Identification of Serum Metabolites for Predicting Chronic Kidney Disease Progression according to Chronic Kidney Disease Cause
Abstract
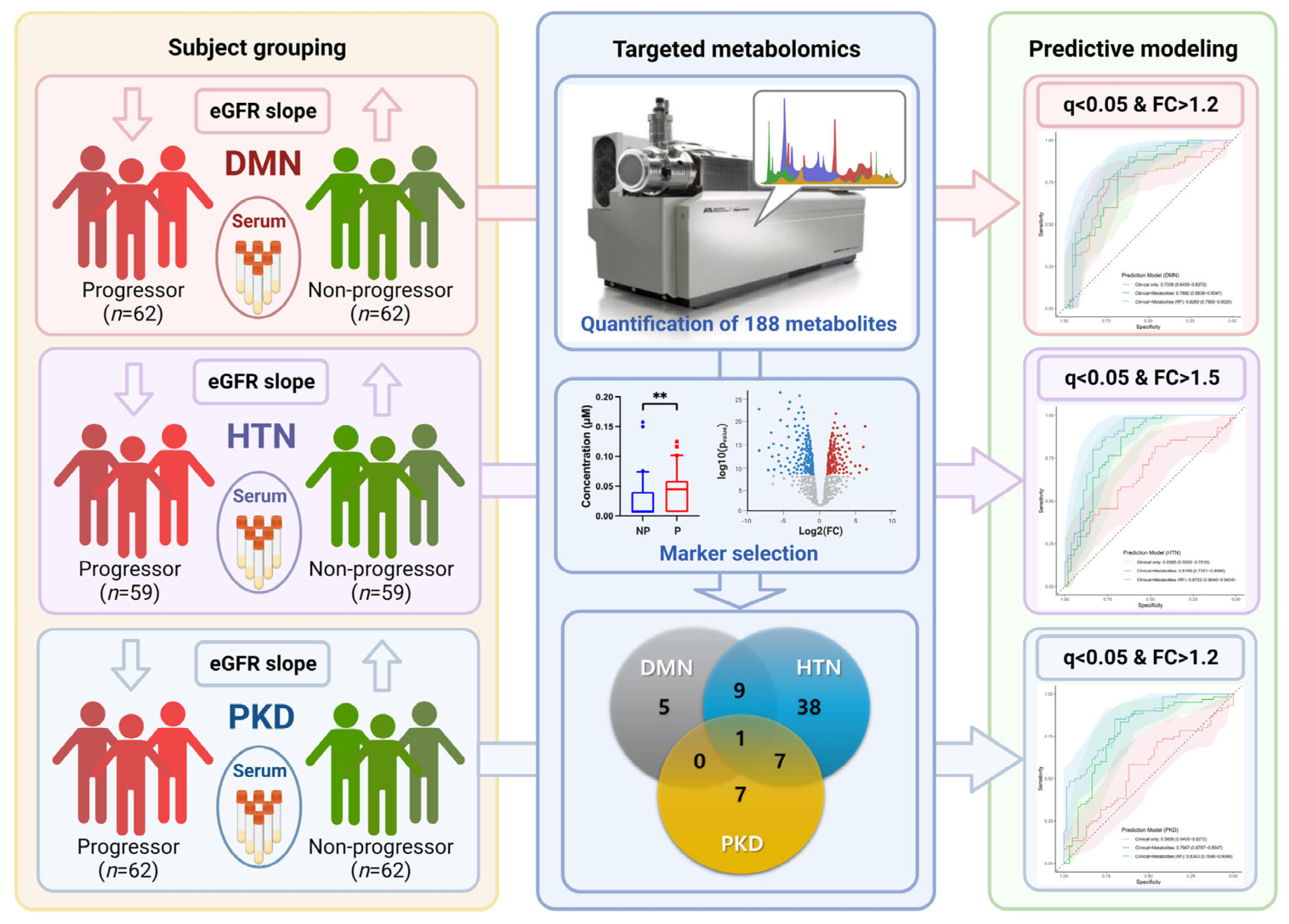
:1. Introduction
2. Materials and Methods
2.1. KNOW-CKD Cohort
2.2. Ethical Approval
2.3. Study Design
2.4. Targeted Metabolomics
2.5. Statistical Analysis
2.6. Prediction Modeling and Network Analysis
3. Results
3.1. Baseline Characteristics of the Study Participants
3.2. Potential Metabolic Biomarkers of Diabetic Nephropathy (DMN)
3.3. Potential Metabolic Biomarkers for Hypertensive Nephropathy (HTN)
3.4. Potential Metabolic Biomarkers for Polycystic Kidney Disease (PKD)
3.5. Metabolite Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stevens, P.E.; Levin, A. Evaluation and management of chronic kidney disease: Synopsis of the kidney disease: Improving global outcomes 2012 clinical practice guideline. Ann. Intern. Med. 2013, 158, 825–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, V.; Garcia-Garcia, G.; Iseki, K.; Li, Z.; Naicker, S.; Plattner, B.; Saran, R.; Wang, A.Y.; Yang, C.W. Chronic kidney disease: Global dimension and perspectives. Lancet 2013, 382, 260–272. [Google Scholar] [CrossRef]
- Hill, N.R.; Fatoba, S.T.; Oke, J.L.; Hirst, J.A.; O’Callaghan, C.A.; Lasserson, D.S.; Hobbs, F.D. Global Prevalence of Chronic Kidney Disease—A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0158765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.L.; Rothenbacher, D. Prevalence of chronic kidney disease in population-based studies: Systematic review. BMC Public Health 2008, 8, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.-W.; Harris, D.C.H.; Luyckx, V.A.; Nangaku, M.; Hou, F.F.; Garcia, G.G.; Abu-Aisha, H.; Niang, A.; Sola, L.; Bunnag, S.; et al. Global case studies for chronic kidney disease/end-stage kidney disease care. Kidney Int. Suppl. 2020, 10, e24–e48. [Google Scholar] [CrossRef]
- KDIGO CKD Work Group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Suppl. 2013, 3, 1–150. [Google Scholar]
- Rosansky, S.J. Renal Function Trajectory Is More Important than Chronic Kidney Disease Stage for Managing Patients with Chronic Kidney Disease. Am. J. Nephrol. 2012, 36, 1–10. [Google Scholar] [CrossRef]
