Different Metabolomic and Proteomic Profiles of Cerebrospinal Fluid in Ventricular and Lumbar Compartments in Relation to Leptomeningeal Metastases
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of the Patients
2.2. CSF Profiles by Comparment and by Presence of Spinal LM
2.3. Metabolomic Profiles for Paired Ventricular and Lumbar CSF Samples
2.4. LMIs for Discriminating Spinal LM (+) from Spinal LM (−) Lumbar CSF
2.5. Metabolic Pathways That Involve the Discriminative LMIs
2.6. Similarities of LMIs between Spinal LM and CSF Compartments
2.7. Proteomic Profiles in Paired Ventricular and Lumbar CSF Samples
2.8. Proteins That Were Highly Expressed in Spinal LM (+) CSF Compared with Spinal LM (−) CSF
2.9. Pathways Associated with Discriminative CSF Proteins
2.10. Similarities of Discriminative Proteins between Spinal LM and CSF Compartments
2.11. Associations between Discriminative LMIs and Proteins
3. Discussion
3.1. CSF Profiles by Compartment
3.2. CSF Micromolecules as Biomarkers of CNS Disease Activity
3.3. Comparison of CSF Protein Expression between Groups
3.4. Limitations of Comparisons of CSF LMIs between Groups and Correlation with CSF Protein Expression
4. Materials and Methods
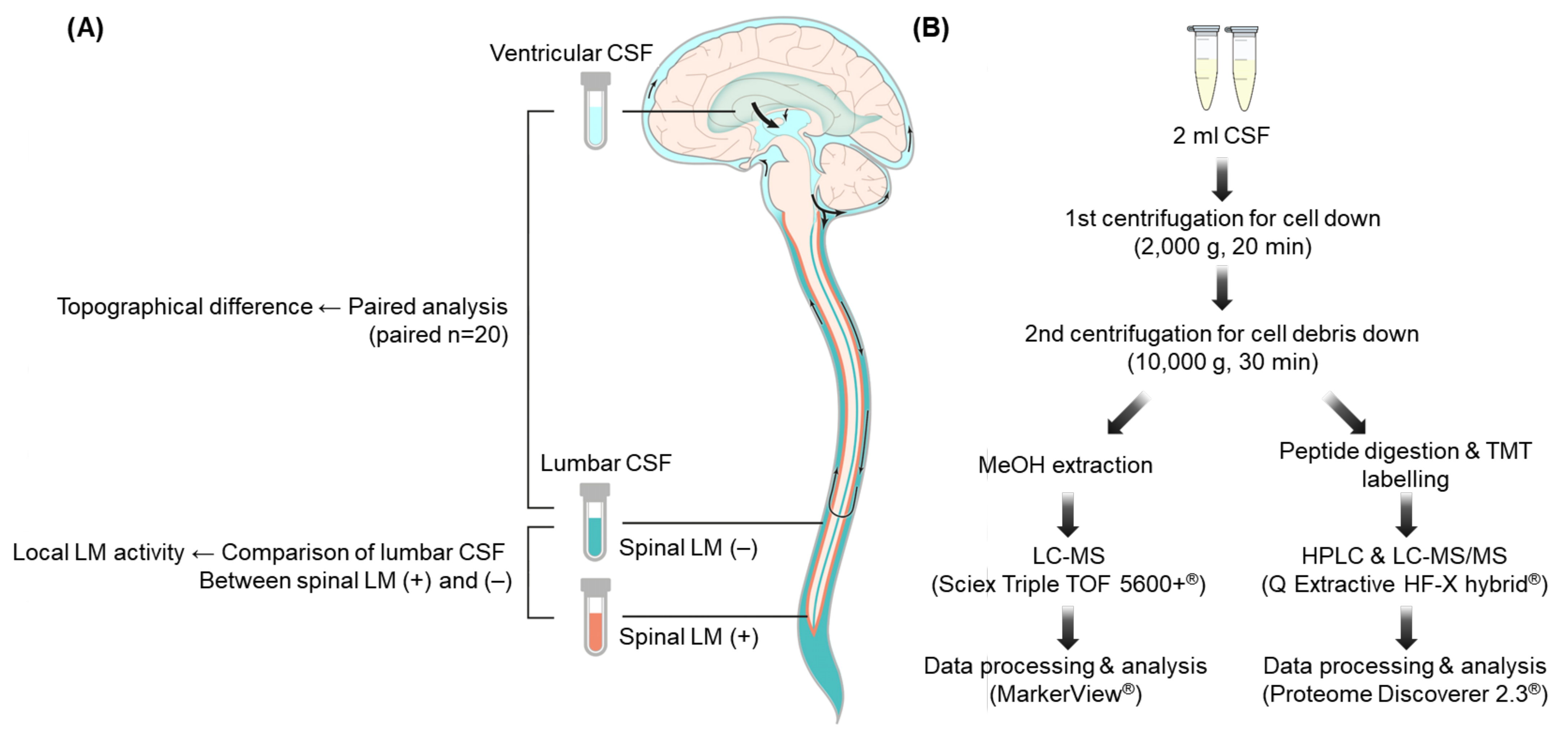
4.1. CSF Samples
4.2. Diagnosis of LM
4.3. CSF Metabolite Profiling Using LC–MS
4.4. CSF Proteomic Profiling Using LC–MS/MS
4.5. Extraction of Discriminating LMIs and Proteins in Pairwise Comparisons of Lumbar and Ventricular CSF Samples
4.6. Extraction of Discriminating LMIs and Proteins in Groupwise Comparisons of Spinal LM (+) and LM (−) Lumbar CSF Samples
4.7. Metabolite and Protein Functional Enrichment Analysis
4.8. Metabolite and Protein Interaction Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chamberlain, M.C. Leptomeningeal metastasis. Curr. Opin. Oncol. 2010, 22, 627–635. [Google Scholar] [CrossRef]
- Taillibert, S.; Laigle-Donadey, F.; Chodkiewicz, C.; Sanson, M.; Hoang-Xuan, K.; Delattre, J.Y. Leptomeningeal metastases from solid malignancy: A review. J. Neurooncol. 2005, 75, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Bruna, J.; Gonzalez, L.; Miro, J.; Velasco, R.; Gil, M.; Tortosa, A. Neuro-Oncology Unit of the Institute of Biomedical Investigation of B: Leptomeningeal carcinomatosis: Prognostic implications of clinical and cerebrospinal fluid features. Cancer 2009, 115, 381–389. [Google Scholar] [CrossRef]
- Grossman, S.A.; Krabak, M.J. Leptomeningeal carcinomatosis. Cancer Treat Rev. 1999, 25, 103–119. [Google Scholar] [CrossRef]
