Dereplication of Cytochalasans and Octaketides in Cytotoxic Extracts of Endophytic Fungi from Casearia arborea (Salicaceae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Isolation of Endophytic Fungi
2.3. Identification of Endophytic Fungi
2.4. Secondary Fungal Metabolite Prospection
2.5. UPLC-ESI-HRMS-MS Data Acquisition
2.6. HRMS-MS Data Organization
2.7. Diaporthe sp. CarGL8 Chromatographic Procedures
Compound II-1, Cytochalasin H (Cyt-H)
2.8. Cell Lines, Cultures Condition, and Viability Assay
3. Results
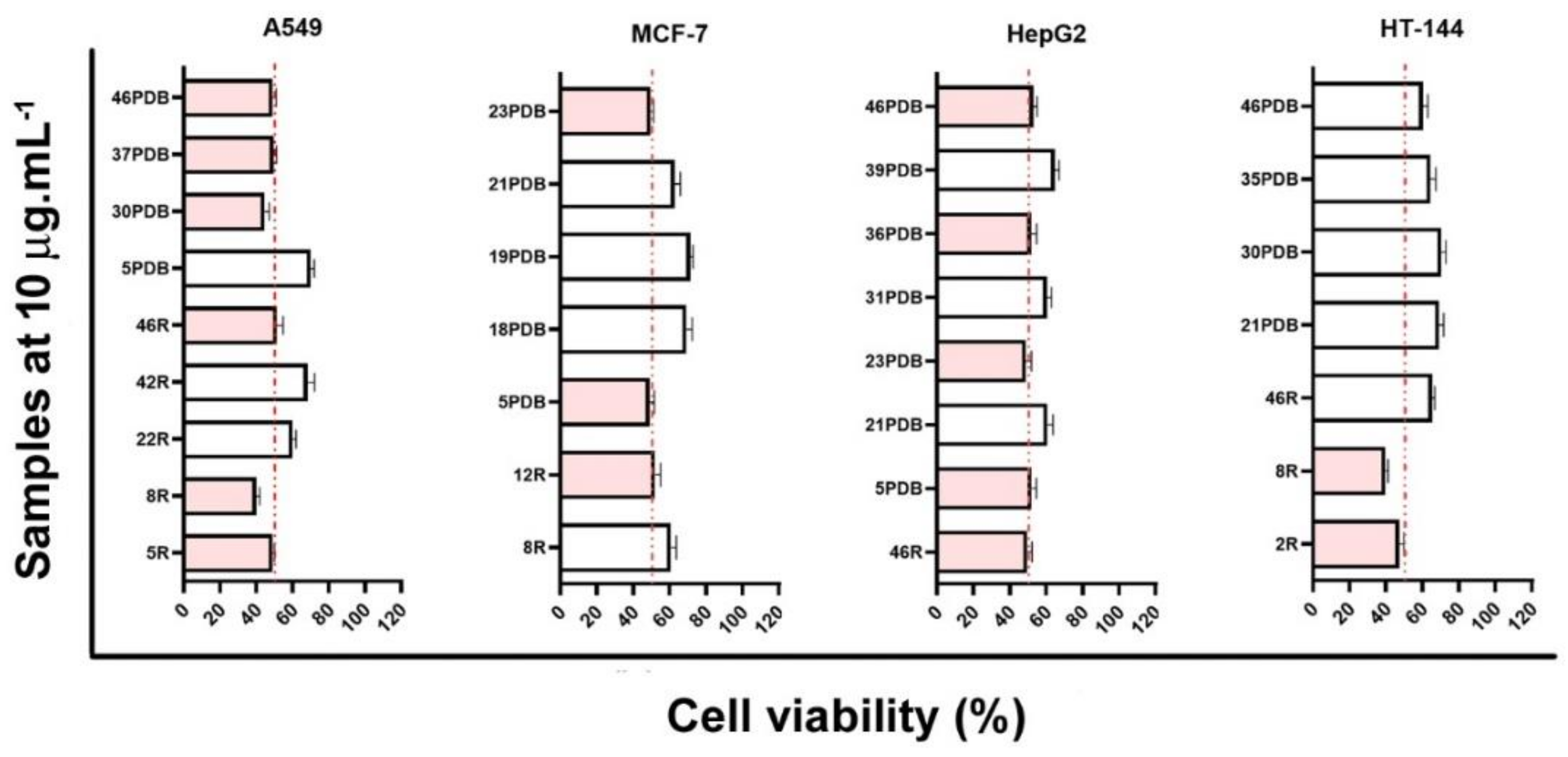
3.1. Cytotoxic Fungi from Casearia arborea Leaves
3.2. Dereplication Based on Molecular Networking Organization from HRMS-MS Data
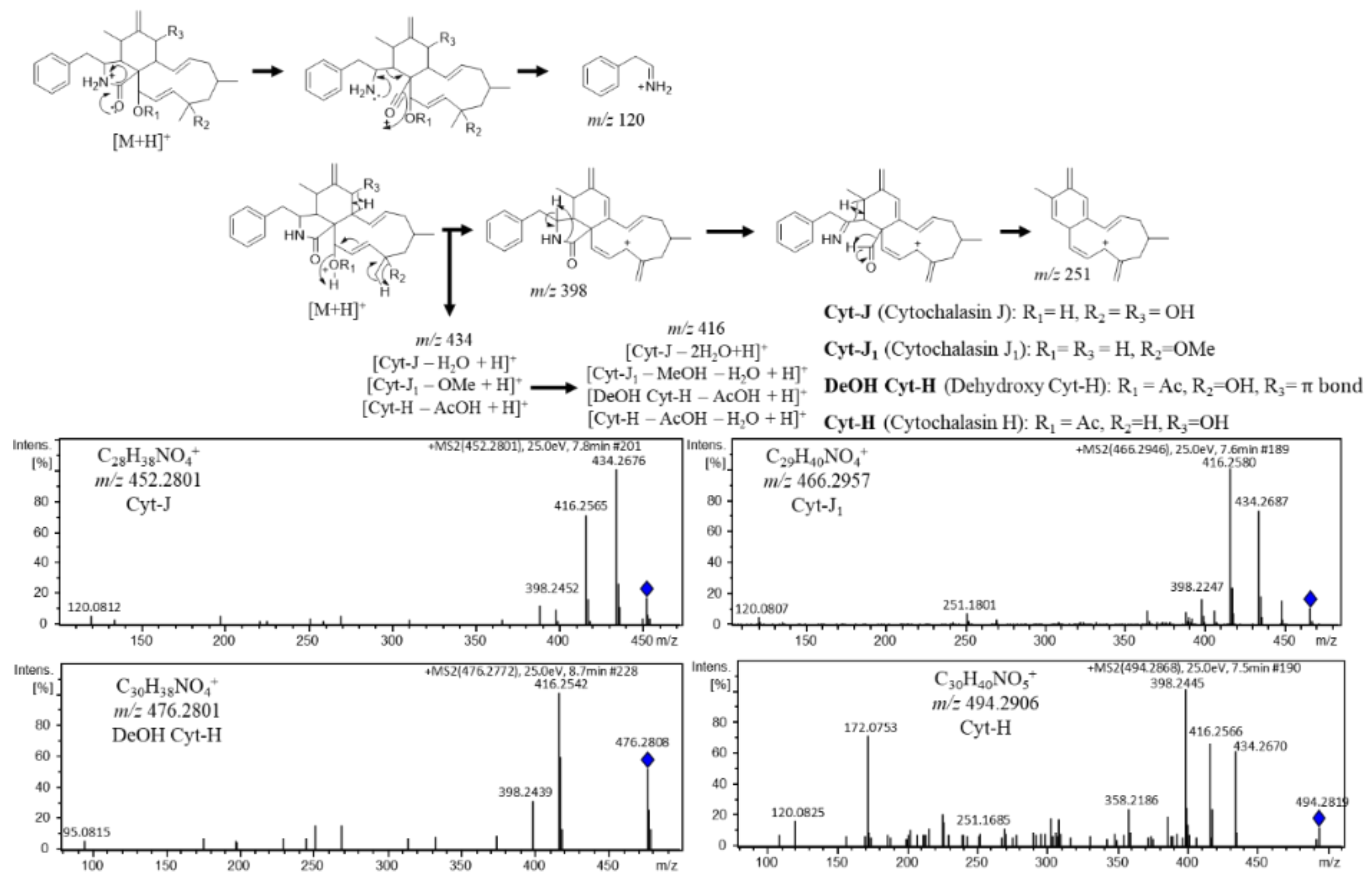
3.2.1. Cytochalasin Dereplication
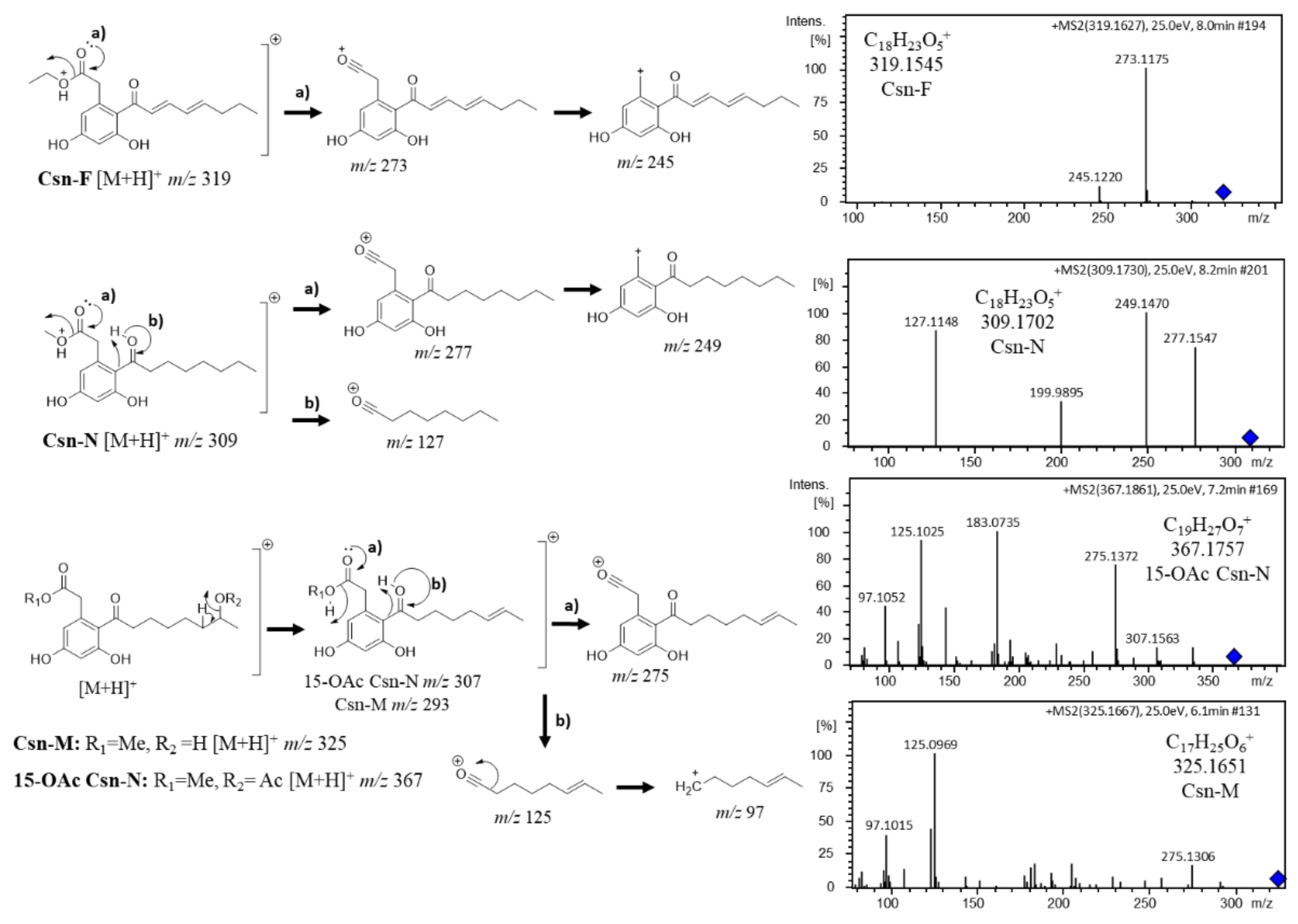
3.2.2. Octaketide Dereplication
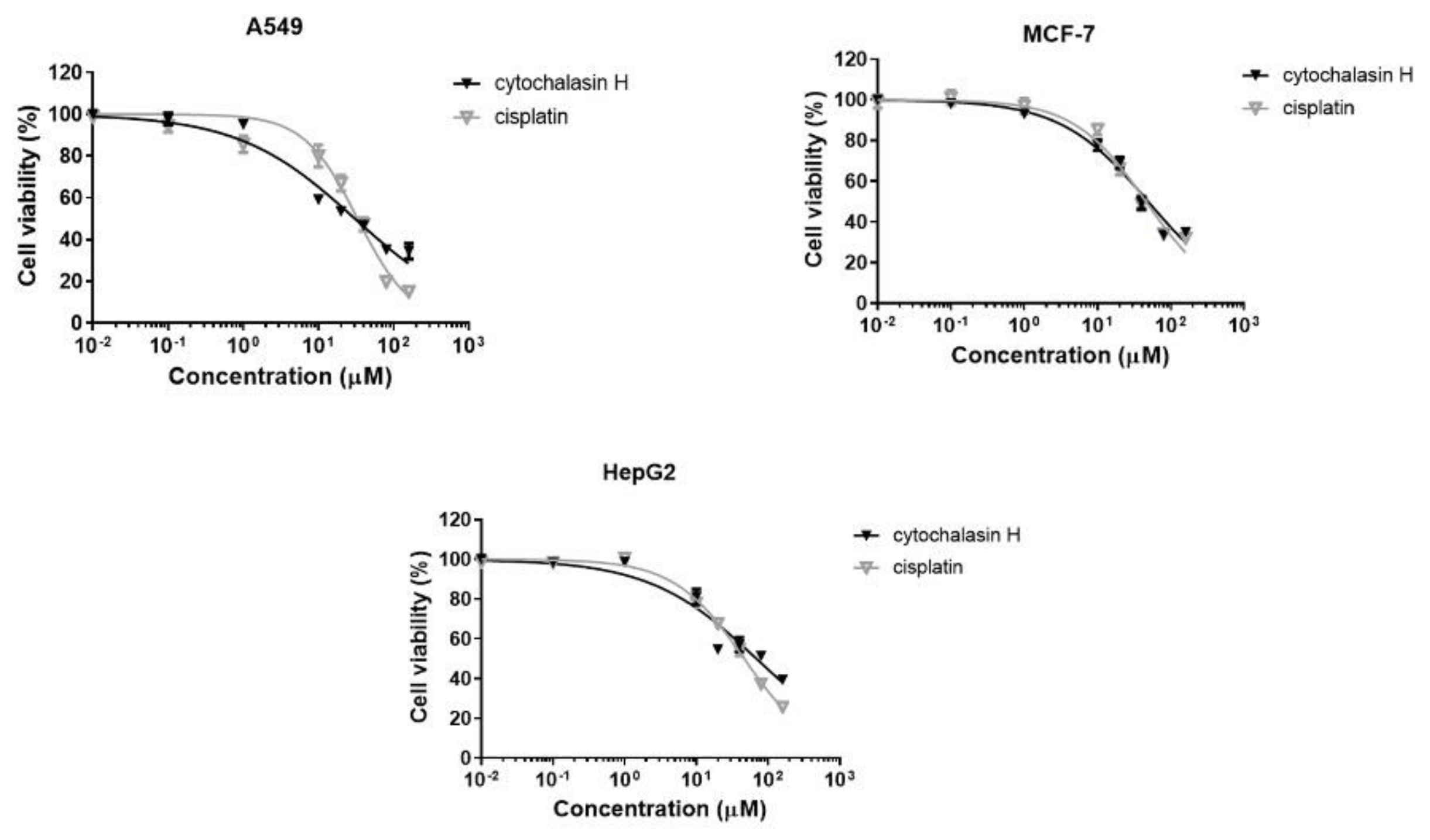
3.3. Cytotoxic Activity of Cytochalasin H
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferreira, P.M.P.; Lotufo, L.V.C.; Moraes, M.O.; Barros, F.W.A.; Martins, A.M.A.; Cavalheiro, A.J.; Bolzani, V.S.; Santos, A.G.; Pessoa, C. Folk uses and pharmacological properties of Casearia sylvestris: A medicinal review. An. Acad. Bras. Ciênc. 2011, 83, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, H.; Matos, F.J.A. Plantas Medicinais No Brasil: Nativas e Exóticas; Instituto Plantarum de Estudos da Flora: Nova Odessa, Brazil, 2002. [Google Scholar]
- Ferreira, P.M.P.; Bezerra, D.P.; Silva, J.D.; da Costa, M.P.; Ferreira, J.R.D.; Alencar, N.M.N.; de Figueiredo, I.S.T.; Cavalheiro, A.J.; Machado, C.M.L.; Chammas, R.; et al. Preclinical anticancer effectiveness of a fraction from Casearia sylvestris and its component casearin X: In vivo and ex vivo methods and microscopy examinations. J. Ethnopharmacol. 2016, 186, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Guo, Q.; Tu, P.; Chai, X. The genus Casearia: A phytochemical and pharmacological overview. Phytochem. Rev. 2015, 14, 99–135. [Google Scholar] [CrossRef]
