Metabolomic Signatures in Doxorubicin-Induced Metabolites Characterization, Metabolic Inhibition, and Signaling Pathway Mechanisms in Colon Cancer HCT116 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. DOX Solution Preparation
2.3. Cell Culture and DOX Treatments
2.4. WST-8 Cell Viability Assay
2.5. Isolation of HCT116 Cells and Quenching Metabolome
2.6. Instrumentation of NMR and Data Files
2.7. Computational Tools and Statistical Analysis
3. Results
3.1. Optimization of the Quenching and Extraction Procedures for DOX-Treated Cells
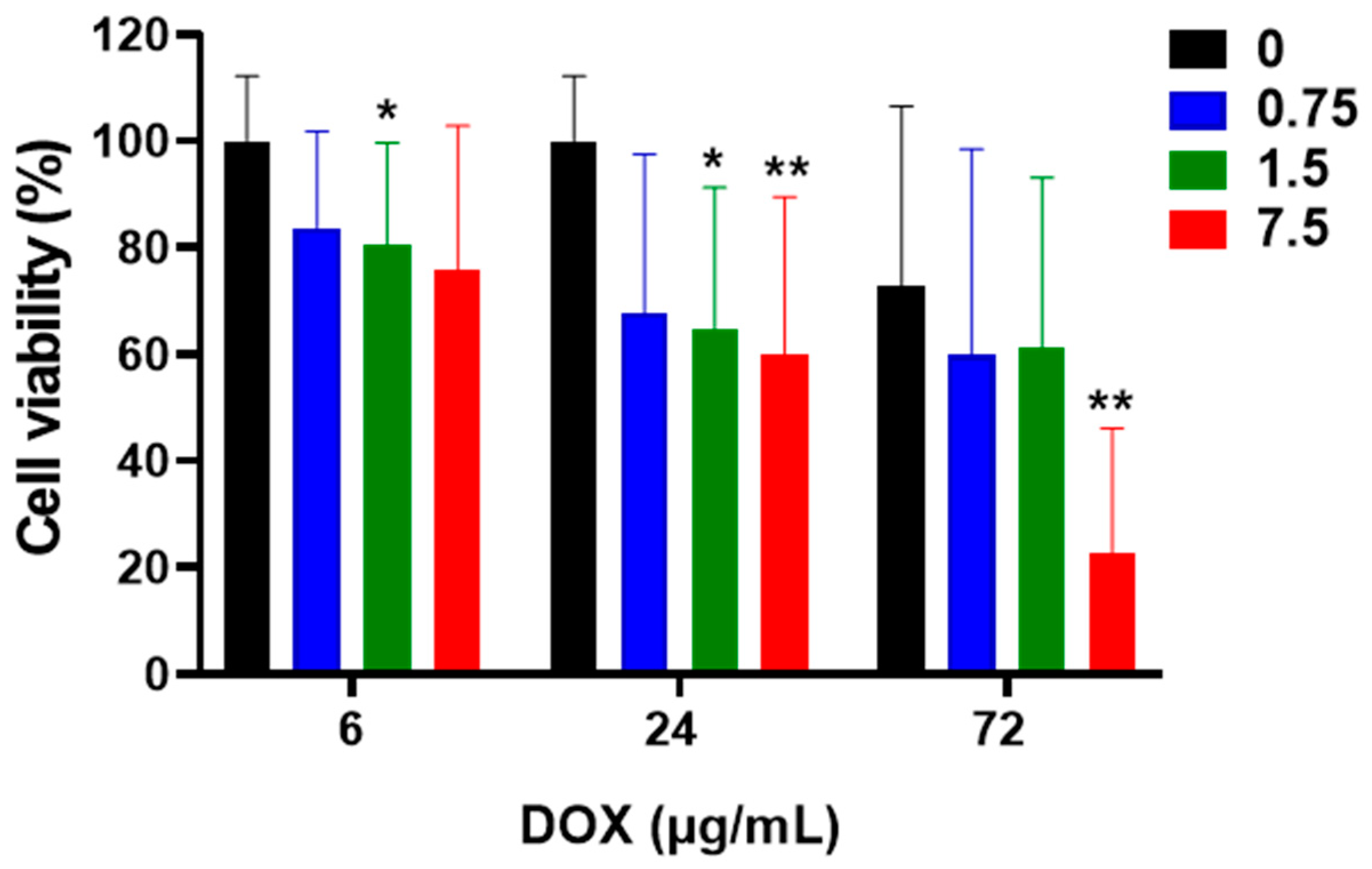
3.2. Cell Viability-Treatment with DOX in HCT116 Cells
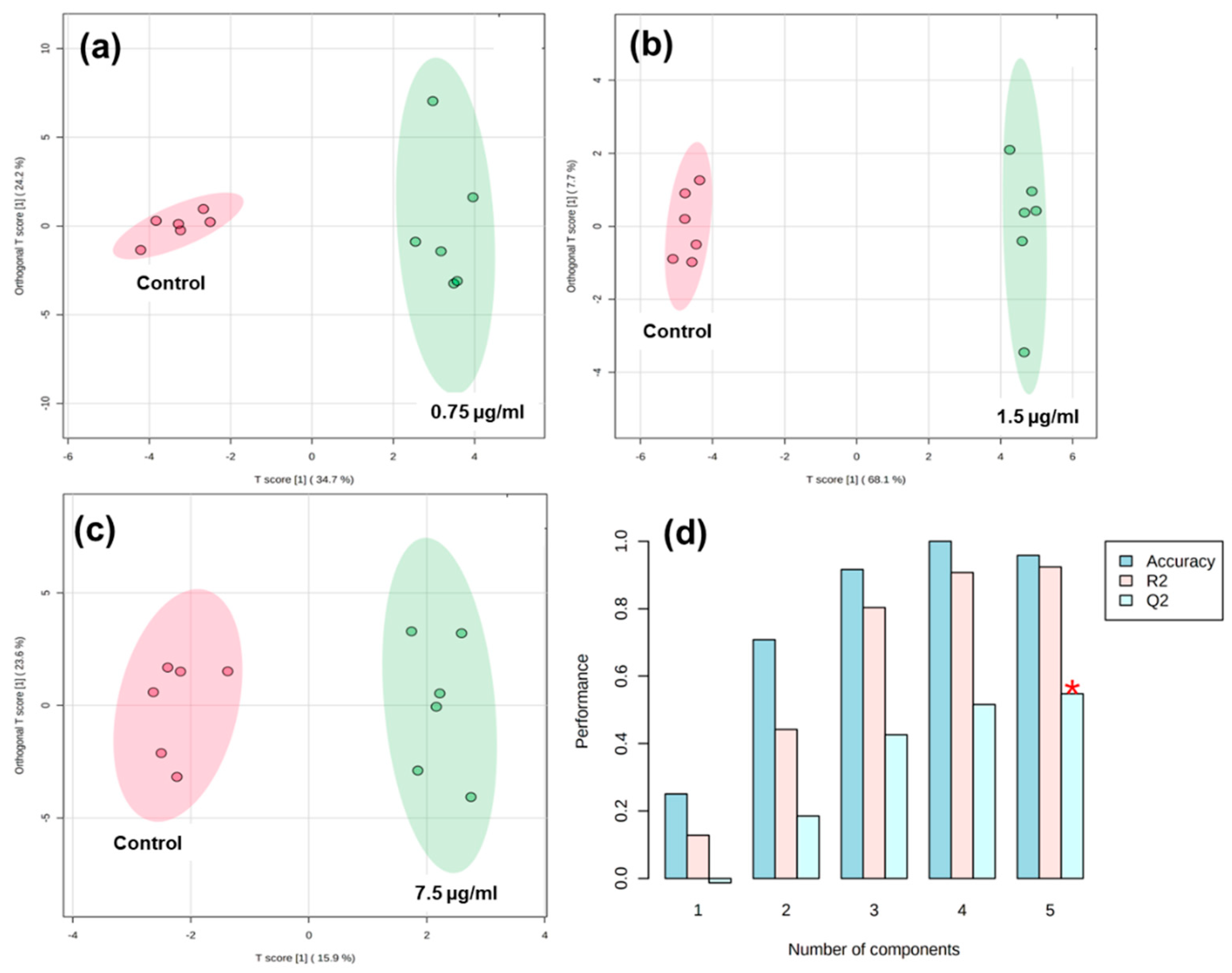
3.3. Integration of Targeted Metabolomics Datasets
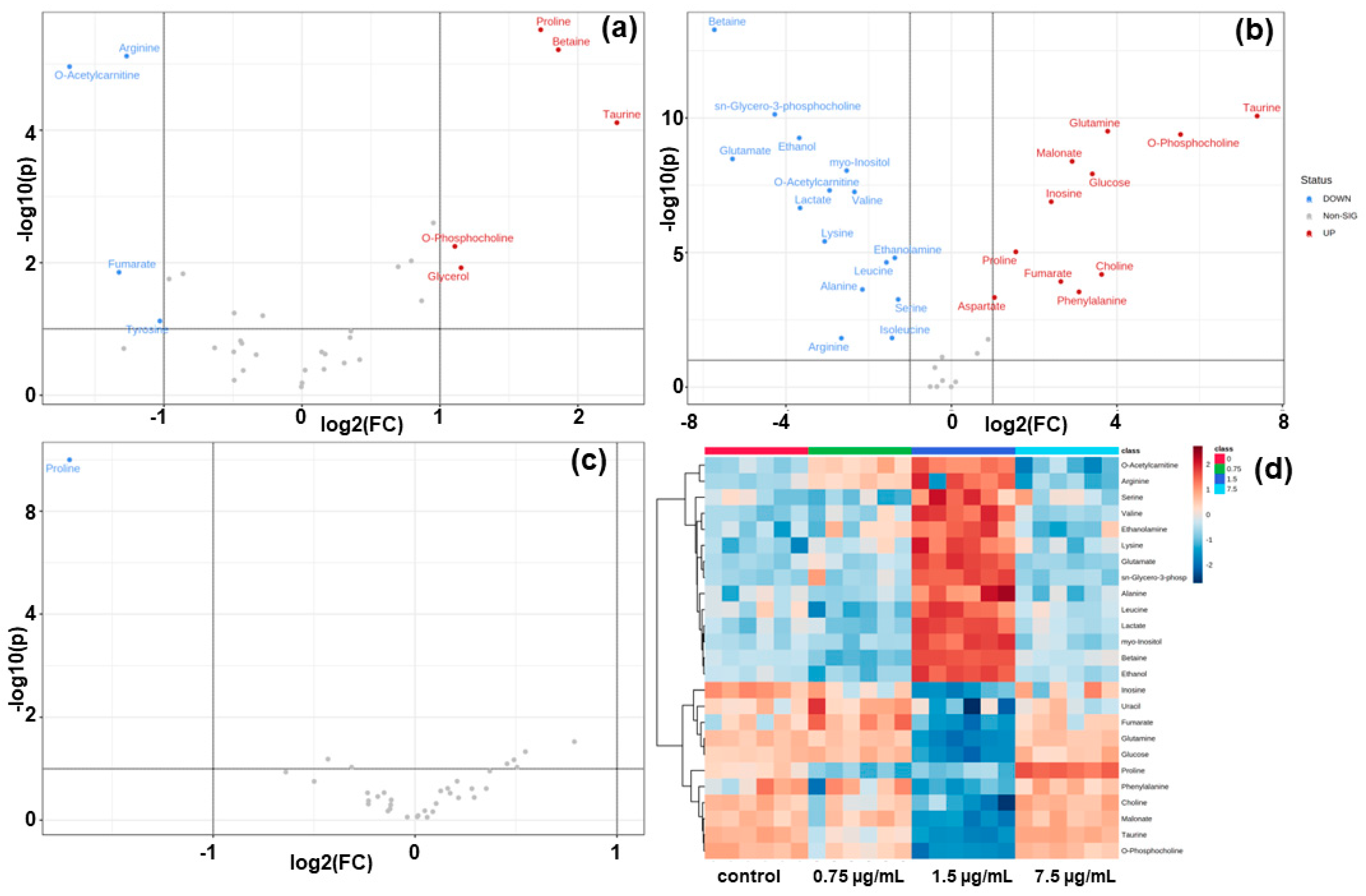
3.4. Volcano Plots and Heatmap Analysis of HCT116 Cells Metabolites
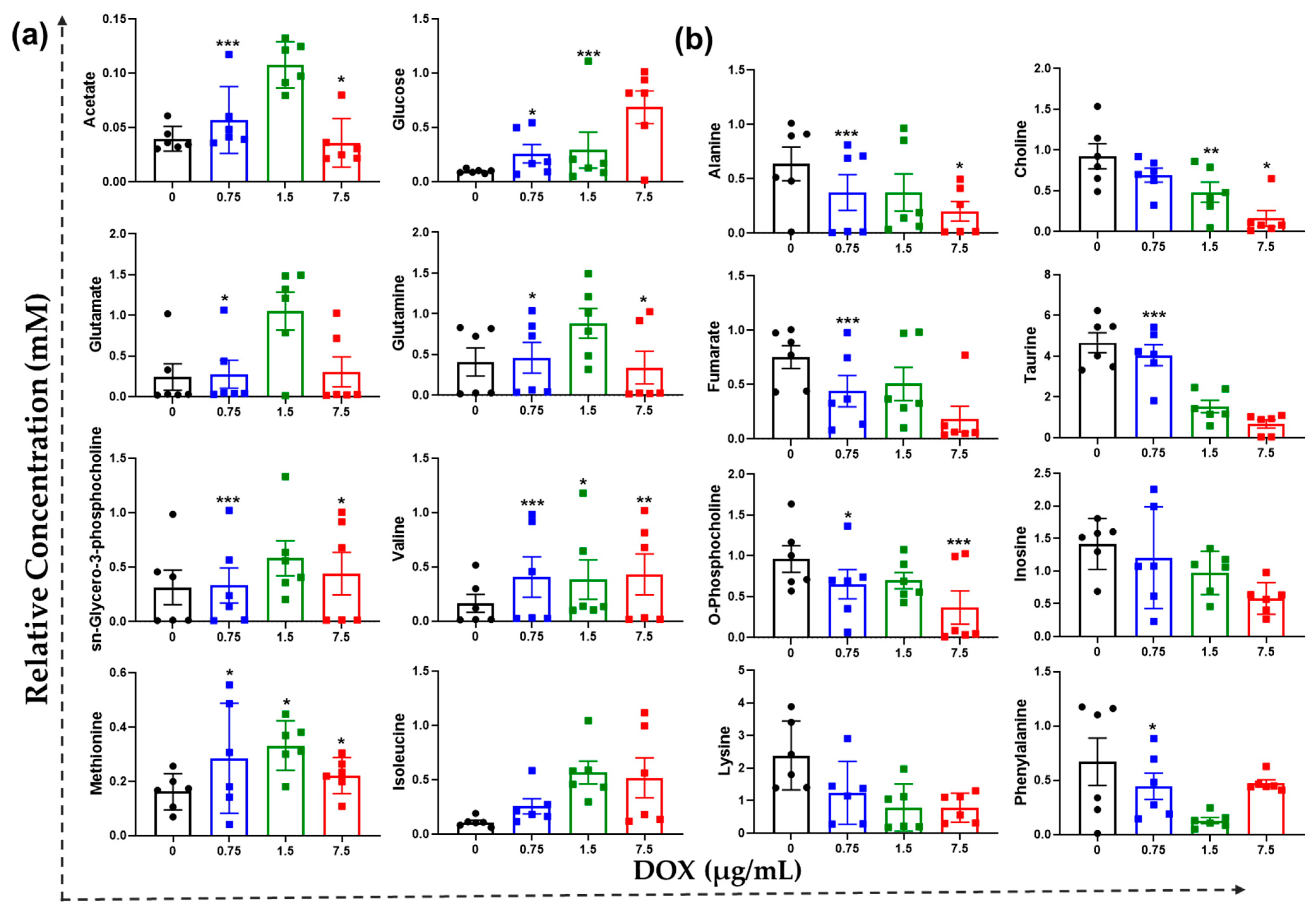
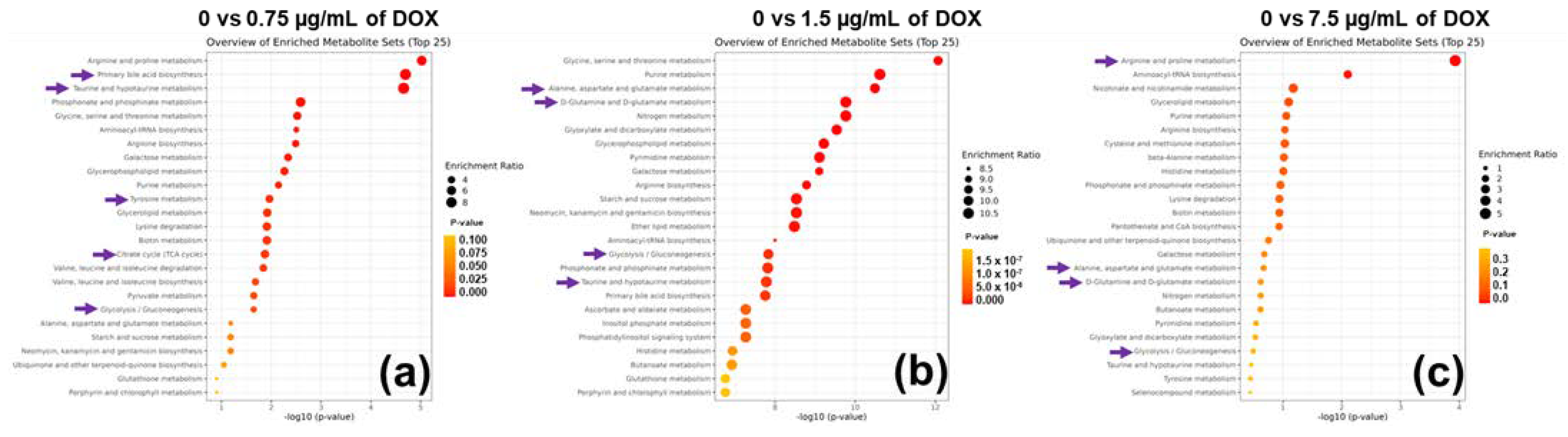
3.5. Tailored Metabolic Adaptations to DOX and Primary Metabolites with Pathways
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.; Gaspar, R. Nanomedicine(s) under the microscope. Mol. Pharm. 2011, 8, 2101–2141. [Google Scholar] [CrossRef] [PubMed]
- Raja, G.; Cao, S.; Kim, D.-H.; Kim, T.-J. Mechanoregulation of titanium dioxide nanoparticles in cancer therapy. Mater. Sci. Eng. C 2020, 107, 110303. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, Y.; Parikh, A.; Joyce, P.; Chung, R.; Liu, L.; Afinjuomo, F.; Hayball, J.D.; Petrovsky, N.; Barclay, T.G.; et al. Doxorubicin-loaded delta inulin conjugates for controlled and targeted drug delivery: Development, characterization, and in vitro evaluation. Pharmaceutics 2019, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Koshkaryev, A.; Sawant, R.; Deshpande, M.; Torchilin, V. Immunoconjugates and long circulating systems: Origins, current state of the art and future directions. Adv. Drug Deliv. Rev. 2013, 65, 24–35. [Google Scholar] [CrossRef]
