A Preliminary Pilot Study: Metabolomic Analysis of Saliva in Oral Candidiasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Study Items
2.3. Candida Detection Tests
2.4. Saliva Collection
2.5. Metabolome Analysis
2.6. Data Analysis
3. Results
3.1. Subjects
3.2. Overview of the Detected Salivary Metabolites
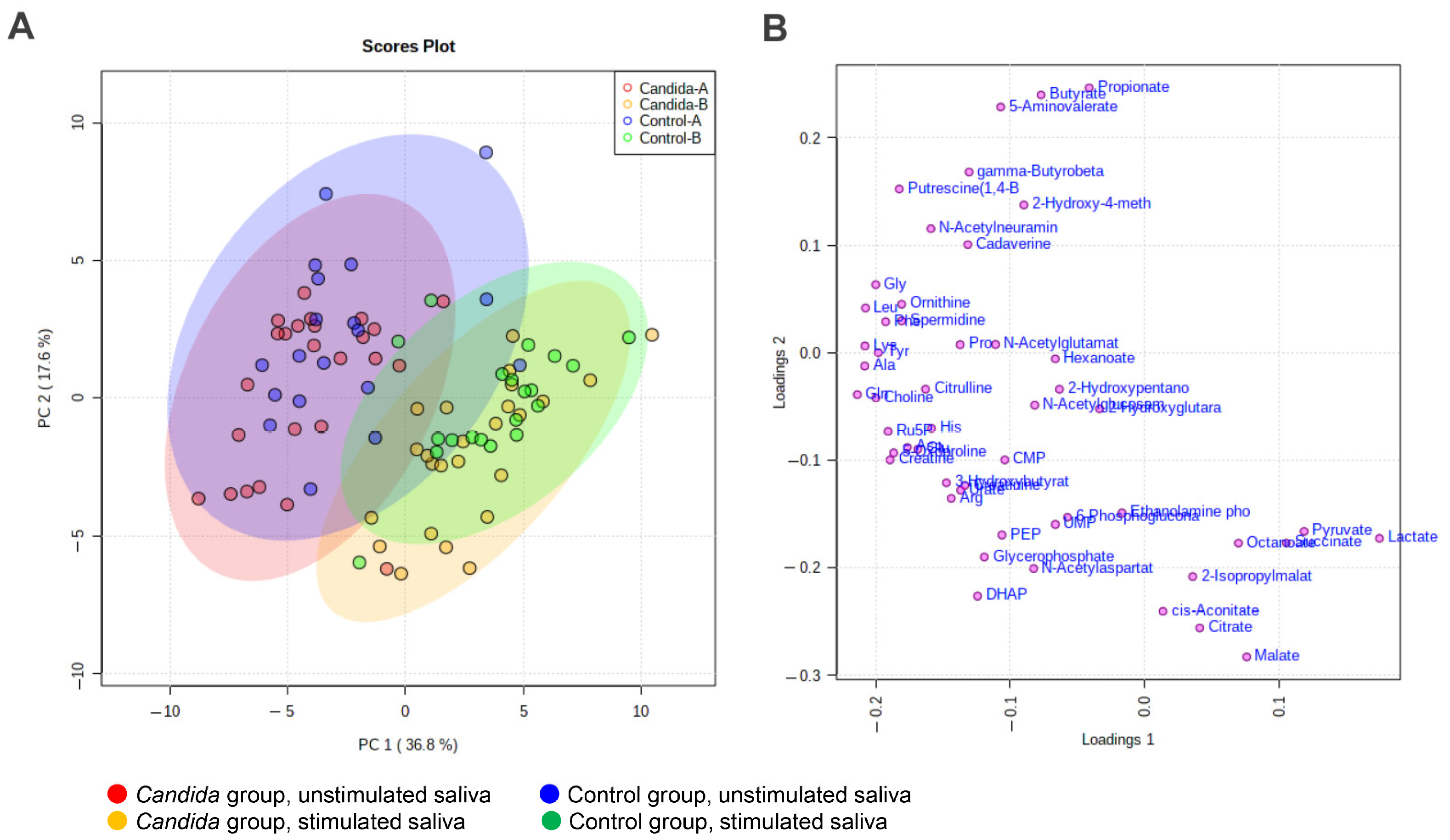
3.3. Principal Component Analysis (PCA)
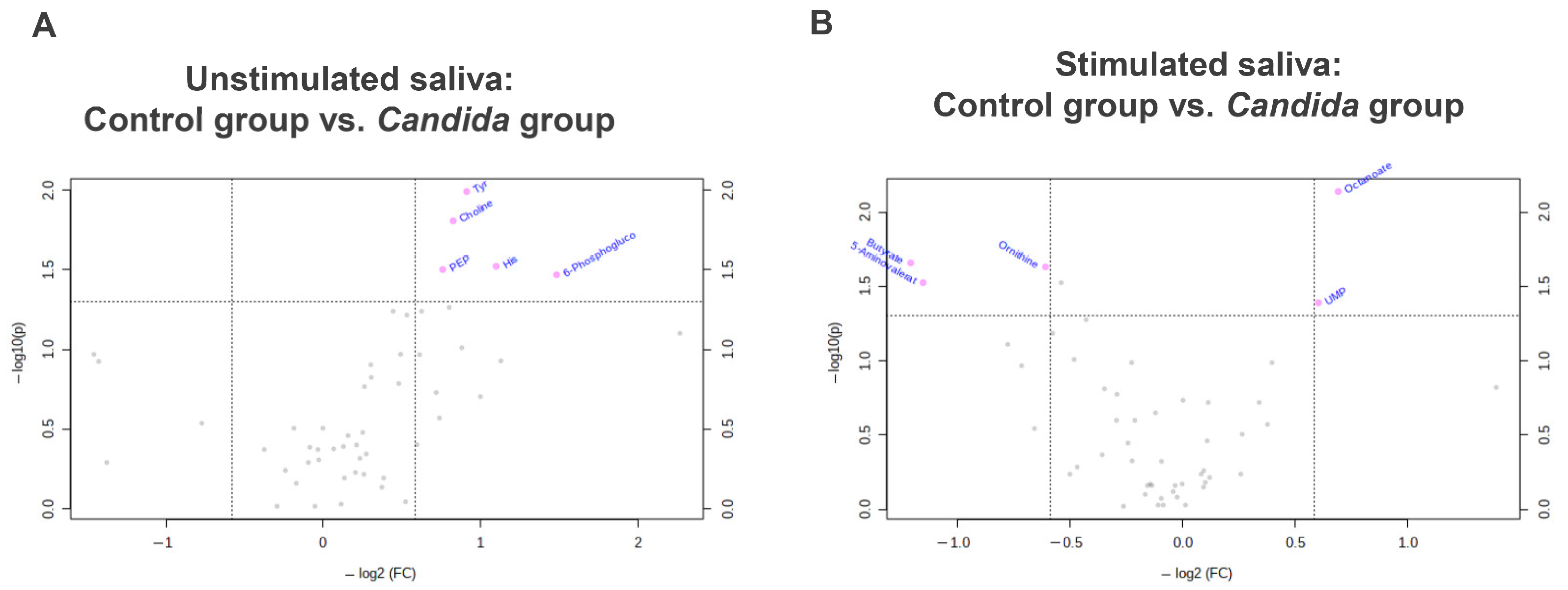