- Bou, D.D.; Santos, A.L.; Figueiredo, C.R.; Farias, C.F.; Matsuo, A.L.; Kitamura, R.O.S.; Gimenes, L.; Lago, J.H.G.; Sartorelli, P. Dinor casearin X, a new cytotoxic clerodane diterpene from Casearia sylvestris. J. Braz. Chem. Soc. 2015, 26, 1725–1729. [Google Scholar] [CrossRef]
- Ioca, L.P.; Allard, P.M.; Berlinck, R.G. Thinking big about small beings: The (yet) underdeveloped microbial natural products chemistry in Brazil. Nat. Prod. Rep. 2014, 31, 646–675. [Google Scholar] [CrossRef]
- Li, S.-J.; Zhang, X.; Wang, X.-H.; Zhao, C.-Q. Novel natural compounds from endophytic fungi with anticancer activity. Eur. J. Med. Chem. 2018, 156, 316–343. [Google Scholar] [CrossRef]
- Kharwar, R.N.; Mishra, A.; Gond, S.K.; Stierle, A.; Stierle, D. Anticancer compounds derived from fungal endophytes: Their importance and future challenges. Nat. Prod. Rep. 2011, 28, 1208–1228. [Google Scholar] [CrossRef]
- Bacon, C.W.; White, J. Microbial Endophytes, Part III; CRC press: New York, NY, USA, 2000. [Google Scholar]
- Chandra, S.; Bandopadhyay, R.; Kumar, V.; Chandra, R. Acclimatization of tissue cultured plantlets: From laboratory to land. Biotechnol. Lett. 2010, 32, 1199–1205. [Google Scholar] [CrossRef]
- Tan, R.X.; Zou, W.X. Endophytes: A rich source of functional metabolites. Nat. Prod. Rep. 2001, 18, 448–459. [Google Scholar] [CrossRef]
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Lima, N.B.; de A. Batista, M.V.; De Morais, M.A.; Barbosa, M.A.G.; Michereff, S.J.; Hyde, K.D.; Câmara, M.P.S. Five Colletotrichum species are responsible for mango anthracnose in northeastern Brazil. Fungal Divers. 2013, 61, 75–88. [Google Scholar] [CrossRef]
- Schena, L.; Mosca, S.; Cacciola, S.O.; Faedda, R.; Sanzani, S.M.; Agosteo, G.E.; Sergeeva, V.; Magnano di San Lio, G. Species of the Colletotrichum gloeosporioides and C. boninense complexes associated with olive anthracnose. Plant Pathol. 2014, 63, 437–446. [Google Scholar] [CrossRef]
