HILIC-MS for Untargeted Profiling of the Free Glycation Product Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
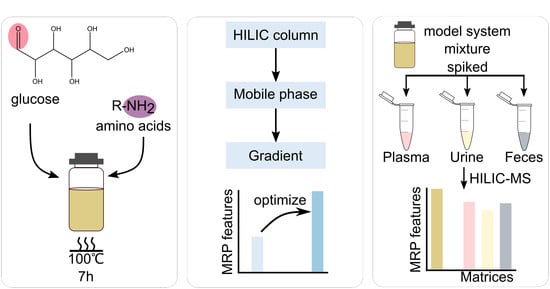
2.2. Maillard Model Systems Preparation
2.3. Biological Sample Preparation
2.4. LC-MS/MS
2.5. Chromatographic Conditions Optimization
2.6. Data Processing
3. Results
3.1. Data Cleaning for Reliable Feature Lists
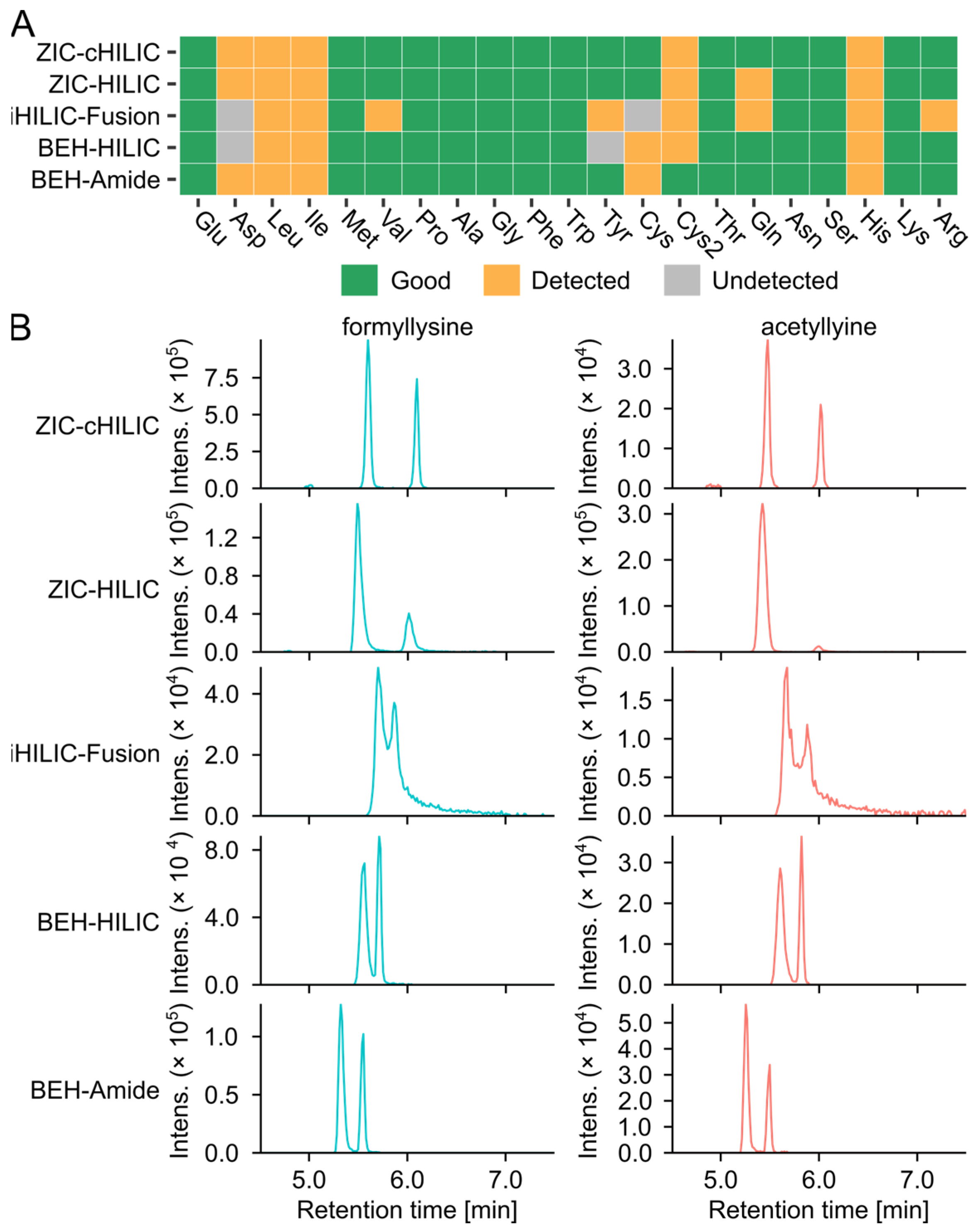
3.2. Selection of HILIC Columns
3.2.1. Non-Targeted Evaluation of the Column Selection
3.2.2. Selectivity of Columns for Analyzing Amino Acids and Glycation Products
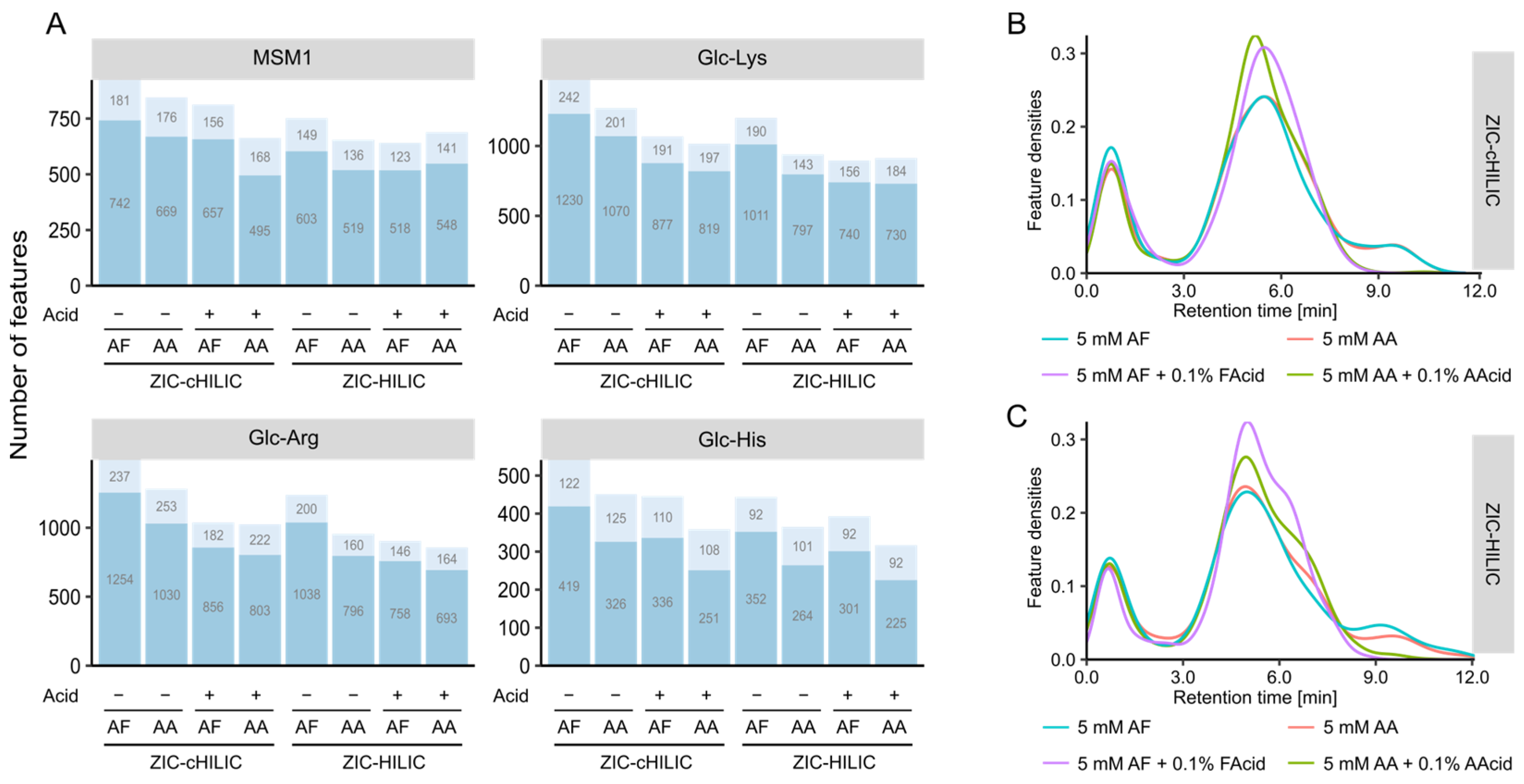
3.3. Mobile Phase Optimization
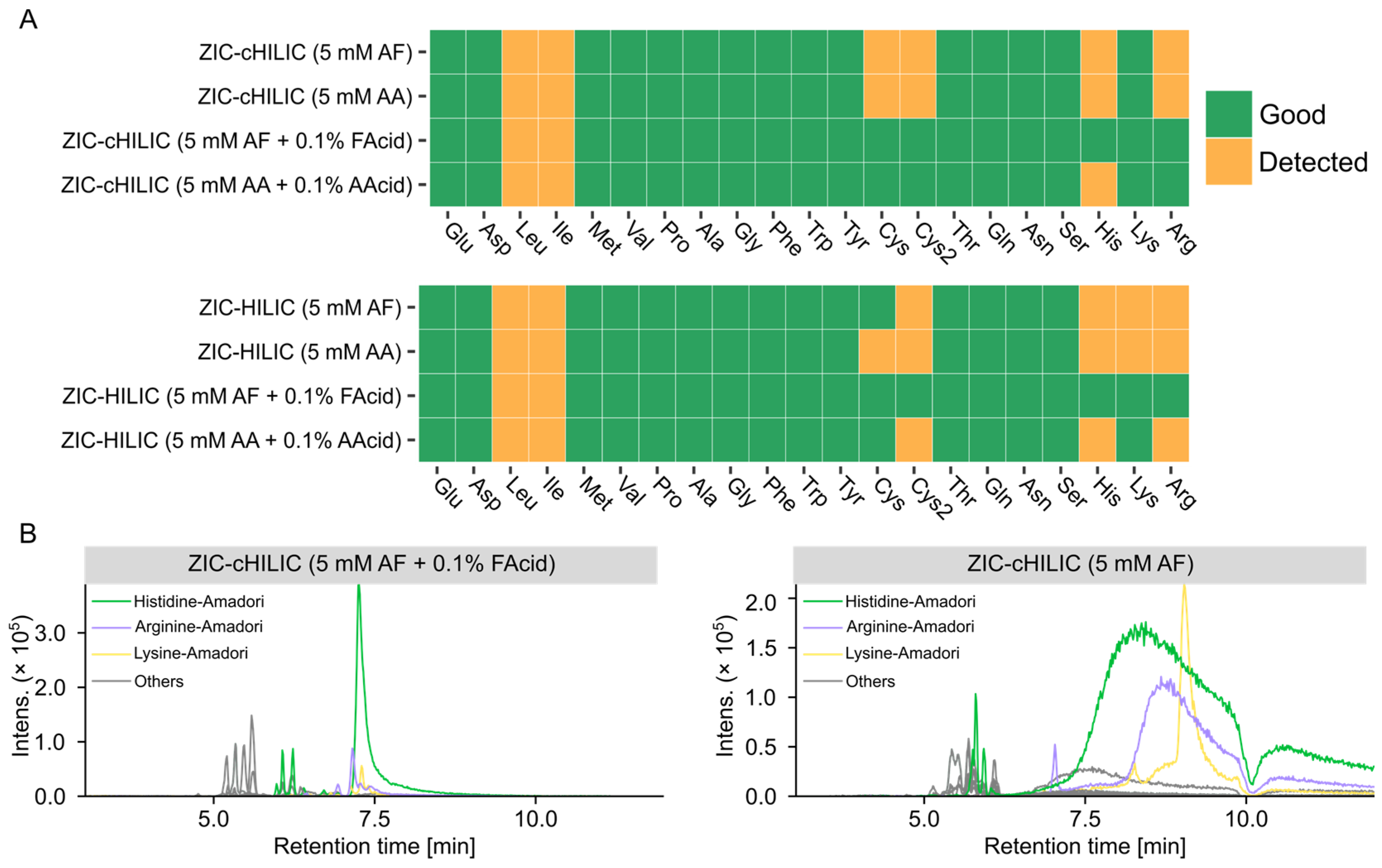
3.3.1. Non-Targeted Evaluation of the Mobile Phase pH and Modifiers
3.3.2. Effect of Mobile Phase on Detections of Amino Acids and Glycation Products
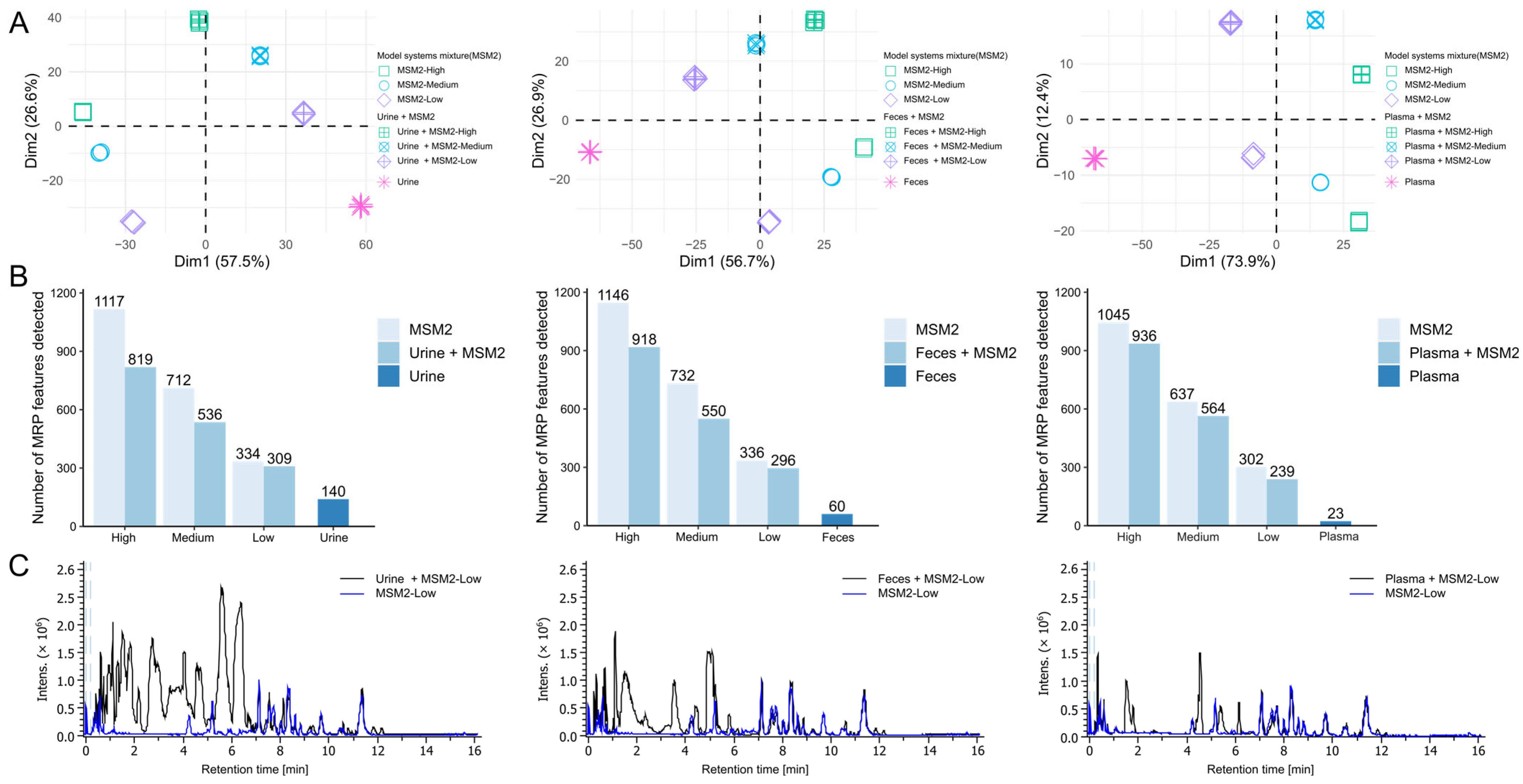
3.4. Evaluation of the Optimized HILIC-MS Method Using Biological Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeng, C.; Li, Y.; Ma, J.; Niu, L.; Tay, F.R. Clinical/Translational Aspects of Advanced Glycation End-Products. Trends Endocrinol. Metab. 2019, 30, 959–973. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, M.; Henle, T. Baking, ageing, diabetes: A short history of the Maillard reaction. Angew. Chem. Int. Ed. 2014, 53, 10316–10329. [Google Scholar] [CrossRef] [PubMed]
- Hodge, J.E. Dehydrated Foods, Chemistry of Browning Reactions in Model Systems. J. Agric. Food Chem. 1953, 1, 928–943. [Google Scholar] [CrossRef]
- Beisswenger, P.J.; Howell, S.K.; Russell, G.B.; Miller, M.E.; Rich, S.S.; Mauer, M. Early progression of diabetic nephropathy correlates with methylglyoxal-derived advanced glycation end products. Diabetes Care 2013, 36, 3234–3239. [Google Scholar] [CrossRef] [PubMed]