- Raja, G.; Jang, Y.-K.; Suh, J.-S.; Kim, H.-S.; Ahn, S.H.; Kim, T.-J. Microcellular environmental regulation of silver nanoparticles in cancer therapy: A critical review. Cancers 2020, 12, 664. [Google Scholar] [CrossRef]
- Frezza, C.; Zheng, L.; Tennant, D.A.; Papkovsky, D.B.; Hedley, B.A.; Kalna, G.; Watson, D.G.; Gottlieb, E. Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS ONE 2011, 6, e24411. [Google Scholar] [CrossRef]
- Kotze, H.L.; Armitage, E.G.; Sharkey, K.J.; Allwood, J.W.; Dunn, W.B.; Williams, K.J.; Goodacre, R. A novel untargeted metabolomics correlation-based network analysis incorporating human metabolic reconstructions. BMC Syst. Biol. 2013, 7, 107. [Google Scholar] [CrossRef]
- Loftus, N.J.; Lai, L.; Wilkinson, R.W.; Odedra, R.; Wilson, I.D.; Barnes, A.J. Global metabolite profiling of human colorectal cancer xenografts in mice using hplc–ms/ms. J. Proteome Res. 2013, 12, 2980–2986. [Google Scholar] [CrossRef]
- Tolstikov, V.; Nikolayev, A.; Dong, S.; Zhao, G.; Kuo, M.-S. Metabolomics analysis of metabolic effects of nicotinamide phosphoribosyltransferase (nampt) inhibition on human cancer cells. PLoS ONE 2014, 9, e114019. [Google Scholar] [CrossRef]
- Jang, C.; Chen, L.; Rabinowitz, J.D. Metabolomics and isotope tracing. Cell 2018, 173, 822–837. [Google Scholar] [CrossRef] [PubMed]
- Aftab, O.; Engskog, M.K.; Haglöf, J.; Elmsjö, A.; Arvidsson, T.; Pettersson, C.; Hammerling, U.; Gustafsson, M.G. Nmr spectroscopy-based metabolic profiling of drug-induced changes in vitro can discriminate between pharmacological classes. J. Chem. Inf. Model. 2014, 54, 3251–3258. [Google Scholar] [CrossRef] [PubMed]