- Talhinhas, P.; Gonçalves, E.; Sreenivasaprasad, S.; Oliveira, H. Virulence diversity of anthracnose pathogens (Colletotrichum acutatum and C. gloeosporioides species complexes) on eight olive cultivars commonly grown in Portugal. Eur. J. Plant Pathol. 2015, 142, 73–83. [Google Scholar] [CrossRef]
- Silva, C.; Michereff, S. Biology of Colletotrichum spp. and epidemiology of the anthracnose in tropical fruit trees. Rev. Caatinga 2014, 26, 130–138. [Google Scholar]
- Da Silva, L.L.; Moreno, H.L.A.; Correia, H.L.N.; Santana, M.F.; de Queiroz, M.V. Colletotrichum: Species complexes, lifestyle, and peculiarities of some sources of genetic variability. Appl. Microbiol. Biotechnol. 2020, 104, 1891–1904. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Chapla, V.M.; Zeraik, M.L.; Cafeu, M.C.; Silva, G.H.; Cavalheiro, A.J.; Bolzani, V.S.; Young, M.; Pfenning, L.H.; Araujo, A.R. Griseofulvin, diketopiperazines and cytochalasins from endophytic fungi Colletotrichum crassipes and Xylaria sp., and their antifungal, antioxidant and anticholinesterase activities. J. Braz. Chem. Soc. 2018, 29, 1707–1713. [Google Scholar] [CrossRef]
- Yodsing, N.; Kanokmedhakul, S.; Mongkolthanaruk, W.; Aimi, T.; Srisapoomi, T.; Boonlue, S. Diversity of endophytic fungi isolated from thai medicinal plants (Casearia grewiifolia) in Khon Kaen, thailand, and their bioactive compounds. Chiang Mai J. Sci. 2018, 45, 45–59. [Google Scholar]
- Santos, A.L.; Ionta, M.; Horvath, R.; Soares, M.G.; de Medeiros, L.S.; Uemi, M.; Kawafune, E.S.; Tangerina, M.M.P.; Ferreira, M.J.P.; Sartorelli, P. Molecular network for accessing polyketide derivatives from Phomopsis sp., an endophytic fungus of Casearia arborea (Salicaceae). Phytochem. Lett. 2021, 42, 1–7. [Google Scholar] [CrossRef]
- da S. Momesso, L.; Kawano, C.Y.; Ribeiro, P.H.; Nomizo, A.; Goldman, G.H.; Pupo, M.T. Chaetoglobosinas produzidas por Chaetomium globosum, fungo endofítico associado a Viguiera robusta Gardn. (Asteraceae). Quím. Nova 2008, 31, 1680–1685. [Google Scholar] [CrossRef]