- Koska, J.; Saremi, A.; Howell, S.; Bahn, G.; De Courten, B.; Ginsberg, H.; Beisswenger, P.J.; Reaven, P.D.; Investigators, V. Advanced Glycation End Products, Oxidation Products, and Incident Cardiovascular Events in Patients with Type 2 Diabetes. Diabetes Care 2018, 41, 570–576. [Google Scholar] [CrossRef] [PubMed]
- van Outersterp, R.E.; Moons, S.J.; Engelke, U.F.H.; Bentlage, H.; Peters, T.M.A.; van Rooij, A.; Huigen, M.; de Boer, S.; van der Heeft, E.; Kluijtmans, L.A.J.; et al. Amadori rearrangement products as potential biomarkers for inborn errors of amino-acid metabolism. Commun. Biol. 2021, 4, 367. [Google Scholar] [CrossRef] [PubMed]
- Heremans, I.P.; Caligiore, F.; Gerin, I.; Bury, M.; Lutz, M.; Graff, J.; Stroobant, V.; Vertommen, D.; Teleman, A.A.; Van Schaftingen, E.; et al. Parkinson’s disease protein PARK7 prevents metabolite and protein damage caused by a glycolytic metabolite. Proc. Natl. Acad. Sci. USA. 2022, 119, e2111338119. [Google Scholar] [CrossRef]
- Poojary, M.M.; Zhang, W.; Greco, I.; De Gobba, C.; Olsen, K.; Lund, M.N. Liquid chromatography quadrupole-Orbitrap mass spectrometry for the simultaneous analysis of advanced glycation end products and protein-derived cross-links in food and biological matrices. J. Chromatogr. A 2020, 1615, 460767. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, C.; Liehr, K.; Henning, C.; Fiedler, R.; Girndt, M.; Gebert, M.; Hulko, M.; Storr, M.; Glomb, M.A. Detection of Free Advanced Glycation End Products In Vivo during Hemodialysis. J. Agric. Food Chem. 2017, 65, 930–937. [Google Scholar] [CrossRef]
- Sillner, N.; Walker, A.; Hemmler, D.; Bazanella, M.; Heinzmann, S.S.; Haller, D.; Schmitt-Kopplin, P. Milk-Derived Amadori Products in Feces of Formula-Fed Infants. J. Agric. Food Chem. 2019, 67, 8061–8069. [Google Scholar] [CrossRef]
