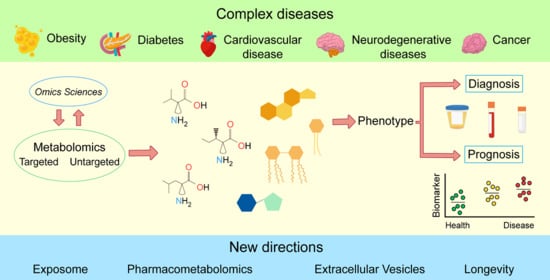
The Potential of Metabolomics in Biomedical Applications
Abstract
:1. Introduction
2. The Potential of Metabolomics in Biomedical Applications
3. General Strategies in Metabolomics
4. Metabolomics in Current Disease Research
4.1. Obesity
4.2. Diabetes
4.3. Cardiovascular Disease
4.4. Cancer
- (i)
- the identification of next-generation prognostic and therapeutic biomarkers by establishing a chemical and morphological mapping of regions of interest,
- (ii)
- the evaluation of the molecular efficacy of chemotherapeutic agents, and
- (iii)
- the classification of tissue types based on molecular patterns to understand their pathways and therapeutic prognoses [120].
4.5. The Metabolomics of Neurodegenerative Diseases
5. New Directions in Metabolomics
5.1. The Exposome
5.2. Pharmacometabolomics
5.3. Metabolomics and Extracellular Vesicles
5.4. Metabolomics and Longevity
6. Discussion
- (i)
- different analytical platforms, such as NMR, gas chromatography, liquid chromatography, and MS;
- (ii)
- various identified metabolites, such as lipids, amino acids, proteomics, glycomics, etc.;
- (iii)
- different matrices, such as blood, plasma, CSF, urine, tissue, tumors, etc.
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Brem, R.B.; Kruglyak, L. The landscape of genetic complexity across 5700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 2005, 102, 1572–1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oresic, M.; McGlinchey, A.; Wheelock, C.E.; Hyotylainen, T. Metabolic Signatures of the Exposome-Quantifying the Impact of Exposure to Environmental Chemicals on Human Health. Metabolites 2020, 10, 454. [Google Scholar] [CrossRef] [PubMed]
- Neavin, D.; Kaddurah-Daouk, R.; Weinshilboum, R. Pharmacometabolomics informs pharmacogenomics. Metabolomics 2016, 12, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J. Systems Biology of Metabolism. Annu. Rev. Biochem. 2017, 86, 245–275. [Google Scholar] [CrossRef] [PubMed]
- Powers, R. NMR metabolomics and drug discovery. Magn. Reson. Chem. 2009, 47 (Suppl 1), S2–S11. [Google Scholar] [CrossRef]
- Zhang, G.F.; Sadhukhan, S.; Tochtrop, G.P.; Brunengraber, H. Metabolomics, pathway regulation, and pathway discovery. J. Biol. Chem. 2011, 286, 23631–23635. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Harrigan, G.G.; Goodacre, R. Introduction. In Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis; Harrigan, G.G., Goodacre, R., Eds.; Springer: Boston, MA, USA, 2003; pp. 1–8. [Google Scholar]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Mussap, M.; Zaffanello, M.; Fanos, V. Metabolomics: A challenge for detecting and monitoring inborn errors of metabolism. Ann. Transl. Med. 2018, 6, 338. [Google Scholar] [CrossRef] [PubMed]
- Spratlin, J.L.; Serkova, N.J.; Eckhardt, S.G. Clinical applications of metabolomics in oncology: A review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, W.J.; Koal, T.; Wang, Y.; Kohl, M.; Enot, D.P.; Deigner, H.P. Targeted metabolomics for biomarker discovery. Angew. Chem. Int. Ed. Engl. 2010, 49, 5426–5445. [Google Scholar] [CrossRef]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Vijayakumar, A.; Kahn, B.B. Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol. 2018, 19, 654–672. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Ortmayr, K.; Dubuis, S.; Zampieri, M. Metabolic profiling of cancer cells reveals genome-wide crosstalk between transcriptional regulators and metabolism. Nat. Commun. 2019, 10, 1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.; Shim, H.; Shin, D.; Shim, J.E.; Ko, Y.; Shin, J.; Kim, H.; Cho, A.; Kim, E.; Lee, T.; et al. TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 2015, 5, 11432. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Simons, K. Lipidomics: Coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010, 11, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Schnackenberg, L.K.; Holland, R.D.; Li, D.; Dragan, Y. Metabonomic models of human pancreatic cancer using 1D proton NMR spectra of lipids in plasma. Metabolomics 2006, 2, 125–134. [Google Scholar] [CrossRef]