3.4. Comparison with Volcano Plot
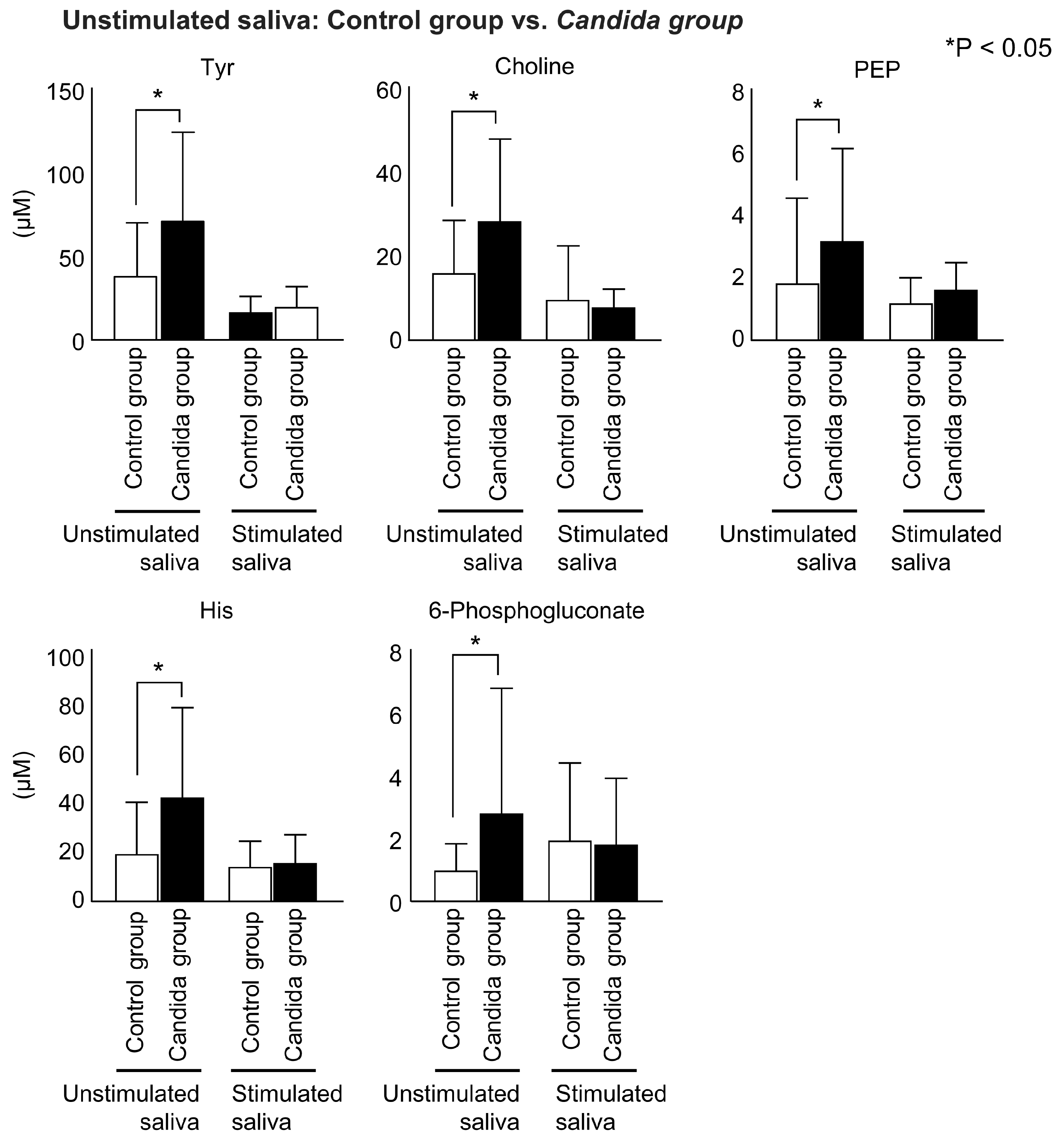
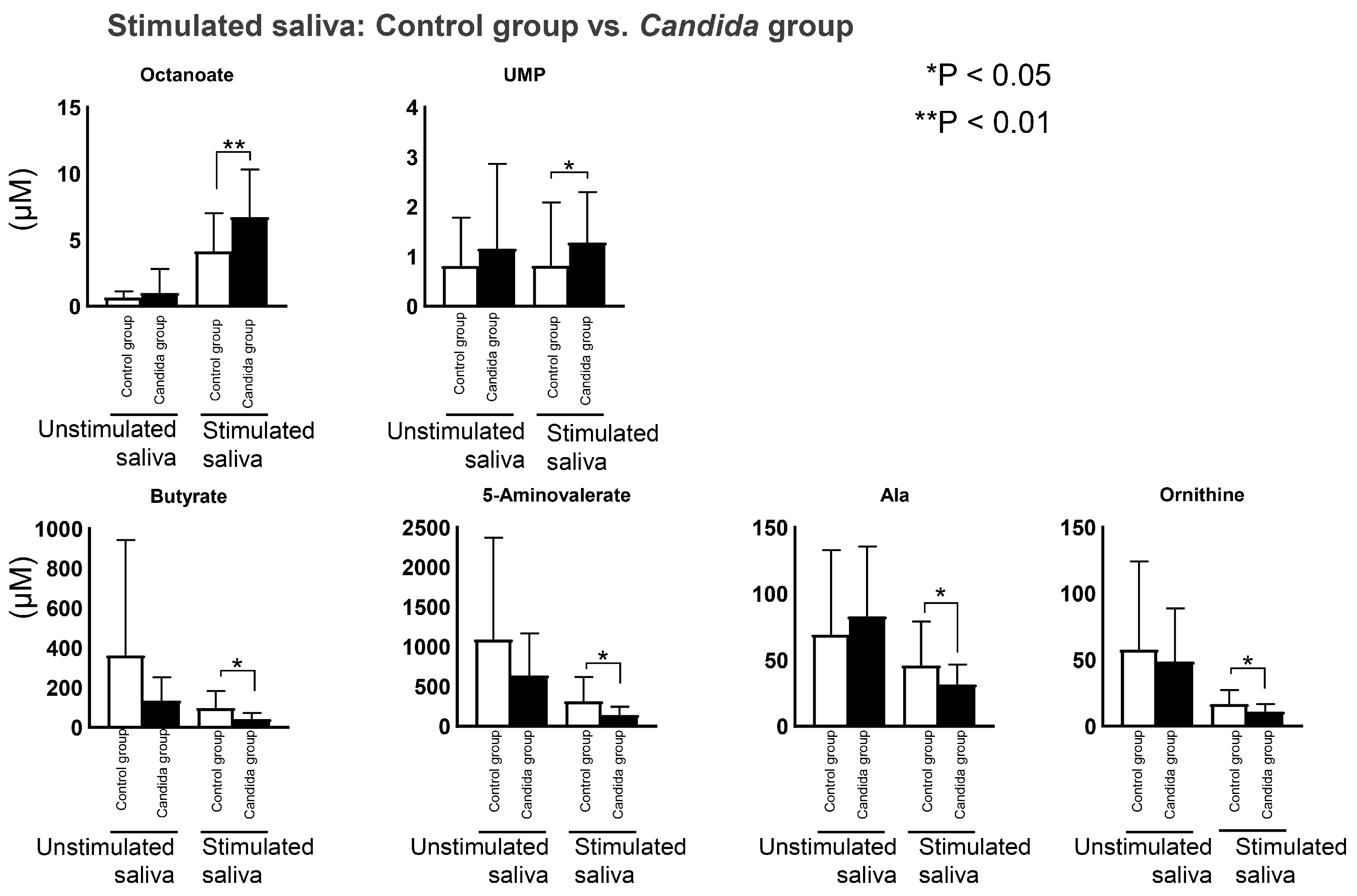
3.5. Comparison between Control and Candida Groups in Connection with Oral Candidiasis
4. Discussion
- Whole-mouth saliva was used, including components from serum, salivary gland secretions, gingival sulcus exudates, mucosal exudates, and intraoral microorganisms [14]. Bacterial metabolites, in particular, have major effects, but in this study, periodontal disease, caries, and other oral diseases were not considered [57].
- The participants’ systemic diseases or medication status were not considered. The subjects in both the control and Candida groups had mean ages over 70 years. In an elderly population, it is difficult to sample subjects who have no systemic diseases and are not currently taking medication. To verify the generalizability of our findings, a large scale study is necessary with more detailed analysis.
- Constructing and unifying a database by including other metabolome profiles would enable more accurate diagnosis.
- Candida species were not identified in this study. The pathogens responsible for candidiasis include C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei. It is essential to evaluate the effects of different species on the salivary metabolites.