- Lin, J.A.; Wu, C.H.; Yen, G.C. Perspective of Advanced Glycation End Products on Human Health. J. Agric. Food Chem. 2018, 66, 2065–2070. [Google Scholar] [CrossRef]
- Nursten, H.E. The Maillard Reaction: Chemistry, Biochemistry, and Implications; Royal Society of Chemistry: London, UK, 2005. [Google Scholar]
- Henning, C.; Glomb, M.A. Pathways of the Maillard reaction under physiological conditions. Glycoconj. J. 2016, 33, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, X.; Li, L.; Li, B.; Yang, Z. The fate of dietary advanced glycation end products in the body: From oral intake to excretion. Crit. Rev. Food Sci. Nutr. 2020, 60, 3475–3491. [Google Scholar] [CrossRef] [PubMed]
- Hemmler, D.; Roullier-Gall, C.; Marshall, J.W.; Rychlik, M.; Taylor, A.J.; Schmitt-Kopplin, P. Evolution of Complex Maillard Chemical Reactions, Resolved in Time. Sci. Rep. 2017, 7, 3227. [Google Scholar] [CrossRef]
- Hemmler, D.; Roullier-Gall, C.; Marshall, J.W.; Rychlik, M.; Taylor, A.J.; Schmitt-Kopplin, P. Insights into the Chemistry of Non-Enzymatic Browning Reactions in Different Ribose-Amino Acid Model Systems. Sci. Rep. 2018, 8, 16879. [Google Scholar] [CrossRef] [PubMed]
- Guilbaud, A.; Howsam, M.; Niquet-Leridon, C.; Delguste, F.; Boulanger, E.; Tessier, F.J. The LepR(db/db) mice model for studying glycation in the context of diabetes. Diabetes/Metab. Res. Rev. 2019, 35, e3103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Troise, A.D.; Zhang, H.; Fogliano, V. Cocoa melanoidins reduce the formation of dietary advanced glycation end-products in dairy mimicking system. Food Chem. 2021, 345, 128827. [Google Scholar] [CrossRef]
