Inhibitory Activity of Bioactive Phloroglucinols from the Rhizomes of Dryopteris crassirhizoma on Escherichia coli β-Glucuronidase: Kinetic Analysis and Molecular Docking Studies
Abstract
:1. Introduction
2. Results
2.1. Inhibitory Effect of Phloroglucinols on β-Glucuronidase
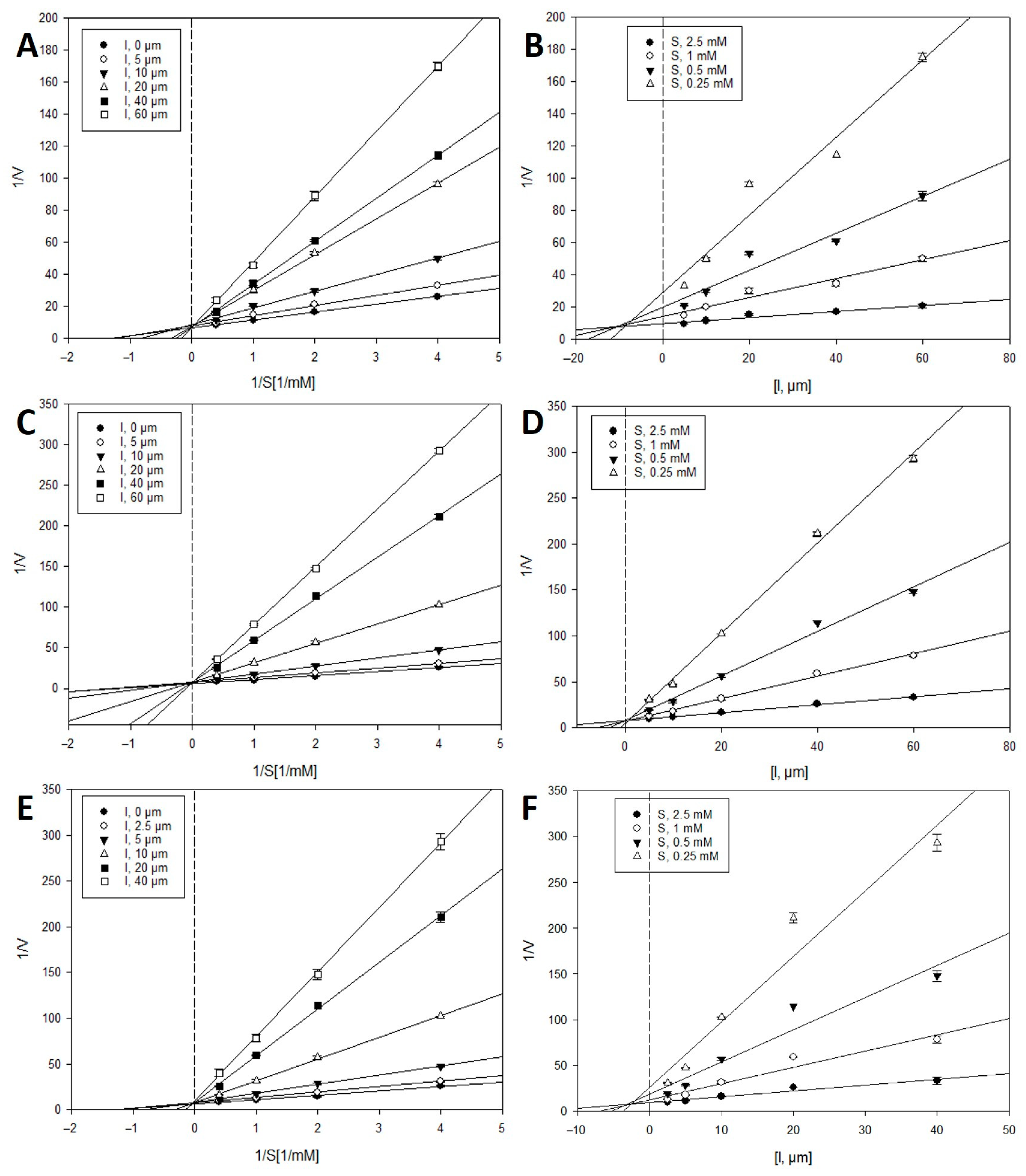
2.2. Inhibition Kinetics of Active Compounds on β-Glucuronidase
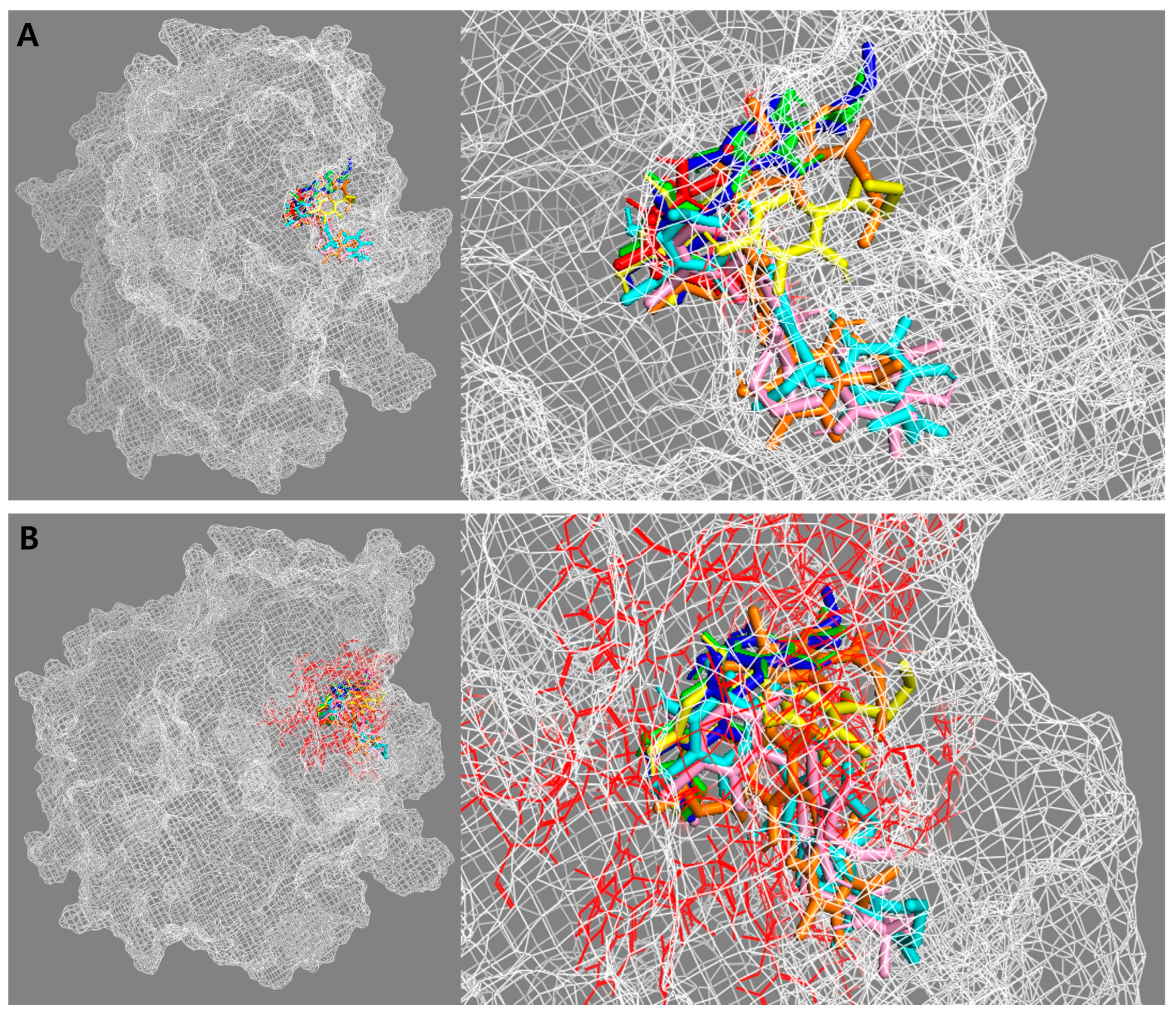
2.3. Molecular Docking Simulation of β-Glucuronidase Inhibition
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Assay for β-Glucuronidase Inhibitory Activity
4.3. Inhibition Kinetic Analysis with β-Glucuronidase
4.4. Molecular Docking Simulation
4.5. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Michikawa, M.; Ichinose, H.; Momma, M.; Biely, P.; Jongkees, S.; Yoshida, M.; Kotake, T.; Tsumuraya, Y.; Withers, S.G.; Fujimoto, Z.; et al. Structural and biochemical characterization of glycoside hydrolase family 79 β-glucuronidase from Acidobacterium capsulatum. J. Biol. Chem. 2012, 287, 14069–14077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-P.; Yan, J.-K.; Yi, J.; Zhang, X.-Y.; Yu, Z.-L.; Huo, X.-K.; Liang, J.-H.; Ning, J.; Feng, L.; Wang, C.; et al. The study of inhibitory effect of natural flavonoids toward β-glucuronidase and interaction of flavonoids with β-glucuronidase. Int. J. Biol. Macromol. 2020, 143, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Drendel, W.B.; Chen, Z.-W.; Mathews, F.S.; Sly, W.S.; Grubb, J.H. Structure of human β-glucuronidase reveals candidate lysosomal targeting and active-site motifs. Nat. Struct. Biol. 1996, 3, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.-A.; Mani, S.; et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef] [Green Version]
