Longitudinal Metabolomic Analysis Reveals Gut Microbial-Derived Metabolites Related to Formula Feeding and Milk Sensitization Development in Infancy
Abstract
:1. Introduction
2. Results
2.1. Population Characteristics
2.2. Identification of Metabolites in Different Breastfeeding Pattern and Milk Sensitization Sets at Different Years of Age
2.3. Metabolites in Different Breastfeeding Pattern and Milk Sensitization Sets from 6 Months to 1 Year of Age
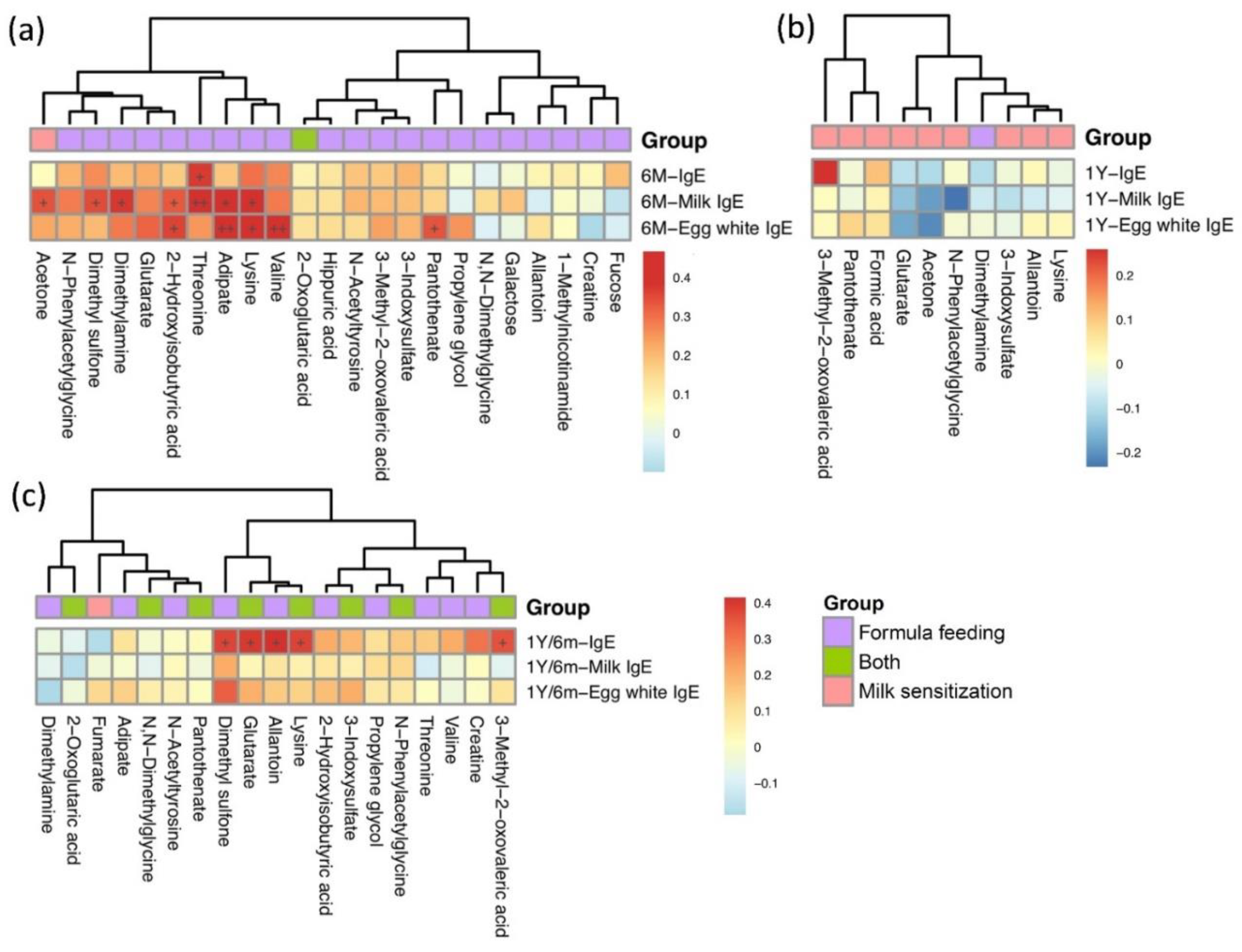
2.4. Association between Metabolites and Food Allergen-Specific IgE Levels in Different Breastfeeding Patterns and Milk Sensitization Sets
2.5. Correlations between Metabolites of Formula Feeding and of Milk Sensitization
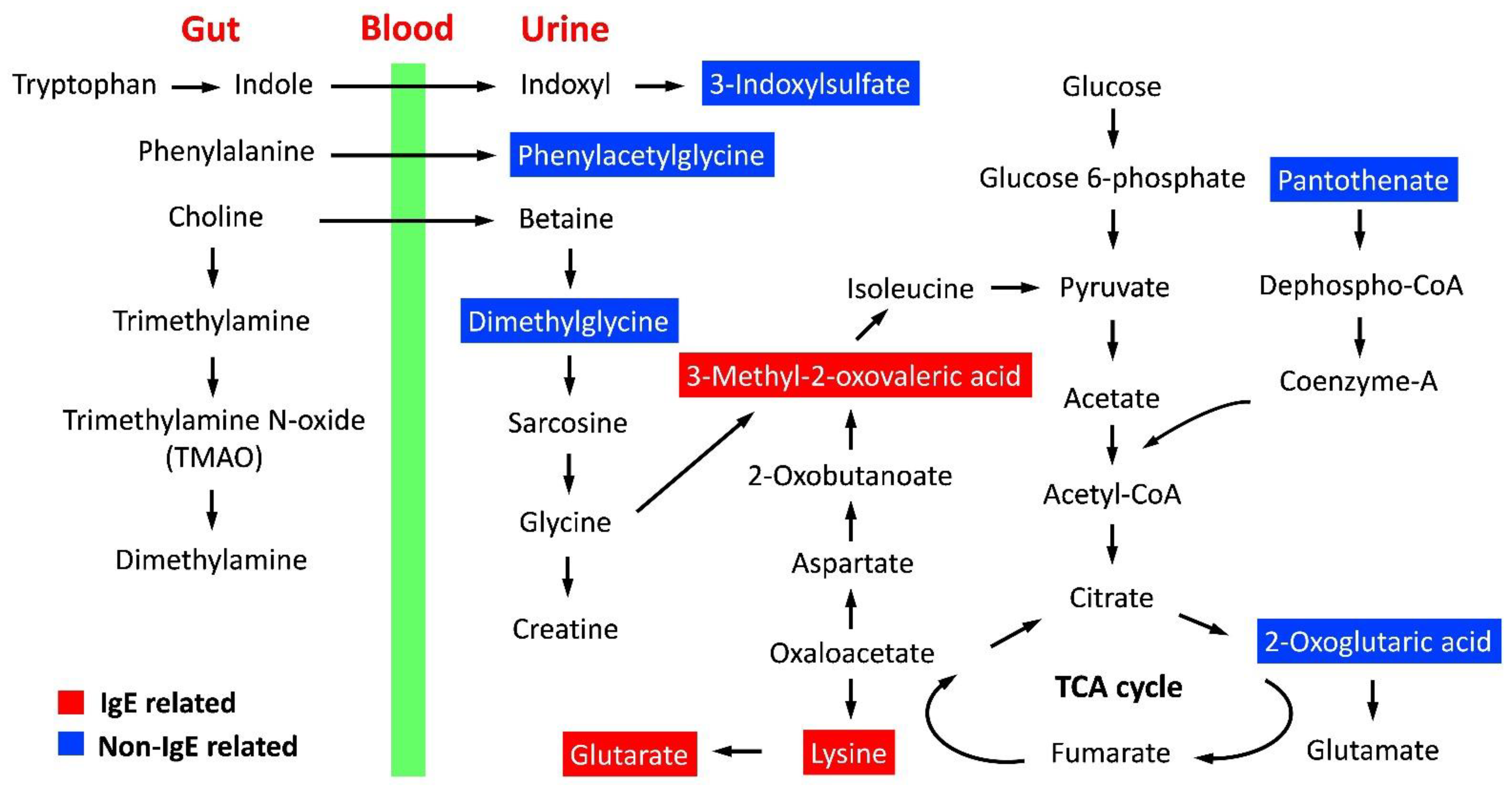
2.6. Metabolic Pathway and Functional Analysis
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Definition of Breastfeeding History
4.3. Total Serum and Food Allergen-Specific IgE Level Measurement
4.4. Urine Sample Preparation
4.5. 1H-Nuclear Magnetic Resonance (NMR) Spectroscopy
4.6. NMR Data Processing and Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, C.R.; Ling, P.-R.; Blackburn, G.L. Review of Infant Feeding: Key Features of Breast Milk and Infant Formula. Nutrients 2016, 8, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Elsen, L.W.J.; Garssen, J.; Burcelin, R.; Verhasselt, V. Shaping the Gut Microbiota by Breastfeeding: The Gateway to Allergy Prevention? Front. Pediatr. 2019, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Walker, M. Formula Supplementation of Breastfed Infants. ICAN 2015, 7, 198–207. [Google Scholar] [CrossRef] [Green Version]
- Hochwallner, H.; Schulmeister, U.; Swoboda, I.; Spitzauer, S.; Valenta, R. Cow’s milk allergy: From allergens to new forms of diagnosis, therapy and prevention. Methods (San Diego Calif.) 2014, 66, 22–33. [Google Scholar] [CrossRef]
- Kulig, M.; Bergmann, R.; Tacke, U.; Wahn, U.; Guggenmoos-Holzmann, I. Long-lasting sensitization to food during the first two years precedes allergic airway disease. The MAS Study Group, Germany. Pediatr. Allergy Immunol. 1998, 9, 61–67. [Google Scholar] [CrossRef]