- Sun, X.; Tang, J.; Wang, J.; Rasco, B.A.; Lai, K.; Huang, Y. Formation of advanced glycation endproducts in ground beef under pasteurisation conditions. Food Chem. 2015, 172, 802–807. [Google Scholar] [CrossRef]
- Akillioglu, H.G.; Lund, M.N. Quantification of advanced glycation end products and amino acid cross-links in foods by high-resolution mass spectrometry: Applicability of acid hydrolysis. Food Chem. 2022, 366, 130601. [Google Scholar] [CrossRef] [PubMed]
- Sillner, N.; Walker, A.; Harrieder, E.M.; Schmitt-Kopplin, P.; Witting, M. Development and application of a HILIC UHPLC-MS method for polar fecal metabolome profiling. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1109, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using Nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Bottcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Shen, S.; Xing, S.; Yu, H.; Huan, T. ISFrag: De Novo Recognition of In-Source Fragments for Liquid Chromatography-Mass Spectrometry Data. Anal. Chem. 2021, 93, 10243–10250. [Google Scholar] [CrossRef]
- Le, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Lommen, A. MetAlign: Interface-Driven, Versatile Metabolomics Tool for Hyphenated Full-Scan Mass Spectrometry Data Preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef] [PubMed]
- Fraisier-Vannier, O.; Chervin, J.; Cabanac, G.; Puech, V.; Fournier, S.; Durand, V.; Amiel, A.; Andre, O.; Benamar, O.A.; Dumas, B.; et al. MS-CleanR: A Feature-Filtering Workflow for Untargeted LC-MS Based Metabolomics. Anal. Chem. 2020, 92, 9971–9981. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lu, Y.; Guo, Y.; Cao, H.; Wang, Q.; Shui, W. Comprehensive evaluation of untargeted metabolomics data processing software in feature detection, quantification and discriminating marker selection. Anal. Chim. Acta 2018, 1029, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. Glycation free adduct accumulation in renal disease: The new AGE. Pediatr. Nephrol. 