- Jayson, G.C.; Howell, A. Carcinomatous meningitis in solid tumours. Ann. Oncol. 1996, 7, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Morris, P.G.; Reiner, A.S.; Szenberg, O.R.; Clarke, J.L.; Panageas, K.S.; Perez, H.R.; Kris, M.G.; Chan, T.A.; DeAngelis, L.M.; Omuro, A.M. Leptomeningeal metastasis from non-small-cell lung cancer: Survival and the impact of whole brain radiotherapy. J. Thorac. Oncol. 2012, 7, 382–385. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, Y.J.; Lee, J.O.; Lee, K.W.; Kim, J.H.; Bang, S.M.; Chung, J.H.; Kim, J.S.; Lee, J.S. Clinical outcomes of leptomeningeal metastasis in patients with non-small-cell lung cancer in the modern chemotherapy era. Lung Cancer 2012, 76, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Gajjar, A.; Fouladi, M.; Walter, A.W.; Thompson, S.J.; Reardon, D.A.; Merchant, T.E.; Jenkins, J.J.; Liu, A.; Boyett, J.M.; Kun, L.E.; et al. Comparison of lumbar and shunt cerebrospinal fluid specimens for cytologic detection of leptomeningeal disease in pediatric patients with brain tumors. J. Clin. Oncol. 1999, 17, 1825–1828. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, M.C.; Kormanik, P.A.; Glantz, M.J. A comparison between ventricular and lumbar cerebrospinal fluid cytology in adult patients with leptomeningeal metastases. Neuro-Oncol. 2001, 3, 42–45. [Google Scholar] [CrossRef]
- Agnihotri, S.; Zadeh, G. Metabolic reprogramming in glioblastoma: The influence of cancer metabolism on epigenetics and unanswered questions. Neuro-Oncol. 2016, 18, 160–172. [Google Scholar] [CrossRef]
- Brandner, S.; Thaler, C.; Lewczuk, P.; Lelental, N.; Buchfelder, M.; Kleindienst, A. Neuroprotein dynamics in the cerebrospinal fluid: Intraindividual concomitant ventricular and lumbar measurements. Eur. Neurol. 2013, 70, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.; Gwak, H.S.; Kim, S.; Joo, J.; Shin, S.H.; Yoo, H. Retrospective Analysis of Cerebrospinal Fluid Profiles in 228 Patients with Leptomeningeal Carcinomatosis: Differences According to the Sampling Site, Symptoms, and Systemic Factors. J. Korean Neurosurg. Soc. 2016, 59, 570–576. [Google Scholar] [CrossRef]
- Chamberlain, M.C. Radioisotope CSF flow studies in leptomeningeal metastases. J. Neurooncol. 1998, 38, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Schiff, D.; Feske, S.K.; Wen, P.Y. Deceptively normal ventricular fluid in lymphomatous meningitis. Arch. Intern. Med. 1993, 153, 389–390. [Google Scholar] [CrossRef] [PubMed]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.; Webb, D.J.; Dear, J.W. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-Oncol. 2012, 14, 689–700. [Google Scholar] [CrossRef]