- Nkuipou-Kenfack, E.; Duranton, F.; Gayrard, N.; Argilés, À.; Lundin, U.; Weinberger, K.M.; Dakna, M.; Delles, C.; Mullen, W.; Husi, H.; et al. Assessment of metabolomic and proteomic biomarkers in detection and prognosis of progression of renal function in chronic kidney disease. PLoS ONE 2014, 9, e96955. [Google Scholar] [CrossRef] [Green Version]
- Van de Poll, M.C.; Soeters, P.B.; Deutz, N.E.; Fearon, K.C.; Dejong, C.H. Renal metabolism of amino acids: Its role in interorgan amino acid exchange. Am. J. Clin. Nutr. 2004, 79, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Lecamwasam, A.R.; Mohebbi, M.; Ekinci, E.I.; Dwyer, K.M.; Saffery, R. Identification of Potential Biomarkers of Chronic Kidney Disease in Individuals with Diabetes: Protocol for a Cross-sectional Observational Study. JMIR Res. Protoc. 2020, 9, e16277. [Google Scholar] [CrossRef]
- Davies, R. The metabolomic quest for a biomarker in chronic kidney disease. Clin. Kidney J. 2018, 11, 694–703. [Google Scholar] [CrossRef]
- Zhong, J.; Yang, H.-C.; Fogo, A.B. A perspective on chronic kidney disease progression. Am. J. Physiol.-Ren. Physiol. 2017, 312, F375–F384. [Google Scholar] [CrossRef] [Green Version]
- Wilson, P.D. Polycystic kidney disease. N. Engl. J. Med. 2004, 350, 151–164. [Google Scholar] [CrossRef]
- Oh, K.H.; Park, S.K.; Park, H.C.; Chin, H.J.; Chae, D.W.; Choi, K.H.; Han, S.H.; Yoo, T.H.; Lee, K.; Kim, Y.S.; et al. KNOW-CKD (KoreaN cohort study for Outcome in patients With Chronic Kidney Disease): Design and methods. BMC Nephrol. 2014, 15, 80. [Google Scholar] [CrossRef] [Green Version]
- Kang, E.; Han, M.; Kim, H.; Park, S.K.; Lee, J.; Hyun, Y.Y.; Kim, Y.S.; Chung, W.; Kim, H.J.; Oh, Y.K.; et al. Baseline General Characteristics of the Korean Chronic Kidney Disease: Report from the KoreaN Cohort Study for Outcomes in Patients With Chronic Kidney Disease (KNOW-CKD). J. Korean Med. Sci. 2017, 32, 221–230. [Google Scholar] [CrossRef]
- Pei, Y.; Obaji, J.; Dupuis, A.; Paterson, A.D.; Magistroni, R.; Dicks, E.; Parfrey, P.; Cramer, B.; Coto, E.; Torra, R.; et al. Unified criteria for ultrasonographic diagnosis of ADPKD. J. Am. Soc. Nephrol. 2009, 20, 205–212. [Google Scholar] [CrossRef] [Green Version]