- Johanningsmeier, S.D.; Harris, G.K.; Klevorn, C.M. Metabolomic technologies for improving the quality of food: Practice and promise. Annu. Rev. Food Sci. Technol. 2016, 7, 413–438. [Google Scholar] [CrossRef] [PubMed]
- Raja, G.; Jung, Y.; Jung, S.H.; Kim, T.-J. 1h-nmr-based metabolomics for cancer targeting and metabolic engineering—A review. Process Biochem. 2020, 99, 112–122. [Google Scholar] [CrossRef]
- Raja, G.; Jang, Y.-K.; Suh, J.-S.; Prabhakaran, V.-S.; Kim, T.-J. Advanced understanding of genetic risk and metabolite signatures in construction workers via cytogenetics and metabolomics analysis. Process Biochem. 2019, 86, 117–126. [Google Scholar] [CrossRef]
- Armiñán, A.; Palomino-Schätzlein, M.; Deladriere, C.; Arroyo-Crespo, J.J.; Vicente-Ruiz, S.; Vicent, M.J.; Pineda-Lucena, A. Metabolomics facilitates the discrimination of the specific anticancer effects of free- and polymer-conjugated doxorubicin in breast cancer models. Biomaterials 2018, 162, 144–153. [Google Scholar] [CrossRef]
- Pramod, P.S.; Shah, R.; Jayakannan, M. Dual stimuli polysaccharide nanovesicles for conjugated and physically loaded doxorubicin delivery in breast cancer cells. Nanoscale 2015, 7, 6636–6652. [Google Scholar] [CrossRef]
- Rosli, N.F.; Fojtů, M.; Fisher, A.C.; Pumera, M. Graphene oxide nanoplatelets potentiate anticancer effect of cisplatin in human lung cancer cells. Langmuir 2019, 35, 3176–3182. [Google Scholar] [CrossRef]
- Raja, G.; Selvaraj, V.; Suk, M.; Suk, K.T.; Kim, T.-J. Metabolic phenotyping analysis of graphene oxide nanosheets exposures in breast cancer cells: Metabolomics profiling techniques. Process Biochem. 2021, 104, 39–45. [Google Scholar] [CrossRef]