- The target age group for this study was 60 years and older. This was established to avoid bias because previous studies have shown that salivary metabolites change with age [58]. However, since there are also young patients with oral candidiasis, analysis of a wide range of age groups is an issue for future research.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, A.; Verma, R.; Murari, A.; Agrawal, A. Oral candidiasis: An overview. J. Oral Maxillofac. Pathol. 2014, 18, S81–S85. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011, 19, 557–563. [Google Scholar] [CrossRef] [Green Version]
- Gulati, M.; Lohse, M.B.; Ennis, C.L.; Gonzalez, R.E.; Perry, A.M.; Bapat, P.; Arevalo, A.V.; Rodriguez, D.L.; Nobile, C.J. In vitro culturing and screening of Candida albicans biofilms. Curr. Protoc. Microbiol. 2018, 50, e60. [Google Scholar] [CrossRef]
- Aragón, F.; Zea-Sevilla, M.A.; Montero, J.; Sancho, P.; Corral, R.; Tejedor, C.; Frades-Payo, B.; Paredes-Gallardo, V.; Albaladejo, A. Oral health in Alzheimer’s disease: A multicenter case-control study. Clin. Oral Investig. 2018, 22, 3061–3070. [Google Scholar] [CrossRef] [PubMed]
- Liotti, F.M.; Posteraro, B.; Mannu, F.; Carta, F.; Pantaleo, A.; De Angelis, G.; Menchinelli, G.; Spanu, T.; Fiori, P.L.; Turrini, F.; et al. Development of a multiplex PCR platform for the rapid detection of bacteria, antibiotic resistance, and candida in human blood samples. Front. Cell. Infect. Microbiol. 2019, 9, 389. [Google Scholar] [CrossRef]
- Hadadi-Fishani, M.; Shakerimoghaddam, A.; Khaledi, A. Candida coinfection among patients with pulmonary tuberculosis in Asia and Africa; A systematic review and meta-analysis of cross-sectional studies. Microb. Pathog. 2020, 139, 103898. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, T.; Yoshida, M.; Matsui, T.; Sasaki, H. Oral care and pneumonia. Oral Care Working Group. Lancet 1999, 354, 515. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.S.; Reyes, C.M.; Stolpman, M.; Speckman, J.; Allen, K.; Beney, J. The direct cost and incidence of systemic fungal infections. Value Health 2002, 5, 26–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korting, H.C.; Ollert, M.; Georgii, A.; Fröschl, M. In vitro susceptibilities and biotypes of Candida albicans isolates from the oral cavities of patients infected with human immunodeficiency virus. J. Clin. Microbiol. 1988, 26, 2626–2631. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.W.; Lewis, M.A. Isolation and identification of Candida from the oral cavity. Oral Dis. 2000, 6, 3–11. [Google Scholar] [CrossRef]
- White, P.L.; Williams, D.W.; Kuriyama, T.; Samad, S.A.; Lewis, M.A.O.; Barnes, R.A. Detection of Candida in concentrated oral rinse cultures by real-time PCR. J. Clin. Microbiol. 2004, 42, 2101–2107. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Khan, Z.; Mustafa, A.S.; Khan, Z.U. Seminested PCR for diagnosis of candidemia: Comparison with culture, antigen detection, and biochemical methods for species identification. J. Clin. Microbiol. 2002, 40, 2483–2489. [Google Scholar] [CrossRef] [Green Version]