- Dekker, L.J.; Boogerd, W.; Stockhammer, G.; Dalebout, J.C.; Siccama, I.; Zheng, P.; Bonfrer, J.M.; Verschuuren, J.J.; Jenster, G.; Verbeek, M.M.; et al. MALDI-TOF mass spectrometry analysis of cerebrospinal fluid tryptic peptide profiles to diagnose leptomeningeal metastases in patients with breast cancer. Mol. Cell Proteom. 2005, 4, 1341–1349. [Google Scholar] [CrossRef]
- Ballester, L.Y.; Lu, G.; Zorofchian, S.; Vantaku, V.; Putluri, V.; Yan, Y.; Arevalo, O.; Zhu, P.; Riascos, R.F.; Sreekumar, A.; et al. Analysis of cerebrospinal fluid metabolites in patients with primary or metastatic central nervous system tumors. Acta Neuropathol. Commun. 2018, 6, r85. [Google Scholar] [CrossRef] [PubMed]
- Nakamizo, S.; Sasayama, T.; Shinohara, M.; Irino, Y.; Nishiumi, S.; Nishihara, M.; Tanaka, H.; Tanaka, K.; Mizukawa, K.; Itoh, T.; et al. GC/MS-based metabolomic analysis of cerebrospinal fluid (CSF) from glioma patients. J. Neurooncol. 2013, 113, r65–r74. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, K.H.; Park, J.W.; Chang, H.J.; Kim, B.C.; Kim, S.Y.; Kim, K.G.; Lee, E.S.; Kim, D.Y.; Oh, J.H.; et al. Low-mass-ion discriminant equation: A new concept for colorectal cancer screening. Int. J. Cancer 2014, 134, r1844–r1853. [Google Scholar] [CrossRef]
- Yoo, B.C.; Lee, J.H.; Kim, K.H.; Lin, W.; Kim, J.H.; Park, J.B.; Park, H.J.; Shin, S.H.; Yoo, H.; Kwon, J.W.; et al. Cerebrospinal fluid metabolomic profiles can discriminate patients with leptomeningeal carcinomatosis from patients at high risk for leptomeningeal metastasis. Oncotarget 2017, 8, r101203–r101214. [Google Scholar] [CrossRef]
- Chiasserini, D.; van Weering, J.R.; Piersma, S.R.; Pham, T.V.; Malekzadeh, A.; Teunissen, C.E.; de Wit, H.; Jimenez, C.R. Proteomic analysis of cerebrospinal fluid extracellular vesicles: A comprehensive dataset. J. Proteom. 2014, 106, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Begcevic, I.; Brinc, D.; Brown, M.; Martinez-Morillo, E.; Goldhardt, O.; Grimmer, T.; Magdolen, V.; Batruch, I.; Diamandis, E.P. Brain-related proteins as potential CSF biomarkers of Alzheimer’s disease: A targeted mass spectrometry approach. J. Proteom. 2018, 182, 12–20. [Google Scholar] [CrossRef]
- Begcevic, I.; Brinc, D.; Dukic, L.; Simundic, A.M.; Zavoreo, I.; Basic Kes, V.; Martinez-Morillo, E.; Batruch, I.; Drabovich, A.P.; Diamandis, E.P. Targeted Mass Spectrometry-Based Assays for Relative Quantification of 30 Brain-Related Proteins and Their Clinical Applications. J. Proteome Res. 2018, 17, r2282–r2292. [Google Scholar] [CrossRef] [PubMed]
- Batruch, I.; Lim, B.; Soosaipillai, A.; Brinc, D.; Fiala, C.; Diamandis, E.P. Mass Spectrometry-Based Assay for Targeting Fifty-Two Proteins of Brain Origin in Cerebrospinal Fluid. J. Proteome Res. 2020, 19, r3060–r3071. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, r1260419. [Google Scholar] [CrossRef] [PubMed]
