | PC | GPC | CC | |
|---|---|---|---|---|
| Initial body weight (g) | 203 ± 5 | 206 ± 2 | 206 ± 2 | 205 ± 4 |
| Final body weight (g) | 420 ± 10 | 432 ± 4 | 422 ± 10 | 428 ± 7 |
| Food intake (g·d−1) | 24.4 ± 0.6 | 24.2 ± 0.4 | 24.5 ± 0.1 | 24.2 ± 0.5 |
| Organ weights (g per 100 g of body weight) | ||||
| Liver | 4.15 ± 0.17 | 4.41 ± 0.1 | 4.15 ± 0.27 | 4.23 ± 0.12 |
| Soleus muscle | 1.04 ± 0.07 | 1.08 ± 0.06 | 1.07 ± 0.03 | 1.01 ± 0.03 |
| White adipose tissue (WAT) weights (g per 100 g of body weight) | ||||
| Epididymal | 1.75 ± 0.10 a | 1.56 ± 0.05 ab | 1.45 ± 0.09 ab | 1.4 ± 0.07 b |
| Perirenal | 2.26 ± 0.17 a | 1.69 ± 0.15 ab | 1.52 ± 0.21 b | 1.77 ± 0.19 ab |
| Mesenteric | 1.32 ± 0.12 | 1.19 ± 0.06 | 1.09 ± 0.08 | 1.05 ± 0.1 |
| Abdominal * | 5.33 ± 0.36 | 4.44 ± 0.19 | 4.07 ± 0.35 | 4.21 ± 0.34 |
| Plasma levels | ||||
| TG (mg·dL−1) | 189 ± 36 | 227 ± 27 | 164 ± 44 | 211 ± 50 |
| NEFA (mEq·dL−1) | 0.404 ± 0.032 | 0.384 ± 0.019 | 0.363 ± 0.053 | 0.388 ± 0.02 |
| Total cholesterol (mg·dL−1) | 77.3 ± 7.9 | 76.3 ± 7 | 93.5 ± 10 | 76.9 ± 3.4 |
| PL (mg·dL−1) | 138 ± 30 | 104 ± 11 | 149 ± 24 | 117 ± 18 |
| Glucose (mg·dL−1) | 259 ± 32 | 232 ± 9 | 237 ± 15 | 241 ± 13 |
| Liver PL contents (mg per whole liver) | 476 ± 33 a | 409 ± 20 ab | 358 ± 18 b | 378 ± 13 b |
| mRNA levels of enzymes related to TMA metabolism in the liver | ||||
| Fmo1 (arbitrary unit) | 1 ± 0.21 | 1.38 ± 0.2 | 1.04 ± 0.19 | 1.03 ± 0.26 |
| Fmo3 (arbitrary unit) | 1 ± 0.16 | 0.894 ± 0.159 | 0.798 ± 0.158 | 0.737 ± 0.107 |
| Feces | ||||
| Weight (wet g·d−1) | 2.32 ± 0.11 | 2.2 ± 0.12 | 2.3 ± 0.16 | 2.26 ± 0.14 |
| Response Variable | Predictor Variable (Fecal Microbe) | β Coefficient | p Value | VIF | Durbin-Watson Ratio | Normality of Unstandardized Residual | Adjusted R2 |
|---|---|---|---|---|---|---|---|
| LN plasma TMAO | Anaerotruncus | −0.725 | p < 0.001 | 1.000 | 1.393 | Yes | 0.656 (p < 0.001) |
| Coprobacter | 0.401 | p < 0.01 | 1.000 |
| Control | PC | GPC | CC | |
|---|---|---|---|---|
| Ingredients | (g·kg−1 diet) | |||
| Sucrose | 479 | 474 | 472.364 | 475.398 |
| Casein | 200 | 200 | 200 | 200 |
| β-Cornstarch | 150 | 150 | 150 | 150 |
| Cellulose | 50 | 50 | 50 | 50 |
| Soybean oil a | 70 | 55 | 70 | 70 |
| Soybean PC b | --- | 20 | --- | --- |
| GPC c | --- | --- | 6.636 | --- |
| Choline chloride (CC) d | --- | --- | --- | 3.602 |
| Mineral mixture (AIN-76) | 35 | 35 | 35 | 35 |
| Vitamin mixture (AIN-76) | 10 | 10 | 10 | 10 |
| DL-Methionine | 3 | 3 | 3 | 3 |
| Choline bitartrate | 2 | 2 | 2 | 2 |
| Cholesterol | 1 | 1 | 1 | 1 |
| (mmol·kg−1 diet) | ||||
| Total choline | 7.9 | 33.7 | 33.7 | 33.7 |
| Total fatty acids | 237.6 | 238.3 | 237.6 | 237.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirouchi, B.; Fukuda, A.; Akasaka, T. Unlike Glycerophosphocholine or Choline Chloride, Dietary Phosphatidylcholine Does Not Increase Plasma Trimethylamine-N-Oxide Levels in Sprague-Dawley Rats. Metabolites 2022, 12, 64. https://doi.org/10.3390/metabo12010064
Shirouchi B, Fukuda A, Akasaka T. Unlike Glycerophosphocholine or Choline Chloride, Dietary Phosphatidylcholine Does Not Increase Plasma Trimethylamine-N-Oxide Levels in Sprague-Dawley Rats. Metabolites. 2022; 12(1):64. https://doi.org/10.3390/metabo12010064
Chicago/Turabian StyleShirouchi, Bungo, Ayano Fukuda, and Taiki Akasaka. 2022. "Unlike Glycerophosphocholine or Choline Chloride, Dietary Phosphatidylcholine Does Not Increase Plasma Trimethylamine-N-Oxide Levels in Sprague-Dawley Rats" Metabolites 12, no. 1: 64. https://doi.org/10.3390/metabo12010064