- Bathe, O.F.; Shaykhutdinov, R.; Kopciuk, K.; Weljie, A.M.; McKay, A.; Sutherland, F.R.; Dixon, E.; Dunse, N.; Sotiropoulos, D.; Vogel, H.J. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol. Biomark. Prev. 2011, 20, 140–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckonert, O.; Coen, M.; Keun, H.C.; Wang, Y.; Ebbels, T.M.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat. Protoc. 2010, 5, 1019–1032. [Google Scholar] [CrossRef]
- Bharti, S.K.; Jaiswal, V.; Ghoshal, U.; Ghoshal, U.C.; Baijal, S.S.; Roy, R.; Khetrapal, C.L. Metabolomic profiling of amoebic and pyogenic liver abscesses: An in vitro NMR study. Metabolomics 2012, 8, 540–555. [Google Scholar] [CrossRef]
- Ramadan, Z.; Jacobs, D.; Grigorov, M.; Kochhar, S. Metabolic profiling using principal component analysis, discriminant partial least squares, and genetic algorithms. Talanta 2006, 68, 1683–1691. [Google Scholar] [CrossRef]
- Monleon, D.; Morales, J.M.; Barrasa, A.; Lopez, J.A.; Vazquez, C.; Celda, B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009, 22, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Auray-Blais, C.; Raiche, E.; Gagnon, R.; Berthiaume, M.; Pasquier, J.-C. Metabolomics and preterm birth: What biomarkers in cervicovaginal secretions are predictive of high-risk pregnant women? Int. J. Mass Spectrom. 2011, 307, 33–38. [Google Scholar] [CrossRef]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, C.; Shi, Y.; Zhang, Y.; Jin, D.; Zhang, M.; Bo, M.; Li, G. A Comprehensive Analysis of Metabolomics and Transcriptomics Reveals Novel Biomarkers and Mechanistic Insights on Lorlatinib Crosses the Blood-Brain Barrier. Front. Pharmacol. 2021, 12, 722627. [Google Scholar] [CrossRef]
- Wang, K.; Gaitsch, H.; Poon, H.; Cox, N.J.; Rzhetsky, A. Classification of common human diseases derived from shared genetic and environmental determinants. Nat. Genet. 2017, 49, 1319–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Povedano, M.M.; Calderon-Santiago, M.; Luque de Castro, M.D.; Priego-Capote, F. Metabolomics analysis of human sweat collected after moderate exercise. Talanta 2018, 177, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.; Carpenter, G.; So, P.W. Salivary Metabolomics: From Diagnostic Biomarker Discovery to Investigating Biological Function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serkova, N.J.; Van Rheen, Z.; Tobias, M.; Pitzer, J.E.; Wilkinson, J.E.; Stringer, K.A. Utility of magnetic resonance imaging and nuclear magnetic resonance-based metabolomics for quantification of inflammatory lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2008, 295, L152–L161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dettmer, K.; Aronov, P.A.; Hammock, B.D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. [Google Scholar] [CrossRef] [PubMed]
- Gebregiworgis, T.; Powers, R. Application of NMR metabolomics to search for human disease biomarkers. Comb. Chem. High Throughput Screen 2012, 15, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30.2.1–30.2.24. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Lopez, E.; Lomonaco, T.; Sugimoto, M.; Qiu, Y.; Campanella, B. Editorial: Metabolomics in the Study of Unconventional Biological Matrices. Front. Chem. 2021, 9, 736661. [Google Scholar] [CrossRef] [PubMed]
- Losacco, G.L.; Ismail, O.; Pezzatti, J.; Gonzalez-Ruiz, V.; Boccard, J.; Rudaz, S.; Veuthey, J.L.; Guillarme, D. Applicability of Supercritical fluid chromatography-Mass spectrometry to metabolomics. II-Assessment of a comprehensive library of metabolites and evaluation of biological matrices. J. Chromatogr. A 2020, 1620, 461021. [Google Scholar] [CrossRef]