- Lauri, I.; Savorani, F.; Iaccarino, N.; Zizza, P.; Pavone, L.M.; Novellino, E.; Engelsen, S.B.; Randazzo, A. Development of an optimized protocol for nmr metabolomics studies of human colon cancer cell lines and first insight from testing of the protocol using DNA g-quadruplex ligands as novel anticancer drugs. Metabolites 2016, 6, 4. [Google Scholar] [CrossRef]
- Niu, Q.Y.; Li, Z.Y.; Du, G.H.; Qin, X.M. (1)h nmr based metabolomic profiling revealed doxorubicin-induced systematic alterations in a rat model. J. Pharm. Biomed. Anal. 2016, 118, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Nicoletto, R.E.; Ofner, C.M. Cytotoxic mechanisms of doxorubicin at clinically relevant concentrations in breast cancer cells. Cancer Chemother. Pharmacol. 2022, 89, 285–311. [Google Scholar] [CrossRef] [PubMed]
- Martineau, E.; Tea, I.; Loaëc, G.; Giraudeau, P.; Akoka, S. Strategy for choosing extraction procedures for nmr-based metabolomic analysis of mammalian cells. Anal. Bioanal. Chem. 2011, 401, 2133–2142. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef]
- Feng, R.; Guo, X.; Kou, Y.; Xu, X.; Hong, C.; Zhang, W.; An, Y.; Philips, C.A.; Mancuso, A.; Qi, X. Association of lipid profile with decompensation, liver dysfunction, and mortality in patients with liver cirrhosis. Postgrad. Med. 2021, 133, 626–638. [Google Scholar] [CrossRef]
- Geng, C.; Cui, C.; Wang, C.; Lu, S.; Zhang, M.; Chen, D.; Jiang, P. Systematic evaluations of doxorubicin-induced toxicity in rats based on metabolomics. ACS Omega 2021, 6, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Altınkaynak, Y.; Kural, B.; Akcan, B.A.; Bodur, A.; Özer, S.; Yuluğ, E.; Munğan, S.; Kaya, C.; Örem, A. Protective effects of l-theanine against doxorubicin-induced nephrotoxicity in rats. Biomed. Pharmacother. 2018, 108, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. CMLS 2019, 76, 473–493. [Google Scholar] [CrossRef]