- Liguori, G.; Lucariello, A.; Colella, G.; De Luca, A.; Marinelli, P. Rapid identification of Candida species in oral rinse solutions by PCR. J. Clin. Pathol. 2007, 60, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Hannig, C.; Hannig, M.; Kensche, A.; Carpenter, G. The mucosal pellicle—An underestimated factor in oral physiology. Arch. Oral Biol. 2017, 80, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Salvatori, O.; Puri, S.; Tati, S.; Edgerton, M. Innate immunity and saliva in Candida albicans–mediated oral diseases. J. Dent. Res. 2016, 95, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Hyvärinen, E.; Savolainen, M.; Mikkonen, J.J.W.; Kullaa, A.M. Salivary metabolomics for diagnosis and monitoring diseases: Challenges and possibilities. Metabolites 2021, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Nadig, S.D.; Ashwathappa, D.T.; Manjunath, M.; Krishna, S.; Annaji, A.G.; Shivaprakash, P.K. A relationship between salivary flow rates and Candida counts in patients with xerostomia. J. Oral Maxillofac. Pathol. 2017, 21, 316. [Google Scholar] [CrossRef] [Green Version]
- Torres, S.R.; Peixoto, C.B.; Caldas, D.M.; Silva, E.B.; Akiti, T.; Nucci, M.; De Uzeda, M. Relationship between salivary flow rates and Candida counts in subjects with xerostomia. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2002, 93, 149–154. [Google Scholar] [CrossRef]
- Kawanishi, N.; Hoshi, N.; Adachi, T.; Ichigaya, N.; Kimoto, K. Positive effects of saliva on oral candidiasis: Basic research on the analysis of salivary properties. J. Clin. Med. 2021, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Martina, E.; Campanati, A.; Diotallevi, F.; Offidani, A. Saliva and oral diseases. J. Clin. Med. 2020, 9, 466. [Google Scholar] [CrossRef]
- Lindon, J.C.; Holmes, E.; Nicholson, J.K. Metabonomics in pharmaceutical R&D. FEBS J. 2007, 274, 1140–1151. [Google Scholar] [CrossRef]
- Mikkonen, J.J.W.; Singh, S.P.; Akhi, R.; Salo, T.; Lappalainen, R.; González-Arriagada, W.A.; Lopes, M.A.; Kullaa, A.M.; Myllymaa, S. Potential role of nuclear magnetic resonance spectroscopy to identify salivary metabolite alterations in patients with head and neck cancer. Oncol. Lett. 2018, 16, 6795–6800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrala, M.; Mikkonen, J.J.W.; Pesonen, P.; Lappalainen, R.; Tjäderhane, L.; Niemelä, R.K.; Seitsalo, H.; Salo, T.; Myllymaa, S.; Kullaa, A.M. Variability of salivary metabolite levels in patients with Sjögren’s syndrome. J. Oral Sci. 2020, 63, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.L.; Duarte, D.; Carneiro, T.J.; Ferreira, S.; Cunha, B.; Soares, D.; Costa, A.L.; Gil, A.M. Saliva NMR metabolomics: Analytical issues in pediatric oral health research. Oral Dis. 2019, 25, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Romano, F.; Meoni, G.; Manavella, V.; Baima, G.; Tenori, L.; Cacciatore, S.; Aimetti, M. Analysis of salivary phenotypes of generalized aggressive and chronic periodontitis through nuclear magnetic resonance-based metabolomics. J. Periodontol. 2018, 89, 1452–1460. [Google Scholar] [CrossRef] [Green Version]
- Okuma, N.; Saita, M.; Hoshi, N.; Soga, T.; Tomita, M.; Sugimoto, M.; Kimoto, K. Effect of masticatory stimulation on the quantity and quality of saliva and the salivary metabolomic profile. PLoS ONE 2017, 12, e0183109. [Google Scholar] [CrossRef] [Green Version]
- Soga, T.; Ohashi, Y.; Ueno, Y.; Naraoka, H.; Tomita, M.; Nishioka, T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003, 2, 488–494. [Google Scholar] [CrossRef]
- Soga, T.; Ishikawa, T.; Igarashi, S.; Sugawara, K.; Kakazu, Y.; Tomita, M. Analysis of nucleotides by pressure-assisted capillary electrophoresis-mass spectrometry using silanol mask technique. J. Chromatogr. A 2007, 1159, 125–133. [Google Scholar] [CrossRef]
- Soga, T.; Baran, R.; Suematsu, M.; Ueno, Y.; Ikeda, S.; Sakurakawa, T.; Kakazu, Y.; Ishikawa, T.; Robert, M.; Nishioka, T.; et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006, 281, 16768–16776. [Google Scholar] [CrossRef] [Green Version]