- Pusztaszeri, M.P.; Genta, R.M.; Cryer, B.L. Drug-induced injury in the gastrointestinal tract: Clinical and pathologic considerations. Nat. Clin. Pract. Gastroenterol. Hepatol. 2007, 4, 442–453. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, D.; Gao, Z.; He, M.; Hou, J.; Zhang, H.; Zhang, G.; Ding, D.; Feng, G. Highly sensitive light-up near-infrared fluorescent probe for detection and imaging of β-glucuronidase in human serum, living cells and tumor-bearing mice. Sci. China Mater. 2022, 65, 836–844. [Google Scholar] [CrossRef]
- Houba, P.H.J.; Boven, E.; van der Meulen-Muileman, I.H.; Leenders, R.G.G.; Scheeren, J.W.; Pinedo, H.M.; Haisma, H.J. Pronounced antitumor efficacy of doxorubicin when given as the prodrug DOX-GA3 in combination with a monoclonal antibody β-glucuronidase conjugate. Int. J. Cancer 2001, 91, 550–554. [Google Scholar] [CrossRef]
- Cao, T.Q.; Phong, N.V.; Kim, J.H.; Gao, D.; Anh, H.L.; Ngo, V.-D.; Vinh, L.B.; Koh, Y.S.; Yang, S.Y. Inhibitory effects of cucurbitane-type triterpenoids from Momordica charantia fruit on lipopolysaccharide-stimulated pro-inflammatory cytokine production in bone marrow-derived dendritic cells. Molecules 2021, 26, 4444. [Google Scholar] [CrossRef]
- Van Cong, P.; Anh, H.L.T.; Trung, N.Q.; Quang Minh, B.; Viet Duc, N.; Van Dan, N.; Trang, N.M.; Phong, N.V.; Vinh, L.B.; Anh, L.T.; et al. Isolation, structural elucidation and molecular docking studies against SARS-CoV-2 main protease of new stigmastane-type steroidal glucosides isolated from the whole plants of Vernonia gratiosa. Nat. Prod. Res. 2022, 1–9. [Google Scholar] [CrossRef]