- Levey, A.S.; Stevens, L.A.; Schmid, C.H.; Zhang, Y.L.; Castro, A.F., 3rd; Feldman, H.I.; Kusek, J.W.; Eggers, P.; van Lente, F.; Greene, T.; et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009, 150, 604–612. [Google Scholar] [CrossRef]
- Inker, L.A.; Heerspink, H.J.L.; Tighiouart, H.; Levey, A.S.; Coresh, J.; Gansevoort, R.T.; Simon, A.L.; Ying, J.; Beck, G.J.; Wanner, C.; et al. GFR Slope as a Surrogate End Point for Kidney Disease Progression in Clinical Trials: A Meta-Analysis of Treatment Effects of Randomized Controlled Trials. J. Am. Soc. Nephrol. 2019, 30, 1735–1745. [Google Scholar] [CrossRef]
- Shou, H.; Hsu, J.Y.; Xie, D.; Yang, W.; Roy, J.; Anderson, A.H.; Landis, J.R.; Feldman, H.I.; Parsa, A.; Jepson, C. Analytic Considerations for Repeated Measures of eGFR in Cohort Studies of CKD. Clin. J. Am. Soc. Nephrol. 2017, 12, 1357–1365. [Google Scholar] [CrossRef] [Green Version]
- KDIGO CKD-MBD Work Group. KDIGO clinical practice guideline for the diagnosis, evaluation, prevention, and treatment of Chronic Kidney Disease-Mineral and Bone Disorder (CKD-MBD). Kidney Int. Suppl. 2009, 113, S1–S130. [Google Scholar]
- Abeysekera, R.A.; Healy, H.G.; Wang, Z.; Cameron, A.L.; Hoy, W.E. Heterogeneity in patterns of progression of chronic kidney disease. Intern. Med. J. 2021, 51, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Fassett, R.G.; Venuthurupalli, S.K.; Gobe, G.C.; Coombes, J.S.; Cooper, M.A.; Hoy, W.E. Biomarkers in chronic kidney disease: A review. Kidney Int. 2011, 80, 806–821. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.A.; Wang, Y.; Zhu, V.; Rupnow, M.F. Chronic kidney disease in US adults with type 2 diabetes: An updated national estimate of prevalence based on Kidney Disease: Improving Global Outcomes (KDIGO) staging. BMC Res. Notes 2014, 7, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saran, R.; Robinson, B.; Abbott, K.C.; Agodoa, L.Y.C.; Bragg-Gresham, J.; Balkrishnan, R.; Bhave, N.; Dietrich, X.; Ding, Z.; Eggers, P.W.; et al. US Renal Data System 2018 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am. J. Kidney Dis. 2019, 73 (Suppl. S1), A7–A8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- 2021 Korean ESRD Registry. Available online: https://ksn.or.kr/bbs/index.php?code=report (accessed on 18 May 2022).