- Kumar, V.; Dwivedi, D.K.; Jagannathan, N.R. High-resolution nmr spectroscopy of human body fluids and tissues in relation to prostate cancer. NMR Biomed. 2014, 27, 80–89. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Lane, A.N. Applications of nmr spectroscopy to systems biochemistry. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 92–93, 18–53. [Google Scholar] [CrossRef]
- Hu, R.; Li, T.; Yang, Y.; Tian, Y.; Zhang, L. Nmr-based metabolomics in cancer research. In Cancer Metabolomics: Methods and Applications; Hu, S., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 201–218. [Google Scholar]
- Nagana Gowda, G.A.; Raftery, D. Recent advances in nmr-based metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef] [PubMed]
- Bingol, K.; Brüschweiler, R. Multidimensional approaches to nmr-based metabolomics. Anal. Chem. 2014, 86, 47–57. [Google Scholar] [CrossRef]
- Ganesan, R.; Vasantha-Srinivasan, P.; Sadhasivam, D.R.; Subramanian, R.; Vimalraj, S.; Suk, K.T. Carbon nanotubes induce metabolomic profile disturbances in zebrafish: Nmr-based metabolomics platform. Front. Mol. Biosci. 2021, 8, 688827. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Oda, A.; Konishi, H. Theanine prevents doxorubicin-induced acute hepatotoxicity by reducing intrinsic apoptotic response. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2015, 78, 147–152. [Google Scholar] [CrossRef]
- Carvalho, C.; Santos, R.X.; Cardoso, S.; Correia, S.; Oliveira, P.J.; Santos, M.S.; Moreira, P.I. Doxorubicin: The good, the bad and the ugly effect. Curr. Med. Chem. 2009, 16, 3267–3285. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Ye, J.; Qin, Z.; Ding, X. Protective effects of madecassoside against doxorubicin induced nephrotoxicity in vivo and in vitro. Sci. Rep. 2015, 5, 18314. [Google Scholar] [CrossRef] [PubMed]