- Bergman, J.; Svenningsson, A.; Liv, P.; Bergenheim, T.; Burman, J. Location matters: Highly divergent protein levels in samples from different CNS compartments in a clinical trial of rituximab for progressive MS. Fluids. Barriers CNS 2020, 17, r49. [Google Scholar] [CrossRef]
- Minta, K.; Jeppsson, A.; Brinkmalm, G.; Portelius, E.; Zetterberg, H.; Blennow, K.; Tullberg, M.; Andreasson, U. Lumbar and ventricular CSF concentrations of extracellular matrix proteins before and after shunt surgery in idiopathic normal pressure hydrocephalus. Fluids Barriers CNS 2021, 18, r23. [Google Scholar] [CrossRef]
- Glantz, M.J.; Cole, B.F.; Glantz, L.K.; Cobb, J.; Mills, P.; Lekos, A.; Walters, B.C.; Recht, L.D. Cerebrospinal fluid cytology in patients with cancer: Minimizing false-negative results. Cancer 1998, 82, r733–r739. [Google Scholar] [CrossRef]
- Cheok, S.K.; Narayan, A.; Arnal-Estape, A.; Gettinger, S.; Goldberg, S.B.; Kluger, H.M.; Nguyen, D.; Patel, A.; Chiang, V. Tumor DNA Mutations from Intraparenchymal Brain Metastases are Detectable in CSF. JCO Precis Oncol. 2021, 5, 163–172. [Google Scholar] [CrossRef]
- Pan, Z.; Yang, G.; He, H.; Gao, P.; Jiang, T.; Chen, Y.; Zhao, G. Identification of Cerebrospinal Fluid MicroRNAs Associated with Leptomeningeal Metastasis from Lung Adenocarcinoma. Front. Oncol. 2020, 10, 387. [Google Scholar] [CrossRef]
- Schmid, D.; Warnken, U.; Latzer, P.; Hoffmann, D.C.; Roth, J.; Kutschmann, S.; Jaschonek, H.; Rubmann, P.; Foltyn, M.; Vollmuth, P.; et al. Diagnostic biomarkers from proteomic characterization of cerebrospinal fluid in patients with brain malignancies. J. Neurochem. 2021, 158, r522–r538. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Im, J.H.; Lin, W.; Gwak, H.S.; Kim, J.H.; Yoo, B.C.; Kim, T.H.; Park, J.B.; Park, H.J.; Kim, H.J.; et al. Nanoparticles in 472 Human Cerebrospinal Fluid: Changes in Extracellular Vesicle Concentration and miR-21 Expression as a Biomarker for Leptomeningeal Metastasis. Cancers 2020, 12, 2745. [Google Scholar] [CrossRef] [PubMed]
- West, E.E.; Kolev, M.; Kemper, C. Complement and the Regulation of T Cell Responses. Annu. Rev. Immunol. 2018, 36, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Boire, A.; Zou, Y.; Shieh, J.; Macalinao, D.G.; Pentsova, E.; Massague, J. Complement Component 3 Adapts the Cerebrospinal Fluid for Leptomeningeal Metastasis. Cell 2017, 168, r1101–r1113.e1113. [Google Scholar] [CrossRef] [PubMed]
- Hladky, S.B.; Barrand, M.A. Mechanisms of fluid movement into, through and out of the brain: Evaluation of the evidence. Fluids Barriers CNS 2014, 11, r26. [Google Scholar] [CrossRef]