2005, 20, 1515–1522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Watson, D.G.; Wang, L.; Westrop, G.D.; Coombs, G.H.; Zhang, T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography-high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. J. Chromatogr. A 2014, 1362, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Iturrospe, E.; Da Silva, K.M.; Talavera Andujar, B.; Cuykx, M.; Boeckmans, J.; Vanhaecke, T.; Covaci, A.; van Nuijs, A.L.N. An exploratory approach for an oriented development of an untargeted hydrophilic interaction liquid chromatography-mass spectrometry platform for polar metabolites in biological matrices. J. Chromatogr. A 2021, 1637, 461807. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Yaylayan, V. Insight into Isomeric Diversity of Glycated Amino Acids in Maillard Reaction Mixtures. Int. J. Mol. Sci. 2022, 23, 3430. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, R.A.; Naz, S.; Cougnaud, L.; Vuckovic, D. Comparison of underivatized silica and zwitterionic sulfobetaine hydrophilic interaction liquid chromatography stationary phases for global metabolomics of human plasma. J. Chromatogr. A 2019, 1608, 460419. [Google Scholar] [CrossRef]
- Wei, J.; Shen, A.; Wan, H.; Yan, J.; Yang, B.; Guo, Z.; Zhang, F.; Liang, X. Highly selective separation of aminoglycoside antibiotics on a zwitterionic Click TE-Cys column. J. Sep. Sci. 2014, 37, 1781–1787. [Google Scholar] [CrossRef] [PubMed]
- Contrepois, K.; Jiang, L.; Snyder, M. Optimized Analytical Procedures for the Untargeted Metabolomic Profiling of Human Urine and Plasma by Combining Hydrophilic Interaction (HILIC) and Reverse-Phase Liquid Chromatography (RPLC)-Mass Spectrometry. Mol. Cell. Proteom. 2015, 14, 1684–1695. [Google Scholar] [CrossRef]