- Oliveira, C.M.; Regasini, L.O.; Silva, G.H.; Pfenning, L.H.; Young, M.C.M.; Berlinck, R.G.S.; Bolzani, V.S.; Araujo, A.R. Dihydroisocoumarins produced by Xylaria sp. and Penicillium sp., endophytic fungi associated with Piper aduncum and Alibertia macrophylla. Phytochem. Lett. 2011, 4, 93–96. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. Proteo Wizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Wang, M.C.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Frank, A.M.; Bandeira, N.; Shen, Z.; Tanner, S.; Briggs, S.P.; Smith, R.D.; Pevzner, P.A. Clustering millions of tandem mass spectra. J. Proteome Res. 2008, 7, 113–122. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Izawa, Y.; Hirose, T.; Shimizu, T.; Koyama, K.; Natori, S. Six new 10-pheynl-[11] cytochalasans, cytochalasins N-S from Phomopsis sp. Tetrahedron 1989, 45, 2323–2335. [Google Scholar] [CrossRef]
- Cory, A.H.; Owen, T.C.; Barltrop, J.A.; Cory, J.G. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991, 3, 207–212. [Google Scholar] [CrossRef]
- Amaral, L.S.; Fill, T.P.; Santos, L.F.A.; Rodrigues-Filho, E. Biosynthesis and mass spectral fragmentation pathways of (13)C and (15)N labeled cytochalasin D produced by Xylaria arbuscula. J. Mass Spectrom. 2017, 52, 239–247. [Google Scholar] [CrossRef]
- Prasain, J.K.; Ueki, M.; Stefanowicz, P.; Osada, H. Rapid screening and identification of cytochalasins by electrospray tandem mass spectrometry. J. Mass Spectrom. 2002, 37, 283–291. [Google Scholar] [CrossRef]
- Chepkirui, C.; Stadler, M. The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycol. Prog. 2017, 16, 477–494. [Google Scholar] [CrossRef]
- Akay, Ş.; Ekiz, G.; Kocabaş, F.; Hameş-Kocabaş, E.E.; Korkmaz, K.S.; Bedir, E. A new 5, 6-dihydro-2-pyrone derivative from Phomopsis amygdali, an endophytic fungus isolated from hazelnut (Corylus avellana). Phytochem. Lett. 2014, 7, 93–96. [Google Scholar] [CrossRef]