- Liao, S.L.; Lai, S.H.; Yeh, K.W.; Huang, Y.L.; Yao, T.C.; Tsai, M.H.; Hua, M.C.; Huang, J.L. Exclusive breastfeeding is associated with reduced cow’s milk sensitization in early childhood. Pediatr. Allergy Immunol. 2014, 25, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.Y.; Liao, S.L.; Su, K.W.; Tsai, M.H.; Hua, M.C.; Lai, S.H.; Chen, L.C.; Yao, T.C.; Yeh, K.W.; Huang, J.L. Exclusive or Partial Breastfeeding for 6 Months Is Associated With Reduced Milk Sensitization and Risk of Eczema in Early Childhood: The PATCH Birth Cohort Study. Medicine (Baltimore) 2016, 95, e3391. [Google Scholar] [CrossRef] [PubMed]
- Urashima, M.; Mezawa, H.; Okuyama, M.; Urashima, T.; Hirano, D.; Gocho, N.; Tachimoto, H. Primary Prevention of Cow’s Milk Sensitization and Food Allergy by Avoiding Supplementation With Cow’s Milk Formula at Birth: A Randomized Clinical Trial. JAMA Pediatr. 2019, 173, 1137–1145. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.Y.; Cheng, M.L.; Chiang, M.H.; Wang, C.J.; Tsai, M.H.; Lin, G. Metabolomic Analysis Reveals Distinct Profiles in the Plasma and Urine Associated with IgE Reactions in Childhood Asthma. J. Clin. Med. 2020, 9, 887. [Google Scholar] [CrossRef] [Green Version]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-H.; Lee, J. Indole as an intercellular signal in microbial community. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef] [PubMed]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.Y.; Hassan, S.A.; Ismail, I.H.; Chong, S.Y.; Raja Ali, R.A.; Amin Nordin, S.; Lee, W.S.; Majid, N.A. Gut microbiota in early life and its influence on health and disease: A position paper by the Malaysian Working Group on Gastrointestinal Health. J. Paediatr. Child Health 2017, 53, 1152–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, L.; Chen, H.; Zhang, S.; Zhuang, J.; Li, Q.; Feng, Z. Intestinal Microbiota in Early Life and Its Implications on Childhood Health. Genom. Proteom. Bioinform. 2019, 17, 13–25. [Google Scholar] [CrossRef]