- Theodoridis, G.A.; Gika, H.G.; Wilson, I.D. (Eds.) Metabolic Profiling; Humana Press: New York, NY, USA, 2018; p. 291. [Google Scholar]
- Kumar, A.; Misra, B.B. Challenges and Opportunities in Cancer Metabolomics. Proteomics 2019, 19, e1900042. [Google Scholar] [CrossRef] [PubMed]
- WHO. World Health Statistics 2020: Monitoring Health for the SDGs, Sustainable Development Goals; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Rangel-Huerta, O.D.; Pastor-Villaescusa, B.; Gil, A. Are we close to defining a metabolomic signature of human obesity? A systematic review of metabolomics studies. Metabolomics 2019, 15, 93. [Google Scholar] [CrossRef] [Green Version]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.H.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Crosslin, D.R.; Haynes, C.; Dungan, J.; Newby, L.K.; Hauser, E.R.; Ginsburg, G.S.; et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ. Cardiovasc. Genet. 2010, 3, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasa, M.; Ishihara, T.; Mifuji-Moroka, R.; Fujita, N.; Kobayashi, Y.; Hasegawa, H.; Iwata, K.; Kaito, M.; Takei, Y. Elevation of branched-chain amino acid levels in diabetes and NAFL and changes with antidiabetic drug treatment. Obes. Res. Clin. Pract. 2015, 9, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Floegel, A.; Stefan, N.; Yu, Z.; Muhlenbruch, K.; Drogan, D.; Joost, H.G.; Fritsche, A.; Haring, H.U.; Hrabe de Angelis, M.; Peters, A.; et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013, 62, 639–648. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.L.; Drysdale, M.; Baimel, C.; Kaur, M.; MacGowan, T.; Pitman, K.A.; Borgland, S.L. Obesity-Induced Structural and Neuronal Plasticity in the Lateral Orbitofrontal Cortex. Neuropsychopharmacology 2017, 42, 1480–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yengo, L.; Arredouani, A.; Marre, M.; Roussel, R.; Vaxillaire, M.; Falchi, M.; Haoudi, A.; Tichet, J.; D.E.S.I.R Study Group; Balkau, B.; et al. Impact of statistical models on the prediction of type 2 diabetes using non-targeted metabolomics profiling. Mol. Metab. 2016, 5, 918–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhre, K.; Meisinger, C.; Doring, A.; Altmaier, E.; Belcredi, P.; Gieger, C.; Chang, D.; Milburn, M.V.; Gall, W.E.; Weinberger, K.M.; et al. Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS ONE 2010, 5, e13953. [Google Scholar] [CrossRef] [Green Version]
- Ibarra-Gonzalez, I.; Cruz-Bautista, I.; Bello-Chavolla, O.Y.; Vela-Amieva, M.; Pallares-Mendez, R.; Ruiz de Santiago, Y.N.D.; Salas-Tapia, M.F.; Rosas-Flota, X.; Gonzalez-Acevedo, M.; Palacios-Penaloza, A.; et al. Optimization of kidney dysfunction prediction in diabetic kidney disease using targeted metabolomics. Acta Diabetol. 2018, 55, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Barrios, C.; Zierer, J.; Wurtz, P.; Haller, T.; Metspalu, A.; Gieger, C.; Thorand, B.; Meisinger, C.; Waldenberger, M.; Raitakari, O.; et al. Circulating metabolic biomarkers of renal function in diabetic and non-diabetic populations. Sci. Rep. 2018, 8, 15249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, C.; Qiu, L.; Wang, Y.; Qin, X.; Liu, Y.; Li, Z. Serum Unsaturated Free Fatty Acids: A Potential Biomarker Panel for Early-Stage Detection of Colorectal Cancer. J. Cancer 2016, 7, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudka, I.; Thysell, E.; Lundquist, K.; Antti, H.; Iglesias-Gato, D.; Flores-Morales, A.; Bergh, A.; Wikstrom, P.; Grobner, G. Comprehensive metabolomics analysis of prostate cancer tissue in relation to tumor aggressiveness and TMPRSS2-ERG fusion status. BMC Cancer 2020, 20, 437. [Google Scholar] [CrossRef]