- Hosseinkhani, F.; Huang, L.; Dubbelman, A.C.; Guled, F.; Harms, A.C.; Hankemeier, T. Systematic Evaluation of HILIC Stationary Phases for Global Metabolomics of Human Plasma. Metabolites 2022, 12, 165. [Google Scholar] [CrossRef] [PubMed]
- Iverson, C.D.; Gu, X.; Lucy, C.A. The hydrophilicity vs. ion interaction selectivity plot revisited: The effect of mobile phase pH and buffer concentration on hydrophilic interaction liquid chromatography selectivity behavior. J. Chromatogr. A 2016, 1458, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Cebo, M.; Ikegami, T.; Lammerhofer, M. Separation of carbohydrate isomers and anomers on poly-N-(1H-tetrazole-5-yl)-methacrylamide-bonded stationary phase by hydrophilic interaction chromatography as well as determination of anomer interconversion energy barriers. J. Chromatogr. A 2020, 1620, 460981. [Google Scholar] [CrossRef] [PubMed]
- Erngren, I.; Nestor, M.; Pettersson, C.; Hedeland, M. Improved Sensitivity in Hydrophilic Interaction Liquid Chromatography-Electrospray-Mass Spectrometry after Removal of Sodium and Potassium Ions from Biological Samples. Metabolites 2021, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Ekdahl, A.; Johansson, M.C.; Ahnoff, M. Tracing and separating plasma components causing matrix effects in hydrophilic interaction chromatography-electrospray ionization mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 923–924, 83–91. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, E.; Van Meulebroek, L.; Rombouts, C.; Huysman, S.; Verplanken, K.; Lapauw, B.; Wauters, J.; Hemeryck, L.Y.; Vanhaecke, L. A validated multi-matrix platform for metabolomic fingerprinting of human urine, feces and plasma using ultra-high performance liquid-chromatography coupled to hybrid orbitrap high-resolution mass spectrometry. Anal. Chim. Acta 2018, 1033, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Roca, M.; Alcoriza, M.I.; Garcia-Canaveras, J.C.; Lahoz, A. Reviewing the metabolome coverage provided by LC-MS: Focus on sample preparation and chromatography-A tutorial. Anal. Chim. Acta 2021, 1147, 38–55. [Google Scholar] [CrossRef] [PubMed]
- Henle, T. Dietary advanced glycation end products--a risk to human health? A call for an interdisciplinary debate. Mol. Nutr. Food Res. 2007, 51, 1075–1078. [Google Scholar] [CrossRef]
- Birlouez-Aragon, I.; Saavedra, G.; Tessier, F.J.; Galinier, A.; Ait-Ameur, L.; Lacoste, F.; Niamba, C.N.; Alt, N.; Somoza, V.; Lecerf, J.M. A diet based on high-heat-treated foods promotes risk factors for diabetes mellitus and cardiovascular diseases. Am. J. Clin. Nutr. 2010, 91, 1220–1226. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Hemmler, D.; Schmitt-Kopplin, P. HILIC-MS for Untargeted Profiling of the Free Glycation Product Diversity. Metabolites 2022, 12, 1179. https://doi.org/10.3390/metabo12121179
Yan Y, Hemmler D, Schmitt-Kopplin P. HILIC-MS for Untargeted Profiling of the Free Glycation Product Diversity. Metabolites. 2022; 12(12):1179. https://doi.org/10.3390/metabo12121179
Chicago/Turabian StyleYan, Yingfei, Daniel Hemmler, and Philippe Schmitt-Kopplin. 2022. "HILIC-MS for Untargeted Profiling of the Free Glycation Product Diversity" Metabolites 12, no. 12: 1179. https://doi.org/10.3390/metabo12121179
APA StyleYan, Y., Hemmler, D., & Schmitt-Kopplin, P. (2022). HILIC-MS for Untargeted Profiling of the Free Glycation Product Diversity. Metabolites, 12(12), 1179. https://doi.org/10.3390/metabo12121179