- Hu, Q.; Yang, Y.; Yang, S.; Cao, H.; Chunyang, M.; Yang, H.; Gao, X.; Du, G. Xanthones from the fermentation products of the endophytic fungus of Phomopsis amygdali. Chem. Nat. Compd. 2015, 51, 456–459. [Google Scholar] [CrossRef]
- Song, H.-C.; Qin, D.; Han, M.-J.; Wang, L.; Zhang, K.; Dong, J.-Y. Bioactive 2-pyrone metabolites from an endophytic Phomopsis asparagi SWUKJ5. 2020 of Kadsura angustifolia. Phytochem. Lett. 2017, 22, 235–240. [Google Scholar] [CrossRef]
- Cui, H.; Ding, M.; Huang, D.; Zhang, Z.; Liu, H.; Huang, H.; She, Z. Chroman-4-one and pyrano [4, 3-b] chromenone derivatives from the mangrove endophytic fungus Diaporthe phaseolorum SKS019. RSC Adv. 2017, 7, 20128–20134. [Google Scholar] [CrossRef]
- Chopra, A.; Khuller, G.K. Lipid metabolism in fungi. Crit. Rev. Microbiol. 1984, 11, 209–271. [Google Scholar] [CrossRef]
- Vance, J.E.; Steenbergen, R. Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 2005, 44, 207–234. [Google Scholar] [CrossRef]
- Weiss, R.C.; Stiller, R.L. Dibutyryl cyclic adenosine 3’,5-monophosphate and brain lipid-metabolism. Lipids 1972, 9, 514–519. [Google Scholar] [CrossRef]
- Gu, Z.; Suburu, J.; Chen, H.; Chen, Y.Q. Mechanisms of omega-3 polyunsaturated fatty acids in prostate cancer prevention. BioMed Res. Int. 2013, 2013, 824563. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T.; Fuchs, C.S.; Ogino, S.; Hu, F.B.; Mozaffarian, D.; Ma, J.; Willett, W.C.; Giovannucci, E.L.; Wu, K. Dietary intake of fish, ω-3 and ω-6 fatty acids and risk of colorectal cancer: A prospective study in US men and women. Int. J. Cancer Res. 2014, 135, 2413–2423. [Google Scholar] [CrossRef]
- Larsson, S.C.; Kumlin, M.; Ingelman-Sundberg, M.; Wolk, A. Dietary long-chain n−3 fatty acids for the prevention of cancer: A review of potential mechanisms. Am. J. Clin. Nutr. 2004, 79, 935–945. [Google Scholar] [CrossRef]