- Fujisaka, S.; Avila-Pacheco, J.; Soto, M.; Kostic, A.; Dreyfuss, J.M.; Pan, H.; Ussar, S.; Altindis, E.; Li, N.; Bry, L.; et al. Diet, Genetics, and the Gut Microbiome Drive Dynamic Changes in Plasma Metabolites. Cell Rep. 2018, 22, 3072–3086. [Google Scholar] [CrossRef] [Green Version]
- Darling, P.B.; Dunn, M.; Gilani, G.S.; Ball, R.O.; Pencharz, P.B. Phenylalanine kinetics differ between formula-fed and human milk-fed preterm infants. J. Nutr. 2004, 134, 2540–2545. [Google Scholar] [CrossRef] [Green Version]
- Lewis, M.C.; Merrifield, C.A.; Berger, B.; Cloarec, O.; Duncker, S.; Mercenier, A.; Nicholson, J.K.; Holmes, E.; Bailey, M. Early intervention with Bifidobacterium lactis NCC2818 modulates the host-microbe interface independent of the sustained changes induced by the neonatal environment. Sci. Rep. 2017, 7, 5310. [Google Scholar] [CrossRef]
- Kendall, R.V.L.; John, W. Recent Findings on N, N-Dimethylglycine (DMG): A Nutrient for the New Millennium. Townsend Lett. Dr. Patients 2000, 202, 75–85. [Google Scholar]
- Shoji, H.; Taka, H.; Kaga, N.; Ikeda, N.; Hisata, K.; Miura, Y.; Shimizu, T. Choline-related metabolites influenced by feeding patterns in preterm and term infants. J. Matern.-Fetal Neonatal Med. 2020, 33, 230–235. [Google Scholar] [CrossRef]
- Mehta, A.K.; Gaur, S.N.; Arora, N.; Singh, B.P. Effect of choline chloride in allergen-induced mouse model of airway inflammation. Eur. Respir. J. 2007, 30, 662–671. [Google Scholar] [CrossRef]
- Platell, C.; Kong, S.E.; McCauley, R.; Hall, J.C. Branched-chain amino acids. J. Gastroenterol. Hepatol. 2000, 15, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Tu, C.; Gong, W.; Jia, J.; Zhang, B.; Mi, S.; Zhang, L.; Xie, X.; Guo, H. Serum Metabolomic Profiling of Piglets Infected with Virulent Classical Swine Fever Virus. Front. Microbiol. 2017, 8, 731. [Google Scholar]
- O’Brien, W.E. Inhibition of glycine synthase by branched-chain α-keto acids: A possible mechanism for abnormal glycine metabolism in ketotic hyperglycinemia. Arch. Biochem. Biophys. 1978, 189, 291–297. [Google Scholar] [CrossRef]