- Hwang, Y.-H.; Ha, H.; Ma, J.Y. Acute oral toxicity and genotoxicity of Dryopteris crassirhizoma. J. Ethnopharmacol. 2013, 149, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, J.; Shibuya, M.; Masuda, K.; Ebizuka, Y. Dammaradiene synthase, a squalene cyclase, from Dryopteris crassirhizoma Nakai. Phytochemistry 2008, 69, 2559–2564. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Gao, L.; Si, J.; Sun, Y.; Liu, J.; Cao, L.; Feng, W.-H. Inhibition of porcine reproductive and respiratory syndrome virus replication by flavaspidic acid AB. Antivir. Res. 2013, 97, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-M.; Na, M.-K.; An, R.-B.; Min, B.-S.; Lee, H.-K. Antioxidant activity of two phloroglucinol derivatives from Dryopteris crassirhizoma. Biol. Pharm. Bull. 2003, 26, 1354–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.B.; Kim, J.C.; Lee, S.M. Antibacterial activity of two phloroglucinols, flavaspidic acids AB and PB, from Dryopteris crassirhizoma. Arch. Pharm. Res. 2009, 32, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Miyashiro, H.; Nakamura, N.; Hattori, M. Two new triterpenes from the rhizome of Dryopteris crassirhizoma, and inhibitory activities of its constituents on human immunodeficiency virus-1 protease. Chem. Pharm. Bull. 2008, 56, 711–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.-H.; Bae, J.-H.; Hong, D.-P.; Choi, K.-D.; Kim, S.-C.; Her, E.; Kim, S.-H.; Kang, C.-D. Dryopteris crassirhizoma has anti-cancer effects through both extrinsic and intrinsic apoptotic pathways and G0/G1 phase arrest in human prostate cancer cells. J. Ethnopharmacol. 2010, 130, 248–254. [Google Scholar] [CrossRef]
- Yim, N.-H.; Lee, J.-J.; Lee, B.; Li, W.; Ma, J.Y. Antiplatelet activity of acylphloroglucinol derivatives isolated from Dryopteris crassirhizoma. Molecules 2019, 24, 2212. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Li, Z.; Li, C.-Y.; Jia, W.-N.; Wang, H.-T.; Wang, C.-H. Phytochemical constituents and biological activities of plants from the genus Dryopteris. Chem. Biodivers. 2015, 12, 1131–1162. [Google Scholar] [CrossRef]