- Liu, J.; Abdelmagid, S.A.; Pinelli, C.J.; Monk, J.M.; Liddle, D.M.; Hillyer, L.M.; Hucik, B.; Silva, A.; Subedi, S.; Wood, G.A. Marine fish oil is more potent than plant-based n-3 polyunsaturated fatty acids in the prevention of mammary tumors. J. Nutr. Biochem. 2018, 55, 41–52. [Google Scholar] [CrossRef]
- Wannous, R.; Bon, E.; Mahéo, K.; Goupille, C.; Chamouton, J.; Bougnoux, P.; Roger, S.; Besson, P.; Chevalier, S. PPARβ mRNA expression, reduced by n-3 PUFA diet in mammary tumor, controls breast cancer cell growth. Biochim. Biophys. Acta 2013, 1831, 1618–1625. [Google Scholar] [CrossRef]
- Meza, A.; Santos, E.D.A.; Gomes, R.D.S.; Lima, D.P.; Beatriz, A. Cytosporones and related compound, a review: Isolation, biossynthesis, synthesis and biological activity of promissing fungal resorcinolic lipids. Curr. Org. Synth. 2015, 12, 618–638. [Google Scholar] [CrossRef]
- Giguère, V. Orphan nuclear receptors: From gene to function. Endocr. Rev. 1999, 20, 689–725. [Google Scholar] [CrossRef]
- Winoto, A.; Littman, D.R. Nuclear hormone receptors in T lymphocytes. Cell 2002, 109, S57–S66. [Google Scholar] [CrossRef]
- Banno, A.; Lakshmi, S.P.; Reddy, A.T.; Kim, S.C.; Reddy, R.C. Key functions and therapeutic prospects of Nur77 in inflammation related lung diseases. Am. J. Clin. Pathol. 2019, 189, 482–491. [Google Scholar] [CrossRef]
- Guan, Y.-F.; Huang, Q.-L.; Ai, Y.-L.; Chen, Q.-T.; Zhao, W.-X.; Wang, X.-M.; Wu, Q.; Chen, H.-Z. Nur77-activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene 2020, 39, 2408–2423. [Google Scholar] [CrossRef]
- Palumbo-Zerr, K.; Zerr, P.; Distler, A.; Fliehr, J.; Mancuso, R.; Huang, J.; Mielenz, D.; Tomcik, M.; Fürnrohr, B.G.; Scholtysek, C. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat. Med. 2015, 21, 150–158. [Google Scholar] [CrossRef]
- Wohlkoenig, C.; Leithner, K.; Olschewski, A.; Olschewski, H.; Hrzenjak, A. TR3 is involved in hypoxia-induced apoptosis resistance in lung cancer cells downstream of HIF-1alpha. Lung Cancer 2017, 111, 15–22. [Google Scholar] [CrossRef]