- Foster, D.A. Phosphatidic acid signaling to mTOR: Signals for the survival of human cancer cells. Biochim. Biophys Acta 2009, 1791, r949–r955. [Google Scholar] [CrossRef]
- Goodsell, D.S. The molecular perspective: Methotrexate. Oncologist 1999, 4, r340–r341. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, r4–r22. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, r33–r50. [Google Scholar] [CrossRef]
- Gwak, H.S.; Joo, J.; Shin, S.H.; Yoo, H.; Han, J.Y.; Kim, H.T.; Yun, T.; Ro, J.; Lee, J.S.; Lee, S.H. Ventriculolumbar perfusion chemotherapy with methotrexate for treating leptomeningeal carcinomatosis: A Phase II Study. Oncologist 2014, 19, r1044–r1045. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Freilich, R.J.; Krol, G.; DeAngelis, L.M. Neuroimaging and cerebrospinal fluid cytology in the diagnosis of leptomeningeal metastasis. Ann. Neurol. 1995, 38, r51–r57. [Google Scholar] [CrossRef] [PubMed]







| Characteristics | No. of Patients (%) |
|---|---|
| Gender | |
| Male | 11 (55) |
| Female | 9 (45) |
| Median age (range) | 56.5 (35–71) |
| Primary cancer | |
| Lung | 10 (50) |
| Breast | 4 (20) |
| Melanoma | 2 (10) |
| Ovary | 1 (5) |
| Colorectal and Lung | 2 (10) |
| Unknown | 1 (5) |
| MRI finding at LM diagnosis | |
| Accompanying brain metastasis | 14 (70) |
| Sulci enhancement | 15 (75) |
| Both ventricle wall and sulci enhancment | 1 (5%) |
| Spinal cord enhancement | 12 (63) a |
| CSF Profiles (Median (Range)) | |||
|---|---|---|---|
| Cell Count (cells/mm3) | Protein (mg/dL) | Glucose (mg/dL) | |
| Sampling site | |||
| Lumbar | 13 (0–160) | 56 (12–294) | 50 (14–143) |
| Ventricular | 2 (0–13) | 11 (5–767) | 72 (18–129) |
| p value | <0.001 | 0.013 | 0.047 |
| Spinal LM | |||
| (+) (n = 12) | 12 (0–160) | 65 (40–294) | 50 (14–143) |
| (−) (n = 7) | 5 (2–60) | 32 (12–82) | 65 (34–115) |
| p value | 0.239 | 0.013 | 0.526 |
| Selected LMI (m/z a) | Compound b | Metabolite Name | Chemical Formula | Class |
|---|---|---|---|---|
| 179.1052 | HMDB0036385 HMDB0040411 | Methyl 4-phenylbutanoate Ethyl 3-phenylpropanoate | C11H14O2 C11H14O2 | Fatty Acyls Fatty Acyls |
| 213.8876 | No matched results | |||
| 241.8833 | No matched results | |||
| 378.9042 | No matched results | |||
| 446.8877 | No matched results | |||
| 473.2008 | HMDB0001534 | 5-Formiminotetrahydrofolic acid | C20H24N8O6 | Pteridines |
| 650.8546 | No matched results | |||