- Gehlhar, K.; Rajashankar, K.R.; Hofmann, E.; Betzel, C.; Weber, W.; Werner, S.; Bufe, A. Lysine as a critical amino acid for IgE binding in Phl p 5b C terminus. Int. Arch. Allergy Immunol. 2006, 140, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, R.; Jackowski, S. Biosynthesis of Pantothenic Acid and Coenzyme A. EcoSal Plus 2007, 2. [Google Scholar] [CrossRef] [Green Version]
- Ryan, D.G.; O’Neill, L.A.J. Krebs cycle rewired for macrophage and dendritic cell effector functions. FEBS Lett. 2017, 591, 2992–3006. [Google Scholar] [CrossRef] [Green Version]
- Agosti, M.; Tandoi, F.; Morlacchi, L.; Bossi, A. Nutritional and metabolic programming during the first thousand days of life. Pediatr. Med. Chir. 2017, 39, 157. [Google Scholar] [CrossRef] [Green Version]
- Ballardini, N.; Nilsson, C.; Nilsson, M.; Lilja, G. ImmunoCAP Phadiatop Infant—A new blood test for detecting IgE sensitisation in children at 2 years of age. Allergy 2006, 61, 337–343. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Lin, G.; Cheng, M.L.; Chiang, M.H.; Tsai, M.H.; Su, K.W.; Hua, M.C.; Liao, S.L.; Lai, S.H.; Yao, T.C.; et al. Longitudinal urinary metabolomic profiling reveals metabolites for asthma development in early childhood. Pediatr. Allergy Immunol. 2018, 29, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.Y.; Yeh, K.W.; Lin, G.; Chiang, M.H.; Yang, S.C.; Chao, W.J.; Yao, T.C.; Tsai, M.H.; Hua, M.C.; Liao, S.L.; et al. Metabolomics Reveals Dynamic Metabolic Changes Associated with Age in Early Childhood. PLoS ONE 2016, 11, e0149823. [Google Scholar]
- Heavner, D.L.; Morgan, W.T.; Sears, S.B.; Richardson, J.D.; Byrd, G.D.; Ogden, M.W. Effect of creatinine and specific gravity normalization techniques on xenobiotic biomarkers in smokers’ spot and 24-h urines. J. Pharm. Biomed. Anal. 2006, 40, 928–942. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Breastfeeding (n = 33) | Formula Feeding (n = 22) | p Value |
|---|---|---|---|
| Family | |||
| Maternal atopy | 16 (48.5%) | 9 (40.9%) | 0.580 |
| Paternal atopy | 18 (54.5%) | 10 (45.5%) | 0.509 |
| Parental smoking | 11 (33.3%) | 12 (54.5%) | 0.118 |
| Household income | |||
| Low, <500,000 NTD | 13 (39.4%) | 12 (54.5%) | 0.517 |
| Medium, 500,000–1,000,000 NTD | 15 (45.5%) | 8 (36.4%) | |
| High, >1,000,000 NTD | 5 (15.2%) | 2 (9.1%) | |