- Klag, M.J.; Whelton, P.K.; Randall, B.L.; Neaton, J.D.; Brancati, F.L.; Ford, C.E.; Shulman, N.B.; Stamler, J. Blood pressure and end-stage renal disease in men. N. Engl. J. Med. 1996, 334, 13–18. [Google Scholar] [CrossRef]
- Haroun, M.K.; Jaar, B.G.; Hoffman, S.C.; Comstock, G.W.; Klag, M.J.; Coresh, J. Risk factors for chronic kidney disease: A prospective study of 23,534 men and women in Washington County, Maryland. J. Am. Soc. Nephrol. 2003, 14, 2934–2941. [Google Scholar] [CrossRef] [Green Version]
- Nelson, R.G.; Grams, M.E.; Ballew, S.H.; Sang, Y.; Azizi, F.; Chadban, S.J.; Chaker, L.; Dunning, S.C.; Fox, C.; Hirakawa, Y.; et al. Development of Risk Prediction Equations for Incident Chronic Kidney Disease. JAMA 2019, 322, 2104–2114. [Google Scholar] [CrossRef]
- Dunkler, D.; Gao, P.; Lee, S.F.; Heinze, G.; Clase, C.M.; Tobe, S.; Teo, K.K.; Gerstein, H.; Mann, J.F.; Oberbauer, R. Risk Prediction for Early CKD in Type 2 Diabetes. Clin. J. Am. Soc. Nephrol. 2015, 10, 1371–1379. [Google Scholar] [CrossRef] [Green Version]
- Jardine, M.J.; Hata, J.; Woodward, M.; Perkovic, V.; Ninomiya, T.; Arima, H.; Zoungas, S.; Cass, A.; Patel, A.; Marre, M.; et al. Prediction of kidney-related outcomes in patients with type 2 diabetes. Am. J. Kidney Dis. 2012, 60, 770–778. [Google Scholar] [CrossRef]
- Lin, C.C.; Niu, M.J.; Li, C.I.; Liu, C.S.; Lin, C.H.; Yang, S.Y.; Li, T.C. Development and validation of a risk prediction model for chronic kidney disease among individuals with type 2 diabetes. Sci. Rep. 2022, 12, 4794. [Google Scholar] [CrossRef]
- An, J.H.; Cho, Y.M.; Yu, H.G.; Jang, H.C.; Park, K.S.; Kim, S.Y.; Lee, H.K. The clinical characteristics of normoalbuminuric renal insufficiency in Korean type 2 diabetic patients: A possible early stage renal complication. J. Korean Med. Sci. 2009, 24 (Suppl. S1), S75–S81. [Google Scholar] [CrossRef] [Green Version]
- Robles-Osorio, M.L.; Sabath, E. Tubular dysfunction and non-albuminuric renal disease in subjects with type 2 diabetes mellitus. Rev. Investig. Clin. 2014, 66, 234–239. [Google Scholar]
- Fiseha, T. Urinary biomarkers for early diabetic nephropathy in type 2 diabetic patients. Biomark. Res. 2015, 3, 16. [Google Scholar] [CrossRef]
- Weisstuch, J.M.; Dworkin, L.D. Does essential hypertension cause end-stage renal disease? Kidney Int. Suppl. 1992, 36, S33–S37. [Google Scholar]
- Luft, F.C. Hypertensive nephrosclerosis-a cause of end-stage renal disease? Nephrol. Dial. Transplant. 2000, 15, 1515–1517. [Google Scholar] [CrossRef] [Green Version]
- Øvrehus, M.A.; Oldereid, T.S.; Dadfar, A.; Bjørneklett, R.; Aasarød, K.I.; Fogo, A.B.; Ix, J.H.; Hallan, S.I. Clinical Phenotypes and Long-term Prognosis in White Patients with Biopsy-Verified Hypertensive Nephrosclerosis. Kidney Int. Rep. 2020, 5, 339–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarif, L.; Covic, A.; Iyengar, S.; Sehgal, A.R.; Sedor, J.R.; Schelling, J.R. Inaccuracy of clinical phenotyping parameters for hypertensive nephrosclerosis. Nephrol. Dial. Transplant. 2000, 15, 1801–1807. [Google Scholar] [CrossRef] [Green Version]