- Hembruff, S.L.; Laberge, M.L.; Villeneuve, D.J.; Guo, B.; Veitch, Z.; Cecchetto, M.; Parissenti, A.M. Role of drug transporters and drug accumulation in the temporal acquisition of drug resistance. BMC Cancer 2008, 8, 318. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Tam, A.; Santi, S.A.; Parissenti, A.M. Role of autophagy and lysosomal drug sequestration in acquired resistance to doxorubicin in mcf-7 cells. BMC Cancer 2016, 16, 762. [Google Scholar] [CrossRef]
- Dubbelboer, I.R.; Pavlovic, N.; Heindryckx, F.; Sjögren, E.; Lennernäs, H. Liver cancer cell lines treated with doxorubicin under normoxia and hypoxia: Cell viability and oncologic protein profile. Cancers 2019, 11, 1024. [Google Scholar] [CrossRef]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2012, 65, 157–170. [Google Scholar] [CrossRef]
- Eom, Y.-W.; Kim, M.A.; Park, S.S.; Goo, M.J.; Kwon, H.J.; Sohn, S.; Kim, W.-H.; Yoon, G.; Choi, K.S. Two distinct modes of cell death induced by doxorubicin: Apoptosis and cell death through mitotic catastrophe accompanied by senescence-like phenotype. Oncogene 2005, 24, 4765–4777. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Surma, M.; Gough, G.; Shi, S.; Lambert-Cheatham, N.; Chang, J.; Shi, J. Dissecting the mechanisms of doxorubicin and oxidative stress-induced cytotoxicity: The involvement of actin cytoskeleton and rock1. PLoS ONE 2015, 10, e0131763. [Google Scholar]
- Piskounova, E.; Agathocleous, M.; Murphy, M.M.; Hu, Z.; Huddlestun, S.E.; Zhao, Z.; Leitch, A.M.; Johnson, T.M.; DeBerardinis, R.J.; Morrison, S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015, 527, 186–191. [Google Scholar] [CrossRef]