- Scognamiglio, T.; Zinchuk, R.; Gumpeni, P.; Larone, D.H. Comparison of inhibitory mold agar to Sabouraud dextrose agar as a primary medium for isolation of fungi. J. Clin. Microbiol. 2010, 48, 1924–1925. [Google Scholar] [CrossRef]
- Dawes, C. Physiological factors affecting salivary flow rate, oral sugar clearance, and the sensation of dry mouth in man. J. Dent. Res. 1987, 66, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Dawes, C.; Watanabe, S. The effect of taste adaptation on salivary flow rate and salivary sugar clearance. J. Dent. Res. 1987, 66, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, N.; Hoshi, N.; Masahiro, S.; Enomoto, A.; Ota, S.; Kaneko, M.; Soga, T.; Tomita, M.; Kimoto, K. Effects of inter-day and intra-day variation on salivary metabolomic profiles. Clin. Chim. Acta 2019, 489, 41–48. [Google Scholar] [CrossRef]
- Ishijima, T.; Koshino, H.; Hirai, T.; Takasaki, H. The relationship between salivary secretion rate and masticatory efficiency. J. Oral Rehabil. 2004, 31, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Navazesh, M. Methods for collecting saliva. Ann. N. Y. Acad. Sci. 1993, 694, 72–77. [Google Scholar] [CrossRef]
- Sugimoto, M.; Saruta, J.; Matsuki, C.; To, M.; Onuma, H.; Kaneko, M.; Soga, T.; Tomita, M.; Tsukinoki, K. Physiological and environmental parameters associated with mass spectrometry-based salivary metabolomic profiles. Metabolomics 2013, 9, 454–463. [Google Scholar] [CrossRef]
- Mikkonen, J.J.W.; Singh, S.P.; Herrala, M.; Lappalainen, R.; Myllymaa, S.; Kullaa, A.M. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J. Periodontal Res. 2016, 51, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Duan, Y. Saliva: A potential media for disease diagnostics and monitoring. Oral Oncol. 2012, 48, 569–577. [Google Scholar] [CrossRef]
- Koneru, S.; Tanikonda, R. Salivaomics—A promising future in early diagnosis of dental diseases. Dent. Res. J. 2014, 11, 11–15. [Google Scholar]
- Cuevas-Córdoba, B.; Santiago-García, J.J. Saliva: A fluid of study for OMICS. OMICS: J. Integr. Biol 2014, 18, 87–97. [Google Scholar] [CrossRef]
- Al-Tarawneh, S.K.; Border, M.B.; Dibble, C.F.; Bencharit, S. Defining salivary biomarkers using mass spectrometry-based proteomics: A systematic review. OMICS: J. Integr. Biol. 2011, 15, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Amerongen, A.V.N.; Ligtenberg, A.J.M.; Veerman, E.C.I. Implications for diagnostics in the biochemistry and physiology of saliva. Ann. N. Y. Acad. Sci. 2007, 1098, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Michishige, F.; Kanno, K.; Yoshinaga, S.; Hinode, D.; Takehisa, Y.; Yasuoka, S. Effect of saliva collection method on the concentration of protein components in saliva. J. Med. Investig. 2006, 53, 140–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belstrøm, D.; Holmstrup, P.; Bardow, A.; Kokaras, A.; Fiehn, N.E.; Paster, B.J. Comparative analysis of bacterial profiles in unstimulated and stimulated saliva samples. J. Oral Microbiol. 2016, 8, 30112. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.C.; Laghi, L.; Parolin, C.; Foschi, C.; Marangoni, A.; Liberatore, A.; Dias, A.L.T.; Cricca, M.; Vitali, B. Metabolic profiling of Candida clinical isolates of diferent species and infection sources. Sci. Rep. 2020, 10, 16716. [Google Scholar] [CrossRef]
- Chen, H.; Fujita, M.; Feng, Q.; Clardy, J.; Fink, G.R. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA. 2004, 101, 5048–5052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gow, N.A.R.; Brown, A.J.P.; Odds, F.C. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 2002, 5, 366–371. [Google Scholar] [CrossRef]