- Moran-Ramos, S.; Ocampo-Medina, E.; Gutierrez-Aguilar, R.; Macias-Kauffer, L.; Villamil-Ramirez, H.; Lopez-Contreras, B.E.; Leon-Mimila, P.; Vega-Badillo, J.; Gutierrez-Vidal, R.; Villarruel-Vazquez, R.; et al. An Amino Acid Signature Associated with Obesity Predicts 2-Year Risk of Hypertriglyceridemia in School-Age Children. Sci. Rep. 2017, 7, 5607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurtz, P.; Makinen, V.P.; Soininen, P.; Kangas, A.J.; Tukiainen, T.; Kettunen, J.; Savolainen, M.J.; Tammelin, T.; Viikari, J.S.; Ronnemaa, T.; et al. Metabolic signatures of insulin resistance in 7098 young adults. Diabetes 2012, 61, 1372–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, X.; Wu, J.; Kim, S.; LoRusso, P.; Li, J. Pharmacometabolomics Reveals Irinotecan Mechanism of Action in Cancer Patients. J. Clin. Pharmacol. 2019, 59, 20–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamaziere, A.; Wolf, C.; Quinn, P.J. How lipidomics provides new insight into drug discovery. Expert Opin. Drug Discov. 2014, 9, 819–836. [Google Scholar] [CrossRef] [PubMed]
- Katsila, T.; Balasopoulou, A.; Tsagaraki, I.; Patrinos, G.P. Pharmacomicrobiomics informs clinical pharmacogenomics. Pharmacogenomics 2019, 20, 731–739. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef]
- Deelen, J.; Kettunen, J.; Fischer, K.; van der Spek, A.; Trompet, S.; Kastenmüller, G.; Boyd, A.; Zierer, J.; van den Akker, E.B.; Ala-Korpela, M.; et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun. 2019, 10, 3346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Domínguez, R.; García-Barrera, T.; Gómez-Ariza, J.L. Metabolomic study of lipids in serum for biomarker discovery in Alzheimer’s disease using direct infusion mass spectrometry. J. Pharm. Biomed. Anal. 2014, 98, 321–326. [Google Scholar] [CrossRef]
- Kim, D.H.; Gim, J.A.; Yoon, D.; Kim, S.; Kim, H.S. Metabolomics and mitochondrial dysfunction in Alzheimer’s disease. Genes Genom. 2017, 39, 295–300. [Google Scholar] [CrossRef]
- Chatterjee, P.; Cheong, Y.J.; Bhatnagar, A.; Goozee, K.; Wu, Y.; McKay, M.; Martins, I.J.; Lim, W.L.F.; Pedrini, S.; Tegg, M.; et al. Plasma metabolites associated with biomarker evidence of neurodegeneration in cognitively normal older adults. J. Neurochem. 2020, 159, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Nho, K.; Kueider-Paisley, A.; MahmoudianDehkordi, S.; Arnold, M.; Risacher, S.L.; Louie, G.; Blach, C.; Baillie, R.; Han, X.; Kastenmüller, G.; et al. Altered bile acid profile in mild cognitive impairment and Alzheimer’s disease: Relationship to neuroimaging and CSF biomarkers. Alzheimers Dement. 2019, 15, 232–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tondo, M.; Wasek, B.; Escola-Gil, J.C.; de Gonzalo-Calvo, D.; Harmon, C.; Arning, E.; Bottiglieri, T. Altered brain metabolome is associated with memory impairment in the rTG4510 mouse model of tauopathy. Metabolites 2020, 10, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picca, A.; Calvani, R.; Landi, G.; Marini, F.; Biancolillo, A.; Gervasoni, J.; Persichilli, S.; Primiano, A.; Urbani, A.; Bossola, M.; et al. Circulating amino acid signature in older people with Parkinson’s disease: A metabolic complement to the EXosomes in PArkiNson Disease (EXPAND) study. Exp. Gerontol. 2019, 128, 110766. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Goyal, V.; Kumaran, S.S.; Dwivedi, S.N.; Srivastava, A.; Jagannathan, N.R. Quantitative metabolomics of saliva using proton NMR spectroscopy in patients with Parkinson’s disease and healthy controls. Neurol. Sci. 2020, 41, 1201–1210. [Google Scholar] [CrossRef]
- Musgrove, R.E.; Horne, J.; Wilson, R.; King, A.E.; Edwards, L.M.; Dickson, T.C. The metabolomics of alpha-synuclein (SNCA) gene deletion and mutation in mouse brain. Metabolomics 2014, 10, 114–122. [Google Scholar] [CrossRef]
- Chen, X.; Xie, C.; Sun, L.; Ding, J.; Cai, H. Longitudinal metabolomics profiling of Parkinson’s disease-related α-synuclein A53T transgenic mice. PLoS ONE 2015, 10, e0136612. [Google Scholar] [CrossRef]