| 189.1129/189.1118 | HMDB0011717 HMDB0059814 HMDB0059783 HMDB0059879 HMDB0059760 HMDB0028700 HMDB0240640 | Nonate Diethyl methylsuccinate 3-Methylsuberic acid Diethyl glutarate 2,4-Dimethylpimelic acid Alanylvaline Indolepropionamide | C9H16O4 C9H16O4 C9H16O4 C9H16O4 C9H16O4 C8H16N2O3 C11H12N2O | Fatty Acyls Fatty Acyls Fatty Acyls Fatty Acyls Fatty Acyls Carboxylic acids Indoles |
| 156.8886 | No matched results |
| Selected LMI (m/z a) | Compound b | Metabolite Name | Chemical Formula | Class |
|---|---|---|---|---|
| 319.2823 | HMDB0001449 HMDB0001455 HMDB0001471 HMDB0006759 HMDB0036740 HMDB0060408 HMDB0062782 HMDB0062594 | Allopregnanolone Alloepipregnanolone Epipregnanolone 3a-Hydroxy-5b-pregnane-20-one 1-Hydroxy-2,5,12,15-heneicosatetraen-4-one 5alpha-Pregnan-20alpha-ol-3-one Pregnanolone Methyl Arachidonate | C21H34O2 C21H34O2 C21H34O2 C21H34O2 C21H34O2 C21H34O2 C21H34O2 C21H34O2 | Steroids Steroids Steroids Steroids Fatty Acyls Steroids Steroids Not classified |
| 394.3519 | No matched results | |||
| 545.2581 | No matched results | |||
| 543.2650/543.2579 | HMDB0010320 | Cortolone-3-glucuronide | C27H42O11 | Steroids |
| 544.2640/544.2631 | HMDB0242178 | Glycocholenate sulfate | C26H41NO9S | Steroids |
| 663.4039 | No matched results | |||
| 556.3920 | No matched results | |||
| 512.4137 | No matched results |
| Selected LMI (m/z a) | Compound b | Metabolite Name | Chemical Formula | Class |
|---|---|---|---|---|
| 509.2978 | HMDB0240600 HMDB0115485 | Lysophosphatidylglycerol (18:2(9Z,12Z)/0:0) Phosphatidic acid (8:0/14:0) | C24H45O9P C25H49O8P | Glycerophospholipids Glycerophospholipids |
| 836.4035 | No matched results | |||
| 302.3049 | HMDB0000269 | Sphinganine | C18H39NO2 | Organonitrogen |
| 376.0097 | No matched results | |||
| 377.2650 | HMDB0012868 HMDB0011580 HMDB0012580 HMDB0003733 HMDB0062284 HMDB0011652 | 9′-Carboxy-gamma-chromanol Monoacylgliceride(20:5/0:0/0:0) 14-Hydroxy-E4-neuroprostane Resolvin D1 11-Hydroxy-E4-neuroprostane 11beta-Hydroxy-3,20-dioxopregn-4-en-21-oic acid | C23H36O4 C23H36O4 C22H32O5 C22H32O5 C22H32O5 C22H32O5 | Benzopyrans Glycerolipids Fatty Acryls Fatty Acryls Fatty Acryls Steroids |
| 525.3722 | No matched results | |||
| 166.0553 | HMDB0002005 HMDB0003454 HMDB0004089 | Methionine sulfoxide 4-Pyridoxolactone Formylanthranilic acid | C5H11NO3S C8H7NO3 C8H7NO3 | Carboxylic acids Pyridines Benzene |
| 229.1560 | HMDB0011174 HMDB0011175 HMDB0241042 | Isoleucylproline Leucylproline 4-Methylheptanoylcarnitine | C11H20N2O3 C11H20N2O3 C15H29NO4 | Carboxylic acids Carboxylic acids Not classified |
| 241.1057 | HMDB0000745 HMDB0005769 | Homocarnosine Balenine | C10H16N4O3 C10H16N4O3 | Peptidomimetics Peptidomimetics |
| 400.3389 | HMDB0000222 | Palmitoylcarnitine | C23H46NO4 | Fatty acyl |
| Lumbar > Ventricular | a Ventricular > Lumbar | Spinal LM (+) > LM (−) | |||