- Fogo, A.; Breyer, J.A.; Smith, M.C.; Cleveland, W.H.; Agodoa, L.; Kirk, K.A.; Glassock, R. Accuracy of the diagnosis of hypertensive nephrosclerosis in African Americans: A report from the African American Study of Kidney Disease (AASK) Trial. Kidney Int. 1997, 51, 244–252. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.J.; Ghosh, S.; Kovalik, J.P.; Ching, J.; Choi, H.W.; Tavintharan, S.; Ong, C.N.; Sum, C.F.; Summers, S.A.; Tai, E.S.; et al. Profiling of Plasma Metabolites Suggests Altered Mitochondrial Fuel Usage and Remodeling of Sphingolipid Metabolism in Individuals With Type 2 Diabetes and Kidney Disease. Kidney Int. Rep. 2017, 2, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Tofte, N.; Suvitaival, T.; Ahonen, L.; Winther, S.A.; Theilade, S.; Frimodt-Møller, M.; Ahluwalia, T.S.; Rossing, P. Lipidomic analysis reveals sphingomyelin and phosphatidylcholine species associated with renal impairment and all-cause mortality in type 1 diabetes. Sci. Rep. 2019, 9, 16398. [Google Scholar] [CrossRef] [Green Version]
- Mäkinen, V.P.; Tynkkynen, T.; Soininen, P.; Forsblom, C.; Peltola, T.; Kangas, A.J.; Groop, P.H.; Ala-Korpela, M. Sphingomyelin is associated with kidney disease in type 1 diabetes (The FinnDiane Study). Metabolomics 2012, 8, 369–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Villavicencio, R.; Flores-Estrada, J.; Franco, M.; Escalante, B.; Pérez-Méndez, O.; Mercado, A.; Bautista-Pérez, R. Effect of Empagliflozin on Sphingolipid Catabolism in Diabetic and Hypertensive Rats. Int. J. Mol. Sci. 2022, 23, 2883. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Amino acids: Metabolism, functions, and nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Duranton, F.; Lundin, U.; Gayrard, N.; Mischak, H.; Aparicio, M.; Mourad, G.; Daurès, J.P.; Weinberger, K.M.; Argilés, A. Plasma and urinary amino acid metabolomic profiling in patients with different levels of kidney function. Clin. J. Am. Soc. Nephrol. 2014, 9, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Garibotto, G.; Sofia, A.; Saffioti, S.; Bonanni, A.; Mannucci, I.; Verzola, D. Amino acid and protein metabolism in the human kidney and in patients with chronic kidney disease. Clin. Nutr. 2010, 29, 424–433. [Google Scholar] [CrossRef]
- Cornec-Le Gall, E.; Audrézet, M.P.; Chen, J.M.; Hourmant, M.; Morin, M.P.; Perrichot, R.; Charasse, C.; Whebe, B.; Renaudineau, E.; Jousset, P.; et al. Type of PKD1 mutation influences renal outcome in ADPKD. J. Am. Soc. Nephrol. 2013, 24, 1006–1013. [Google Scholar] [CrossRef] [Green Version]
- Hateboer, N.; v Dijk, M.A.; Bogdanova, N.; Coto, E.; Saggar-Malik, A.K.; San Millan, J.L.; Torra, R.; Breuning, M.; Ravine, D.; European PKD1-PKD2 Study Group. Comparison of phenotypes of polycystic kidney disease types 1 and 2. Lancet 1999, 353, 103–107. [Google Scholar] [CrossRef]
- Dicks, E.; Ravani, P.; Langman, D.; Davidson, W.S.; Pei, Y.; Parfrey, P.S. Incident renal events and risk factors in autosomal dominant polycystic kidney disease: A population and family-based cohort followed for 22 years. Clin. J. Am. Soc. Nephrol. 2006, 1, 710–717. [Google Scholar] [CrossRef] [Green Version]