- Zhan, Y.; Du, X.; Chen, H.; Liu, J.; Zhao, B.; Huang, D.; Li, G.; Xu, Q.; Zhang, M.; Weimer, B.C.; et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat. Chem. Biol. 2008, 4, 548–556. [Google Scholar] [CrossRef]
- Cho, S.D.; Yoon, K.; Chintharlapalli, S.; Abdelrahim, M.; Lei, P.; Hamilton, S.; Khan, S.; Ramaiah, S.K.; Safe, S. Nur77 agonists induce proapoptotic genes and responses in colon cancer cells through nuclear receptor–dependent and nuclear receptor–independent pathways. Cancer Res. 2007, 67, 674–683. [Google Scholar] [CrossRef]
- Gao, H.; Chen, Z.; Fu, Y.; Yang, X.; Weng, R.; Wang, R.; Lu, J.; Pan, M.; Jin, K.; McElroy, C. Nur77 exacerbates PC12 cellular injury in vitro by aggravating mitochondrial impairment and endoplasmic reticulum stress. Sci. Rep. 2016, 6, 34403. [Google Scholar] [CrossRef]
- Jiang, Y.; Zeng, Y.; Huang, X.; Qin, Y.; Luo, W.; Xiang, S.H.; Sooranna, S.R.; Pinhu, L. Real-time visualization of lung function: From micro to macro: Nur77 attenuates endothelin-1 expression via downregulation of NF-κB and p38 MAPK in A549 cells and in an ARDS rat model. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 311, L1023–L1035. [Google Scholar] [CrossRef]
- Liu, J.J.; Zeng, H.N.; Zhang, L.R.; Zhan, Y.Y.; Chen, Y.; Wang, Y.; Wang, J.; Xiang, S.H.; Liu, W.J.; Wang, W.J.; et al. A unique pharmacophore for activation of the nuclear orphan receptor Nur77 in vivo and in vitro. Cancer Res. 2010, 70, 3628–3637. [Google Scholar] [CrossRef]
- Spudich, J.A.; Lin, S. Cytochalasin B, its interaction with actin and actomyosin from muscle. Proc. Natl. Acad. Sci. USA 1972, 69, 442–446. [Google Scholar] [CrossRef]
- Cooper, J.A. Effects of cytochalasin and phalloidin on actin. Int. J. Cell Biol. 1987, 105, 1473–1478. [Google Scholar] [CrossRef]
- Chen, H.; Daletos, G.; Okoye, F.; Lai, D.; Dai, H.; Proksch, P. A new cytotoxic cytochalasin from the endophytic fungus Trichoderma harzianum. Nat. Prod. Commun. 2015, 10, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Kongprapan, T.; Rukachaisirikul, V.; Saithong, S.; Phongpaichit, S.; Poonsuwan, W.; Sakayaroj, J. Cytotoxic cytochalasins from the endophytic fungus Eutypella scoparia PSU-H267. Phytochem. Lett. 2015, 13, 171–176. [Google Scholar] [CrossRef]