- Tasdogan, A.; Faubert, B.; Ramesh, V.; Ubellacker, J.M.; Shen, B.; Solmonson, A.; Murphy, M.M.; Gu, Z.; Gu, W.; Martin, M.; et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 2020, 577, 115–120. [Google Scholar] [CrossRef] [PubMed]


| δ 1H (ppm) and Multiplicity a | Targeted Metabolites | Chemical Formula | MW (Da) |
|---|---|---|---|
| 1.90 (s) | Acetate | C2H4O2 | 60.05 |
| 3.89 (dd); 2.80 (dd); 2.66 (dd) | Aspartate | C4H7NO4 | 133.10 |
| 3.78 (q); 1.47 (d) | Alanine | C3H7NO2 | 89.09 |
| 3.76 (t); 1.90 (m); 1.68 (m) | Asparagine | C6H14N4O2 | 132.12 |
| 8.52 (s); 8.12 (d); 4.50 (m); 4.21 (m) | ATP | C10H16N5O13P3 | 507.18 |
| 8.54 (s); 5.94 (m); 4.11 (m); 4.00 (m) | ADP | C10H15N5O10P2 | 427.201 |
| 8.22 (s); 6.16 (s); 4.53 (dd); 4.34 (d) | AMP | C10H14N5O7P | 347.221 |
| 3.89 (s); 3.25 (s) | Betaine | C24H26N2O13 | 550.45 |
| 2.65 (d); 2.53 (d) | Citrate | C6H8O7 | 192.12 |
| 4.05 (dd); 3.50 (dd); 3.18 (s) | Choline | C5H14NO | 104.17 |
| 6.51 (s) | Fumarate | C4H4O4 | 116.072 |
| 5.22 (d); 4.64 (d); 3.88 (dd); 3.72 (m); 3.40 (m) | Glucose | C6H12O6 | 180.16 |
| 4.20 (q); 3.78 (m); 2.97 (dd); 2.15 (m) | Glutathione | C10H17N3O6S | 307.32 |
| 3.76 (t); 2.44 (m); 2.12 (m) | Glutamate | C5H9NO4 | 147.129 |
| 5.75 (m); 7.80 (m); 6.16 (t) | Glutamine | C5H10N2O3 | 146.144 |
| 4.10 (q); 1.32 (d) | Lactate | C3H6O3 | 342.3 |
| 3.66 (d); 1.96 (m); 0.99 (d); 0.92 (t) | Isoleucine | C6H13NO2 | 131.17 |
| 3.60 (d); 2.261 (m); 0.97 (d) | Valine | C5H11NO2 | 117.146 |
| 3.72 (m); 1.70 (m); 0.94 (t) | Leucine | C6H13NO2 | 131.17 |
| 8.30 (s); 6.05 (d); 4.42 (dd); 3.82 (dd) | Inosine | C10H12N4O5 | 268.23 |
| 2.37 (s) | Oxalacetate | C4H4O5 | 132.071 |
| 2.61 (m); 2.51 (dd); 2.11 (dd); 1.07 (d) | Methylsuccinate | C5H8O4 | 132.11 |
| 4.05 (t); 3.61 (t); 3.52 (dd); 3.26 (t) | Myo-Inositol | C6H12O6 | 180.16 |
| 7.33 (d); 7.37 (m); 7.43 (m) | Phenylalanine | C9H11NO2 | 165.19 |