|---|---|---|---|---|---|
| Accession | Description | Accession | Description | Accession | Description |
| P01024 | Complement C3 | P12259 | Coagulation factor V | P11684 | Uteroglobin |
| P01009-1 | Alpha-1-antitrypsin | P02766 | Transthyretin | P43652 | Afamin |
| *Q14624-1 | Inter-alpha-trypsin inhibitor heavy chain H4 | Q14515 | SPARC-like protein 1 | P04217-2 | Isoform 2 of Alpha-1B-glycoprotein |
| P19827-1 | Inter-alpha-trypsin inhibitor heavy chain | Q13822-3 | Isoform 3 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2 | *P07360 | Complement component C8 gamma chain |
| P01042-2 | Isoform LMW of Kininogen-1 | P13591-2 | Neural cell adhesion molecule 1 | Q9NPH3 | Interleukin-1 receptor accessory protein |
| P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 | P02788 | Lactotransferrin | P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 |
| P02748 | Complement component C9 | P36955 | Pigment epithelium-derived factor | P02774 | vitamin D-binding protein |
| P01834 | Immunoglobulin kappa constant | O94985-2 | Isoform 2 of Calsyntenin-1 | Q15063-1 | Periostin |
| *Q96PD5 | N-acetylmuramoyl-L-alanine amidase | O94985-1 | Calsyntenin-1 | P00748 | Coagulation factor XII |
| *P07357 | Complement component C8 alpha chain | P05067-1 | Amyloid-beta A4 protein | Q13231-1 | Chitotriosidase-1 |
| *P07358 | Complement component C8 beta chain | P13521 | Secretogranin-2 | *Q14624-1 | Inter-alpha-trypsin inhibitor heavy chain H4 |
| *P07360 | Complement component C8 gamma chain | Q02246 | Contactin-2 | *Q96PD5 | N-acetylmuramoyl-L-alanine amidase |
| P35542 | Serum amyloid A-4 protein | Q16270-1 | Insulin-like growth factor-binding protein 7 | P08185 | Corticosteroid-binding globulin |
| Q4LDE5 | Sushi, von Willebrand factor type A, EGF, and pentraxin domain-containing protein 1 | P23471-1 | Receptor-type tyrosine-protein phosphatase zeta | P02768-1 | Serum albumin |
| P01700 | Immunoglobulin lambda variable 1–47 | Q92876-1 | Kallikrein-6 | P00734 | Prothrombin |
| Q9HDC9 | Adipocyte plasma membrane-associated | Q8TEU8 | WAP, kazal, immunoglobulin, kunitz, and NTR domain-containing protein 2 | P00740 | Coagulation factor IX |
| A0A0C4DH68 | Immunoglobulin kappa variable 2–24 | Q96GW7 | Brevican core protein | P01009-1 | Alpha-1-antitrypsin |
| A0A0B4J1Y8 | Immunoglobulin lambda variable 9- | P09486 | Sparc | P21741 | Midkine |
| Q8NFZ8 | Cell adhesion molecule 4 | P08697-1 | Alpha-2-antiplasmin | ||
| O94919 | Endonuclease domain-containing 1 protein | P01031 | Complement C5 | ||
| P31150 | Rab GDP dissociation inhibitor alpha | *P07358 | Complement component C8 beta chain | ||