- Apostolopoulos, V.; Bojarska, J.; Chai, T.T.; Elnagdy, S.; Kaczmarek, K.; Matsoukas, J.; New, R.; Parang, K.; Lopez, O.P.; Parhiz, H.; et al. A global review on short peptides: Frontiers and perspectives. Molecules 2021, 26, 430. [Google Scholar] [CrossRef] [PubMed]
| A549 | MCF-7 | HepG2 | |
|---|---|---|---|
| Cytochalasin H | 31.0 ± 3.2 | 49.2 ± 4.2 | 71.2 ± 11.0 |
| Cisplatin | 33.2 ± 1.2 | 45.8 ± 0.8 | 46.6 ± 0.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, A.L.; Ionta, M.; Horvath, R.O.; Soares, M.G.; Silva, D.O.; Kawafune, E.S.; Ferreira, M.J.P.; Sartorelli, P. Dereplication of Cytochalasans and Octaketides in Cytotoxic Extracts of Endophytic Fungi from Casearia arborea (Salicaceae). Metabolites 2022, 12, 903. https://doi.org/10.3390/metabo12100903
Santos AL, Ionta M, Horvath RO, Soares MG, Silva DO, Kawafune ES, Ferreira MJP, Sartorelli P. Dereplication of Cytochalasans and Octaketides in Cytotoxic Extracts of Endophytic Fungi from Casearia arborea (Salicaceae). Metabolites. 2022; 12(10):903. https://doi.org/10.3390/metabo12100903
Chicago/Turabian StyleSantos, Augusto L., Marisa Ionta, Renato O. Horvath, Marisi G. Soares, Daniele O. Silva, Eunizinis S. Kawafune, Marcelo J. P. Ferreira, and Patricia Sartorelli. 2022. "Dereplication of Cytochalasans and Octaketides in Cytotoxic Extracts of Endophytic Fungi from Casearia arborea (Salicaceae)" Metabolites 12, no. 10: 903. https://doi.org/10.3390/metabo12100903
APA StyleSantos, A. L., Ionta, M., Horvath, R. O., Soares, M. G., Silva, D. O., Kawafune, E. S., Ferreira, M. J. P., & Sartorelli, P. (2022). Dereplication of Cytochalasans and Octaketides in Cytotoxic Extracts of Endophytic Fungi from Casearia arborea (Salicaceae). Metabolites, 12(10), 903. https://doi.org/10.3390/metabo12100903