| Infant | |||
| Sex, male (%) | 19 (57.6%) | 15 (68.2%) | 0.428 |
| Maternal age (yr) | 31.2 ± 4.3 | 30.3 ± 4.9 | 0.442 |
| Gestational age (wk) | 38.5 ± 1.6 | 38.2 ± 1.8 | 0.578 |
| Birth BMI (kg/m2) | 12.1 ± 1.1 | 13.6 ± 4.3 | 0.123 |
| Milk sensitization | |||
| 6 mo | 4 (15.4%) | 3 (23.1%) | 0.666 |
| 1 yr | 7 (26.9%) | 11 (57.9%) | 0.036 |
| 2 yr | 11 (33.3%) | 13 (59.1%) | 0.106 |
| Formula Feeding | p | Milk Sensitization | p | ||||
|---|---|---|---|---|---|---|---|
| Metabolites | Chemical Shift, ppm (Multiplicity a) | VIP Score b | Fold change c | VIP Score | Fold Change | ||
| Allantoin | 5.39–5.40 (s) | 2.22 | 0.48 | 0.001 | 1.77 | 0.75 | 0.060 |
| 2-Hydroxyisobutyric acid | 1.35–1.37 (s) | 1.01 | 0.83 | 0.004 | 0.22 | 0.99 | 0.658 |
| Threonine | 4.25–4.27 (d) | 1.16 | 0.79 | 0.004 | 0.58 | 0.88 | 0.327 |
| Dimethylamine | 2.71–2.73 (s) | 0.92 | 0.85 | 0.006 | 0.48 | 1.07 | 0.321 |
| Valine | 1.04–1.05 (d) | 0.87 | 0.85 | 0.014 | 0.42 | 0.96 | 0.406 |
| Dimethyl sulfone | 3.15–3.16 (s) | 1.13 | 0.77 | 0.015 | 0.24 | 0.93 | 0.714 |
| Adipate | 1.55–1.56 (m) | 1.15 | 0.72 | 0.018 | 0.39 | 1.19 | 0.576 |
| N-Acetyltyrosine | 7.16–7.18 (d) | 1.63 | 0.51 | 0.020 | 1.56 | 0.59 | 0.114 |
| Creatine | 3.93–3.94 (s) | 1.34 | 0.75 | 0.043 | 0.92 | 0.98 | 0.325 |
| Propylene glycol | 1.14–1.15 (d) | 1.47 | 1.29 | 0.049 | 1.76 | 1.51 | 0.091 |
| Glutarate | 1.76–1.80 (tt) | 2.11 | 0.63 | <0.001 | 1.26 | 0.81 | 0.039 |
| Lysine | 1.89–1.91 (m) | 1.75 | 0.70 | <0.001 | 1.14 | 0.83 | 0.039 |
| 3-Methyl-2-oxovaleric acid | 1.10–1.11 (d) | 2.07 | 0.58 | <0.001 | 1.81 | 0.68 | 0.009 |
| N-Phenylacetylglycine | 7.41–7.45 (s) | 2.54 | 0.38 | <0.001 | 2.59 | 0.43 | 0.005 |
| N,N-Dimethylglycine | 2.93–2.93 (s) | 1.66 | 1.52 | 0.001 | 1.94 | 1.44 | 0.004 |
| 3-Indoxysulfate | 7.50–7.52 (d) | 2.06 | 0.45 | 0.003 | 2.25 | 0.48 | 0.024 |
| 2-Oxoglutaric acid | 3.00–3.01 (t) | 1.32 | 1.45 | 0.008 | 1.95 | 1.56 | 0.004 |
| Pantothenate | 0.93–0.94 (d) | 1.04 | 0.81 | 0.010 | 1.17 | 0.85 | 0.039 |
| Fumarate | 6.52–6.53 (s) | 1.58 | 1.41 | 0.054 | 2.41 | 1.55 | 0.033 |
| Pathway | Metabolites | Pathway Name | Total | Hits | Raw p | FDR | Function |