- Yuk, H.J.; Kim, J.-Y.; Sung, Y.-Y.; Kim, D.-S. Phloroglucinol derivatives from Dryopteris crassirhizoma as potent xanthine oxidase inhibitors. Molecules 2021, 26, 122. [Google Scholar] [CrossRef]
- Pham, V.C.; Kim, O.; Lee, J.-H.; Min, B.S.; Kim, J.A. Inhibitory effects of phloroglucinols from the roots of Dryopteris crassirhizoma on melanogenesis. Phytochem. Lett. 2017, 21, 51–56. [Google Scholar] [CrossRef]
- Wang, J.; Yan, Y.-T.; Fu, S.-Z.; Peng, B.; Bao, L.-L.; Zhang, Y.-L.; Hu, J.-H.; Zeng, Z.-P.; Geng, D.-H.; Gao, Z.-P. Anti-influenza virus (H5N1) activity screening on the phloroglucinols from rhizomes of Dryopteris crassirhizoma. Molecules 2017, 22, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phong, N.V.; Oanh, V.T.; Yang, S.Y.; Choi, J.S.; Min, B.S.; Kim, J.A. PTP1B inhibition studies of biological active phloroglucinols from the rhizomes of Dryopteris crassirhizoma: Kinetic properties and molecular docking simulation. Int. J. Biol. Macromol. 2021, 188, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Dixon, M. The determination of enzyme inhibitor constants. Biochem. J. 1953, 55, 170–171. [Google Scholar] [CrossRef]
- Taha, M.; Ismail, N.H.; Imran, S.; Selvaraj, M.; Rashwan, H.; Farhanah, F.U.; Rahim, F.; Kesavanarayanan, K.S.; Ali, M. Synthesis of benzimidazole derivatives as potent β-glucuronidase inhibitors. Bioorg. Chem. 2015, 61, 36–44. [Google Scholar] [CrossRef]
- Roberts, A.B.; Wallace, B.D.; Venkatesh, M.K.; Mani, S.; Redinbo, M.R. Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 2013, 84, 208–217. [Google Scholar] [CrossRef] [Green Version]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef]