| P00441 | Superoxide dismutase [Cu-Zn] | *P07357 | Complement component C8 alpha chain | ||
| P05023 | Sodium/potassium-transporting ATPase subunit alpha-1 | P02671-1 | Fibrinogen alpha chain | ||
| P07686 | Beta-hexosaminidase subunit beta | Q14520-1 | Hyaluronan-binding protein 2 | ||
| P05408-2 | Isoform 2 of Neuroendocrine protein 7B2 | P00450 | Ceruloplasmin | ||
| O00584 | Ribonuclease T2 | O75636-1 | Ficolin-3 | ||
| P30086 | Phosphatidylethanolamine-binding protein 1 | P17813 | Endoglin | ||
| Q9P121-4 | Isoform 4 of Neurotrimin | P04114 | Apolipoprotein B-100 | ||
| Q08629 | Testican-1 | Q06033-1 | Inter-alpha-trypsin inhibitor heavy chain H3 | ||
| P62987 | Ubiquitin-60S ribosomal protein L40 | P02655 | Apolipoprotein C-II | ||
| Q9H3G5 | Probable serine carboxypeptidase CPVL | P26927 | Hepatocyte growth factor-like protein | ||
| Q9UHL4 | Dipeptidyl peptidase 2 | P13671 | Complement component c6 | ||
| Q92563 | Testican-2 | P04004 | Vitronectin | ||
| P12277 | Creatine kinase B-type | P19652 | Alpha-1-acid glycoprotein 2 | ||
| Q9BQT9 | Calsyntenin-3 | P01008 | Antithrombin-III | ||
| P01210 | Proenkephalin-A | P02675 | Fibrinogen beta chain | ||
| Q96S96 | Phosphatidylethanolamine-binding protein 4 | P04196 | Histidine-rich glycoprotein | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, J.-W.; Im, J.H.; Lee, K.-Y.; Yoo, B.C.; Lee, J.H.; Kim, K.-H.; Kim, J.H.; Shin, S.H.; Yoo, H.; Gwak, H.-S. Different Metabolomic and Proteomic Profiles of Cerebrospinal Fluid in Ventricular and Lumbar Compartments in Relation to Leptomeningeal Metastases. Metabolites 2022, 12, 80. https://doi.org/10.3390/metabo12010080
Kwon J-W, Im JH, Lee K-Y, Yoo BC, Lee JH, Kim K-H, Kim JH, Shin SH, Yoo H, Gwak H-S. Different Metabolomic and Proteomic Profiles of Cerebrospinal Fluid in Ventricular and Lumbar Compartments in Relation to Leptomeningeal Metastases. Metabolites. 2022; 12(1):80. https://doi.org/10.3390/metabo12010080
Chicago/Turabian StyleKwon, Ji-Woong, Ji Hye Im, Kyue-Yim Lee, Byong Chul Yoo, Jun Hwa Lee, Kyung-Hee Kim, Jong Heon Kim, Sang Hoon Shin, Heon Yoo, and Ho-Shin Gwak. 2022. "Different Metabolomic and Proteomic Profiles of Cerebrospinal Fluid in Ventricular and Lumbar Compartments in Relation to Leptomeningeal Metastases" Metabolites 12, no. 1: 80. https://doi.org/10.3390/metabo12010080
APA StyleKwon, J.-W., Im, J. H., Lee, K.-Y., Yoo, B. C., Lee, J. H., Kim, K.-H., Kim, J. H., Shin, S. H., Yoo, H., & Gwak, H.-S. (2022). Different Metabolomic and Proteomic Profiles of Cerebrospinal Fluid in Ventricular and Lumbar Compartments in Relation to Leptomeningeal Metastases. Metabolites, 12(1), 80. https://doi.org/10.3390/metabo12010080