- Albuquerque, P.; Casadevall, A. Quorum sensing in fungi—A review. Med. Mycol. 2012, 50, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Lai, R.Y.; Huang, S.; Fenwick, M.K.; Hazra, A.; Zhang, Y.; Rajashankar, K.; Philmus, B.; Kinsland, C.; Sanders, J.M.; Ealick, S.E.; et al. Thiamin pyrimidine biosynthesis in Candida albicans: A remarkable reaction between histidine and pyridoxal phosphate. J. Am. Chem. Soc. 2012, 134, 9157–9159. [Google Scholar] [CrossRef] [Green Version]
- Costa, C.P.; Bezerra, A.R.; Almeida, A.; Rocha, S.M. Candida species (volatile) metabotyping through advanced comprehensive two-dimensional gas chromatography. Microorganisms 2020, 8, 1911. [Google Scholar] [CrossRef]
- D’Enfert, C.; Diaquin, M.; Delit, A.; Wuscher, N.; Debeaupuis, J.P.; Huerre, M.; Latge, J.P. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect. Immun. 1996, 64, 4401–4405. [Google Scholar] [CrossRef] [PubMed]
- Kuboniwa, M.; Sakanaka, A.; Hashino, E.; Bamba, T.; Fukusaki, E.; Amano, A. Prediction of periodontal inflammation via metabolic profiling of saliva. J. Dent. Res. 2016, 95, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.; Eftimiadi, C.; Damiani, G.; Buffa, P.; Buffa, D.; Botta, G.A. Short chain fatty acids present in periodontal pockets may play a role in human periodontal diseases. J. Periodontal Res. 1987, 22, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Niederman, R.; Buyle-Bodin, Y.; Lu, B.Y.; Robinson, P.; Naleway, C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J. Dent. Res. 1997, 76, 575–579. [Google Scholar] [CrossRef]
- Thein, Z.M.; Samaranayake, Y.H.; Samaranayake, L.P. Effect of oral bacteria on growth and survival of Candida albicans biofilms. Arch. Oral Biol. 2006, 51, 672–680. [Google Scholar] [CrossRef]
- Grainha, T.; Jorge, P.; Alves, D.; Lopes, S.P.; Pereira, M.O. Unraveling Pseudomonas aeruginosa and Candida albicans communication in coinfection scenarios: Insights through network analysis. Front. Cell. Infect. Microbiol. 2020, 10, 550505. [Google Scholar] [CrossRef]
- Zaura, E.; Brandt, B.W.; Prodan, A.; de Mattos, M.J.T.; Imangaliyev, S.; Kool, J.; Buijs, M.J.; Jagers, F.L.; Hennequin-Hoenderdos, N.L.; Slot, D.E.; et al. On the ecosystemic network of saliva in healthy young adults. ISME J. 2017, 11, 1218–1231. [Google Scholar] [CrossRef] [PubMed]
- Bosman, P.; Pichon, V.; Acevedo, A.C.; Le Pottier, L.; Pers, J.O.; Chardin, H.; Combès, A. Untargeted Metabolomic Approach to Study the Impact of Aging on Salivary Metabolome in Women. Metabolites 2022, 12, 986. [Google Scholar] [CrossRef] [PubMed]

| Control Group | Candida Group | |
|---|---|---|
| Age (mean ± SD) | 76.2 ± 6.9 | 76.6 ± 6.4 |
| Sex (men/women) | 7/13 | 8/17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adachi, T.; Kawanishi, N.; Ichigaya, N.; Sugimoto, M.; Hoshi, N.; Kimoto, K. A Preliminary Pilot Study: Metabolomic Analysis of Saliva in Oral Candidiasis. Metabolites 2022, 12, 1294. https://doi.org/10.3390/metabo12121294
Adachi T, Kawanishi N, Ichigaya N, Sugimoto M, Hoshi N, Kimoto K. A Preliminary Pilot Study: Metabolomic Analysis of Saliva in Oral Candidiasis. Metabolites. 2022; 12(12):1294. https://doi.org/10.3390/metabo12121294
Chicago/Turabian StyleAdachi, Takuya, Norishige Kawanishi, Narumi Ichigaya, Masahiro Sugimoto, Noriyuki Hoshi, and Katsuhiko Kimoto. 2022. "A Preliminary Pilot Study: Metabolomic Analysis of Saliva in Oral Candidiasis" Metabolites 12, no. 12: 1294. https://doi.org/10.3390/metabo12121294