- Viet Phong, N.; Thi Nguyet Anh, D.; Yeong Chae, H.; Young Yang, S.; Jeong Kwon, M.; Sun Min, B.; Ah Kim, J. Anti-inflammatory activity and cytotoxicity against ovarian cancer cell lines by amide alkaloids and piperic esters isolated from Piper longum fruits: In vitro assessments and molecular docking simulation. Bioorg. Chem. 2022, 128, 106072. [Google Scholar] [CrossRef]
- Torres, P.H.M.; Sodero, A.C.R.; Jofily, P.; Silva-Jr, F.P. Key topics in molecular docking for drug design. Int. J. Mol. Sci. 2019, 20, 4574. [Google Scholar] [CrossRef]
- Kim, J.H.; Vinh, L.B.; Hur, M.; Koo, S.-C.; Park, W.T.; Moon, Y.-H.; Lee, Y.J.; Kim, Y.H.; Huh, Y.-C.; Yang, S.Y. Inhibitory activity of 4-O-benzoyl-3′-O-(O-methylsinapoyl) sucrose from Polygala tenuifolia on Escherichia coli β-glucuronidase. J. Microbiol. Biotechnol. 2021, 31, 1576–1582. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.-Y.; Chen, C.-Y.; Lin, T.-C.; Yeh, L.-F.; Hsieh, W.-C.; Gao, S.; Burnouf, P.-A.; Chen, B.-M.; Hsieh, T.-J.; Dashnyam, P.; et al. Entropy-driven binding of gut bacterial β-glucuronidase inhibitors ameliorates irinotecan-induced toxicity. Commun. Biol. 2021, 4, 280. [Google Scholar] [CrossRef] [PubMed]
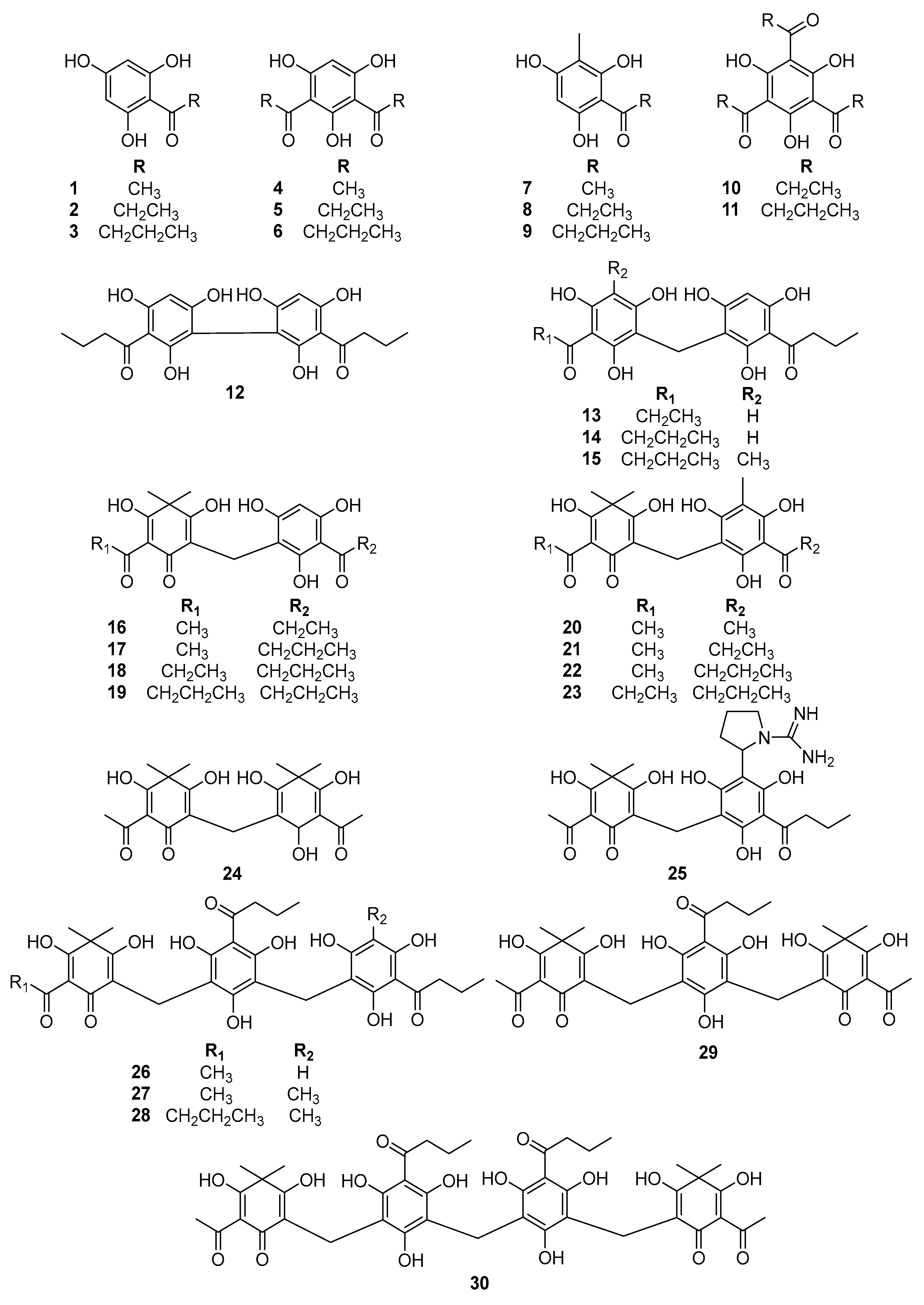




| Compounds | Inhibition of Compounds on β-glucuronidase | |
|---|---|---|
| Inhibition at 100 μM (%) | IC50 (μM) 1 | |
| 1 | 11.6 ± 3.9 | >100 |
| 2 | 24.4 ± 0.2 | >100 |
| 3 | 22.0 ± 2.1 | >100 |
| 4 | 22.1 ± 2.4 | >100 |
| 5 | 27.1 ± 0.4 | >100 |
| 6 | 33.7 ± 2.3 | >100 |
| 7 | 26.7 ± 1.5 | >100 |
| 8 | 24.9 ± 5.5 | >100 |
| 9 | 34.7 ± 2.4 | >100 |
| 10 | 41.4 ± 1.7 | >100 |
| 11 | 87.3 ± 3.4 | 43.0 ± 2.9 |
| 12 | 34.1 ± 0.5 | >100 |
| 13 | 93.7 ± 0.3 | 18.1 ± 1.5 |
| 14 | 97.1 ± 0.7 | 14.4 ± 0.6 |
| 15 | 89.1 ± 0.3 | 17.0 ± 2.5 |
| 16 | 68.3 ± 0.2 | 64.4 ± 3.5 |
| 17 | 65.1 ± 0.1 | 67.0 ± 1.3 |
| 18 | 42.2 ± 0.1 | >100 |
| 19 | 41.0 ± 1.4 | >100 |
| 20 | 76.1 ± 3.4 | 75.7 ± 1.2 |