|---|---|---|---|---|---|---|---|
| Non-IgE related | 2-Oxoglutaric acid | D-glutamine and D-glutamate metabolism | 6 | 1 | 0.015 | 0.85 | Amino acid metabolism |
| Arginine biosynthesis | 14 | 1 | 0.036 | 0.85 | Amino acid metabolism | ||
| Butanoate metabolism | 15 | 1 | 0.038 | 0.85 | Carbohydrate metabolism | ||
| Pantothenate | Pantothenate and CoA biosynthesis | 19 | 1 | 0.048 | 0.85 | Metabolism of cofactors and vitamins | |
| IgE-related | Lysine | Biotin metabolism | 10 | 1 | 0.019 | 1.00 | Metabolism of cofactors and vitamins |
| Lysine degradation | 25 | 1 | 0.048 | 1.00 | Amino acid metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, C.-M.; Lin, G.; Chiang, M.-H.; Yeh, K.-W.; Huang, J.-L.; Su, K.-W.; Tsai, M.-H.; Hua, M.-C.; Liao, S.-L.; Lai, S.-H.; et al. Longitudinal Metabolomic Analysis Reveals Gut Microbial-Derived Metabolites Related to Formula Feeding and Milk Sensitization Development in Infancy. Metabolites 2022, 12, 127. https://doi.org/10.3390/metabo12020127
Tang C-M, Lin G, Chiang M-H, Yeh K-W, Huang J-L, Su K-W, Tsai M-H, Hua M-C, Liao S-L, Lai S-H, et al. Longitudinal Metabolomic Analysis Reveals Gut Microbial-Derived Metabolites Related to Formula Feeding and Milk Sensitization Development in Infancy. Metabolites. 2022; 12(2):127. https://doi.org/10.3390/metabo12020127
Chicago/Turabian StyleTang, Ching-Min, Gigin Lin, Meng-Han Chiang, Kuo-Wei Yeh, Jing-Long Huang, Kuan-Wen Su, Ming-Han Tsai, Man-Chin Hua, Sui-Ling Liao, Shen-Hao Lai, and et al. 2022. "Longitudinal Metabolomic Analysis Reveals Gut Microbial-Derived Metabolites Related to Formula Feeding and Milk Sensitization Development in Infancy" Metabolites 12, no. 2: 127. https://doi.org/10.3390/metabo12020127
APA StyleTang, C. -M., Lin, G., Chiang, M. -H., Yeh, K. -W., Huang, J. -L., Su, K. -W., Tsai, M. -H., Hua, M. -C., Liao, S. -L., Lai, S. -H., & Chiu, C. -Y. (2022). Longitudinal Metabolomic Analysis Reveals Gut Microbial-Derived Metabolites Related to Formula Feeding and Milk Sensitization Development in Infancy. Metabolites, 12(2), 127. https://doi.org/10.3390/metabo12020127