| 2.46 (s) | Pyruvate | C3H4O3 | 88.0621 |
| 2.39 (s) | Succinate | C4H6O4 | 118.088 |
| 3.43 (t); 3.42 (t); 3.25 (t) | Taurine | C2H7NO3S | 125.15 |
| 4.24 (m); 3.57 (d); 1.31 (d) | Threonine | C4H9NO3 | 119.12 |
| 7.72 (d); 7.31 (s); 4.04 (dd); 3.29 (dd) | Tryptophan | C11H12N2O2 | 204.26 |
| 2.89 (s) | Trimethylamine | C3H9N | 59.1103 |
| 7.24 (d); 6.94 (m); 3.34 (dd); 3.30 (dd) | Tyrosine | C9H11NO3 | 181.188 |
| 3.25 (s) | TMAO | C3H9NO | 75.11 |
| 7.892 (s) | Xanthine | C5H4N4O2 | 152.110 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganesan, R.; Prabhakaran, V.-S.; Valsala Gopalakrishnan, A. Metabolomic Signatures in Doxorubicin-Induced Metabolites Characterization, Metabolic Inhibition, and Signaling Pathway Mechanisms in Colon Cancer HCT116 Cells. Metabolites 2022, 12, 1047. https://doi.org/10.3390/metabo12111047
Ganesan R, Prabhakaran V-S, Valsala Gopalakrishnan A. Metabolomic Signatures in Doxorubicin-Induced Metabolites Characterization, Metabolic Inhibition, and Signaling Pathway Mechanisms in Colon Cancer HCT116 Cells. Metabolites. 2022; 12(11):1047. https://doi.org/10.3390/metabo12111047
Chicago/Turabian StyleGanesan, Raja, Vasantha-Srinivasan Prabhakaran, and Abilash Valsala Gopalakrishnan. 2022. "Metabolomic Signatures in Doxorubicin-Induced Metabolites Characterization, Metabolic Inhibition, and Signaling Pathway Mechanisms in Colon Cancer HCT116 Cells" Metabolites 12, no. 11: 1047. https://doi.org/10.3390/metabo12111047
APA StyleGanesan, R., Prabhakaran, V.-S., & Valsala Gopalakrishnan, A. (2022). Metabolomic Signatures in Doxorubicin-Induced Metabolites Characterization, Metabolic Inhibition, and Signaling Pathway Mechanisms in Colon Cancer HCT116 Cells. Metabolites, 12(11), 1047. https://doi.org/10.3390/metabo12111047