| 21 | 47.9 ± 4.7 | >100 |
| 22 | 48.7 ± 4.8 | >100 |
| 23 | 73.5 ± 0.7 | 76.5 ± 1.5 |
| 24 | 28.1 ± 1.2 | >100 |
| 25 | 30.7 ± 1.7 | >100 |
| 26 | 98.09 ± 2.5 | 8.0 ± 1.8 |
| 27 | 101.2 ± 0.6 | 7.1 ± 2.6 |
| 28 | 106.2 ± 3.4 | 5.6 ± 1.1 |
| 29 | 53.4 ± 2.3 | 90.5 ± 2.7 |
| 30 | 51.8 ± 2.7 | 94.9 ± 2.5 |
| DSA 2 | 79.0 ± 0.3 | 23.4 ± 1.5 |
| Compounds | Inhibition Type 1 | Ki (μM) 2 |
|---|---|---|
| 13 | Competitive | 6.3 |
| 14 | Competitive | 4.3 |
| 15 | Competitive | 25.8 |
| 26 | Competitive | 7.5 |
| 27 | Competitive | 0.5 |
| 28 | Competitive | 2.8 |
| Compounds | Binding Energy (kcal/mol) | Hydrogen Bonds | van der Waals Interactions | Hydrophobic Interactions | Electrostatic Interactions |
|---|---|---|---|---|---|
| 13 | −8.22 | Asn412, Glu413, Tyr468, Tyr472 | Phe161, His162, His330, Gly356, Asn358, Val446, Asn466, Trp549, Arg562, Asn566, Lys568 | Leu361 (π-σ) Val355 (alkyl) Met447 (π-alkyl) Phe448 (π-alkyl) | Asp163 (π-anion) Glu504 (π-anion) |
| 14 | −8.49 | Asp163, His330, Glu413, Tyr468, Tyr472 | Phe161, His162, Phe164, Gly356, Asn358, Asn412, Val446, Asn466, Trp549, Asn566, Lys568 | Val355 (alkyl) Leu361 (alkyl) Met447 (π-alkyl) Phe448 (π-alkyl) | Glu504 (π-anion) Glu562 (π-cation) |
| 15 | −8.02 | Asp163, His330, Leu361, Glu413, Tyr468, Tyr472 | His162, Phe164, His296, Gly356, Asn358, Gly362, Ile363, Asn412, Trp549, Phe554, Arg562, Asn566, Lys568 | Val355 (alkyl) Leu561 (alkyl) | Glu504 (π-anion) |
| 26 | −10.67 | Phe161, Asp163, His330, Leu361, Lys370, Glu504 | Ser159, Tyr160, Phe164, Gly356, Asn358, Leu359, Gly362, Asn412, Tyr468, Tyr472, Trp549, Thr556, Ser557, Gly559, Leu561 | His162 (amide-π) Ile363 (alkyl) Val355 (alkyl) Arg562 (alkyl) Phe365 (π-alkyl) | Glu413 (π-anion) |