- Chapman, A.B.; Bost, J.E.; Torres, V.E.; Guay-Woodford, L.; Bae, K.T.; Landsittel, D.; Li, J.; King, B.F.; Martin, D.; Wetzel, L.H.; et al. Kidney volume and functional outcomes in autosomal dominant polycystic kidney disease. Clin. J. Am. Soc. Nephrol. 2012, 7, 479–486. [Google Scholar] [CrossRef] [Green Version]
- Sriwi, D.; Alabdaljabar, M.S.; Jacob, M.; Mujamammi, A.H.; Gu, X.; Sabi, E.M.; Li, L.; Hussein, M.H.; Dasouki, M.; Abdel Rahman, A.M. Metabolomics Profiling of Cystic Renal Disease towards Biomarker Discovery. Biology 2021, 10, 770. [Google Scholar] [CrossRef]
- Hwang, V.J.; Kim, J.; Rand, A.; Yang, C.; Sturdivant, S.; Hammock, B.; Bell, P.D.; Guay-Woodford, L.M.; Weiss, R.H. The cpk model of recessive PKD shows glutamine dependence associated with the production of the oncometabolite 2-hydroxyglutarate. Am. J. Physiol. Ren. Physiol. 2015, 309, F492–F498. [Google Scholar] [CrossRef]




| Group | Characteristic | Non-Progressor | Progressor | p |
|---|---|---|---|---|
| DMN | Subject | 62 (50%) | 62 (50%) | 0.999 |
| Male sex | 43 (69.4%) | 43 (69.4%) | 0.999 | |
| Age (years) | 61.0 [51.0; 65.0] | 60.0 [53.0; 64.0] | 0.622 | |
| Baseline eGFR (mL/min/1.73 m2) | 41.8 [35.2; 49.6] | 42.2 [34.7; 49.9] | 0.871 | |
| eGFR slope (mL/min/1.73 m2/year) | 0.2 ± 1.3 | −3.2 ± 1.3 | <0.001 | |
| Systolic BP (mmHg) | 129.3 ± 16.3 | 133.0 ± 16.8 | 0.207 | |
| Diastolic BP (mmHg) | 75.1 ± 9.5 | 75.1 ± 9.8 | 0.985 | |
| BMI (kg/m2) | 24.8 [23.0; 27.1] | 24.8 [23.1; 26.6] | 0.928 | |
| uPCR (g/g) | 0.38 [0.23; 0.76] | 2.17 [1.04; 4.28] | <0.001 | |
| HTN | Subject | 59 (50%) | 59 (50%) | 0.999 |
| Male | 46 (77.97%) | 37 (62.7%) | 0.107 | |
| Age (years) | 62.00 [55.0; 67.5] | 60.0 [56.0; 68.0] | 0.948 | |
| Baseline eGFR (mL/min/1.73 m2) | 33.30 [24.4; 45.0] | 30.4 [25.9; 41.2] | 0.823 | |
| eGFR slope (mL/min/1.73 m2/year) | 0.6 [0.1; 1.3] | −1.5 [−2.7; −1.2] | <0.001 | |
| Systolic BP (mmHg) | 122.7 ± 13.8 | 124.1 ± 14.8 | 0.603 | |
| Diastolic BP (mmHg) | 75.1 ± 10.9 | 74.9 ± 9.8 | 0.951 | |
| BMI (kg/m2) | 25.2 ± 3.4 | 24.9 ± 3.2 | 0.684 | |
| uPCR (g/g) | 0.1 [0.05; 0.4] | 0.6 [0.2; 1.1] | <0.001 | |
| PKD | Subject | 62 (50%) | 62 (50%) | 0.999 |
| Male | 40 (64.52%) | 40 (64.5%) | 0.999 | |
| Age (years) | 46.5 ± 11.1 | 45.7 ± 8.6 | 0.679 | |
| Baseline eGFR (mL/min/1.73 m2) | 81.3 [63.5; 106.6] | 71.3 [60.9; 100.0] | 0.202 | |
| eGFR slope (mL/min/1.73 m2/year) | 0.4 [−0.3; 1.2] | −2.8 [−4.2; −1.7] | <0.001 | |
| Systolic BP (mmHg) | 127.6 ± 12.6 | 129.9 ± 11.8 | 0.302 | |
| Diastolic BP (mmHg) | 80.7 ± 9.3 | 81.5 ± 9.9 | 0.615 | |
| BMI (kg/m2) | 23.2 [21.7; 25.2] | 23.1 [21.6; 25.8] | 0.656 | |
| uPCR (g/g) | 0.06 [0.04; 0.15] | 0.1 [ 0.0; 0.3] | 0.020 |
| Group | Metabolite | Category | q | FC |
|---|---|---|---|---|
| DMN | SM C26:1 | Sphingomyelin | 0.002 | 0.567 |
| L-2-Aminoadipic acid (alpha-AAA) | Biogenic amine | 0.003 | 0.666 | |
| PC ae C36:5 | Phosphatidylcholine | 0.004 | 0.805 | |
| PC aa C40:4 | Phosphatidylcholine | 0.008 | 0.785 | |
| PC aa C34:1 | Phosphatidylcholine | 0.019 | 0.790 | |
| Asymmetric dimethylarginine (ADMA) | Biogenic amine | 0.019 | 0.808 | |
| PC ae C34:1 | Phosphatidylcholine | 0.028 | 0.830 | |
| SM (OH) C24:1 | Sphingomyelin | 0.028 | 1.323 | |
| PC ae C34:3 | Phosphatidylcholine | 0.029 | 0.816 | |
| PC ae C32:2 | Phosphatidylcholine | 0.030 | 0.821 | |
| HTN | Dodecenoylcarnitine (C12:1) | Acylcarnitine | <0.001 | 2.257 |
| PC aa C34:4 | Phosphatidylcholine | <0.001 | 0.580 | |
| PC ae C34:0 | Phosphatidylcholine | <0.001 | 0.631 | |
| PC ae C44:6 | Phosphatidylcholine | <0.001 | 0.653 | |
| PC aa C32:3 | Phosphatidylcholine | <0.001 | 0.565 | |
| PC ae C30:1 | Phosphatidylcholine | <0.001 | 1.726 | |
| SM C22:3 | Sphingomyelin | 0.007 | 2.085 | |
| Pimelylcarnitine (C7-DC) | Acylcarnitine | 0.010 | 0.658 | |
| SM C26:0 | Sphingomyelin | 0.034 | 1.834 | |
| PKD | PC aa C42:5 | Phosphatidylcholine | <0.001 | 1.846 |
| PC aa C36:6 | Phosphatidylcholine | <0.001 | 1.449 | |
| PC ae C30:1 | Phosphatidylcholine | <0.001 | 1.407 | |
| Pimelylcarnitine (C7-DC) | Acylcarnitine | 0.007 | 1.666 | |
| PC aa C32:3 | Phosphatidylcholine | 0.010 | 1.227 | |
| PC aa C36:0 | Phosphatidylcholine | 0.013 | 1.225 | |
| Creatinine | Biogenic amine | 0.015 | 1.200 | |
| PC aa C34:4 | Phosphatidylcholine | 0.022 | 1.201 | |
| Hexadecenoylcarnitine (C16:1) | Acylcarnitine | 0.025 | 1.316 |
| Group | Model | Mean AUC | Accuracy | Sensitivity | Specificity | F1 Score |
|---|---|---|---|---|---|---|
| DMN | Model 1 | 0.734 | 0.71 | 0.80 | 0.62 | 0.73 |
| Model 2 | 0.770 | 0.72 | 0.68 | 0.75 | 0.71 | |
| Model 3 | 0.826 | 0.74 | 0.67 | 0.82 | 0.71 | |
| HTN | Model 1 | 0.659 | 0.62 | 0.65 | 0.58 | 0.63 |
| Model 2 | 0.817 | 0.75 | 0.67 | 0.82 | 0.72 | |
| Model 3 | 0.872 | 0.79 | 0.71 | 0.87 | 0.77 | |
| PKD | Model 1 | 0.561 | 0.58 | 0.58 | 0.58 | 0.58 |
| Model 2 | 0.767 | 0.72 | 0.72 | 0.73 | 0.72 | |
| Model 3 | 0.834 | 0.72 | 0.70 | 0.73 | 0.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, E.; Li, Y.; Kim, B.; Huh, K.Y.; Han, M.; Ahn, J.-H.; Sung, H.Y.; Park, Y.S.; Lee, S.E.; Lee, S.; et al. Identification of Serum Metabolites for Predicting Chronic Kidney Disease Progression according to Chronic Kidney Disease Cause. Metabolites 2022, 12, 1125. https://doi.org/10.3390/metabo12111125
Kang E, Li Y, Kim B, Huh KY, Han M, Ahn J-H, Sung HY, Park YS, Lee SE, Lee S, et al. Identification of Serum Metabolites for Predicting Chronic Kidney Disease Progression according to Chronic Kidney Disease Cause. Metabolites. 2022; 12(11):1125. https://doi.org/10.3390/metabo12111125
Chicago/Turabian StyleKang, Eunjeong, Yufei Li, Bora Kim, Ki Young Huh, Miyeun Han, Jung-Hyuck Ahn, Hye Youn Sung, Yong Seek Park, Seung Eun Lee, Sangjun Lee, and et al. 2022. "Identification of Serum Metabolites for Predicting Chronic Kidney Disease Progression according to Chronic Kidney Disease Cause" Metabolites 12, no. 11: 1125. https://doi.org/10.3390/metabo12111125