| 27 | −10.26 | Tyr160, Phe161, Asp163, Leu361, Gly362, Lys370, Glu504 | His162, Phe164, His296, His330, Val355, Asn358, Leu359, Ser360, Gly362, Asn412, Glu413, Tyr472, Thr556, Ser557, Gly559, Leu561, Asn566 | Ile363 (alkyl) Arg562 (alkyl) Lys568 (alkyl) Phe365 (π-alkyl) Tyr468 (π-alkyl) Trp549 (π-alkyl) | |
| 28 | −9.56 | Tyr160, Phe161, Asp163, Leu361, Gly362, Ile363, Glu413, Tyr472 | His162, Phe164, Gly356, Leu359, Lys370, Phe448, Asn466, Thr556, Ser557, Leu561, Arg562 | Val355 (alkyl) Val446 (alkyl) Met447 (alkyl) His330 (π-alkyl) Tyr468 (π-alkyl) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phong, N.V.; Zhao, Y.; Min, B.S.; Yang, S.Y.; Kim, J.A. Inhibitory Activity of Bioactive Phloroglucinols from the Rhizomes of Dryopteris crassirhizoma on Escherichia coli β-Glucuronidase: Kinetic Analysis and Molecular Docking Studies. Metabolites 2022, 12, 938. https://doi.org/10.3390/metabo12100938
Phong NV, Zhao Y, Min BS, Yang SY, Kim JA. Inhibitory Activity of Bioactive Phloroglucinols from the Rhizomes of Dryopteris crassirhizoma on Escherichia coli β-Glucuronidase: Kinetic Analysis and Molecular Docking Studies. Metabolites. 2022; 12(10):938. https://doi.org/10.3390/metabo12100938
Chicago/Turabian StylePhong, Nguyen Viet, Yan Zhao, Byung Sun Min, Seo Young Yang, and Jeong Ah Kim. 2022. "Inhibitory Activity of Bioactive Phloroglucinols from the Rhizomes of Dryopteris crassirhizoma on Escherichia coli β-Glucuronidase: Kinetic Analysis and Molecular Docking Studies" Metabolites 12, no. 10: 938. https://doi.org/10.3390/metabo12100938
APA StylePhong, N. V., Zhao, Y., Min, B. S., Yang, S. Y., & Kim, J. A. (2022). Inhibitory Activity of Bioactive Phloroglucinols from the Rhizomes of Dryopteris crassirhizoma on Escherichia coli β-Glucuronidase: Kinetic Analysis and Molecular Docking Studies. Metabolites, 12(10), 938. https://doi.org/10.3390/metabo12100938