- Huang, W.; Xu, Y.; Zhang, Y.; Zhang, P.; Zhang, Q.; Zhang, Z.; Xu, F. Metabolomics-driven identification of adenosine deaminase as therapeutic target in a mouse model of Parkinson’s disease. J. Neurochem. 2019, 150, 282–295. [Google Scholar] [CrossRef] [PubMed]
- Bocarsly, M.E.; Fasolino, M.; Kane, G.A.; LaMarca, E.A.; Kirschen, G.W.; Karatsoreos, I.N.; McEwen, B.S.; Gould, E. Obesity diminishes synaptic markers, alters microglial morphology, and impairs cognitive function. Proc. Natl. Acad. Sci. USA 2015, 112, 15731–15736. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Covarrubias, V. Lipidomics in longevity and healthy aging. Biogerontology 2013, 14, 663–672. [Google Scholar] [CrossRef]
- Kiliaan, A.J.; Arnoldussen, I.A.; Gustafson, D.R. Adipokines: A link between obesity and dementia? Lancet Neurol. 2014, 13, 913–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parimisetty, A.; Dorsemans, A.C.; Awada, R.; Ravanan, P.; Diotel, N.; Lefebvre d’Hellencourt, C. Secret talk between adipose tissue and central nervous system via secreted factors-an emerging frontier in the neurodegenerative research. J. Neuroinflamm. 2016, 13, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tynkkynen, J.; Chouraki, V.; van der Lee, S.J.; Hernesniemi, J.; Yang, Q.; Li, S.; Beiser, A.; Larson, M.G.; Saaksjarvi, K.; Shipley, M.J.; et al. Association of branched-chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer’s disease: A prospective study in eight cohorts. Alzheimers Dement. 2018, 14, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Arnoldussen, I.A.; Kiliaan, A.J.; Gustafson, D.R. Obesity and dementia: Adipokines interact with the brain. Eur. Neuropsychopharmacol. 2014, 24, 1982–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Castaner, O.; Goday, A.; Park, Y.M.; Lee, S.H.; Magkos, F.; Shiow, S.T.E.; Schroder, H. The Gut Microbiome Profile in Obesity: A Systematic Review. Int. J. Endocrinol. 2018, 2018, 4095789. [Google Scholar] [CrossRef] [PubMed]
- Stanislawski, M.A.; Dabelea, D.; Lange, L.A.; Wagner, B.D.; Lozupone, C.A. Gut microbiota phenotypes of obesity. NPJ Biofilms Microbiomes 2019, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Yun, K.E.; Kim, J.; Park, E.; Chang, Y.; Ryu, S.; Kim, H.L.; Kim, H.N. Gut microbiota and metabolic health among overweight and obese individuals. Sci. Rep. 2020, 10, 19417. [Google Scholar] [CrossRef] [PubMed]
- Arnoriaga-Rodriguez, M.; Mayneris-Perxachs, J.; Burokas, A.; Contreras-Rodriguez, O.; Blasco, G.; Coll, C.; Biarnes, C.; Miranda-Olivos, R.; Latorre, J.; Moreno-Navarrete, J.M.; et al. Obesity Impairs Short-Term and Working Memory through Gut Microbial Metabolism of Aromatic Amino Acids. Cell Metab. 2020, 32, 548–560.e7. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, A.; Di Cesare Mannelli, L.; Lucarini, E.; Man, A.L.; Le Gall, G.; Branca, J.J.V.; Ghelardini, C.; Amedei, A.; Bertelli, E.; Regoli, M.; et al. Faecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity- and neurotransmission-related proteins in young recipients. Microbiome 2020, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.H.; Crosslin, D.R.; Haynes, C.S.; Nelson, S.; Turer, C.B.; Stevens, R.D.; Muehlbauer, M.J.; Wenner, B.R.; Bain, J.R.; Laferrere, B.; et al. Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia 2012, 55, 321–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knebel, B.; Strassburger, K.; Szendroedi, J.; Kotzka, J.; Scheer, M.; Nowotny, B.; Mussig, K.; Lehr, S.; Pacini, G.; Finner, H.; et al. Specific Metabolic Profiles and Their Relationship to Insulin Resistance in Recent-Onset Type 1 and Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 2130–2140. [Google Scholar] [CrossRef] [PubMed]
- Haukka, J.K.; Sandholm, N.; Forsblom, C.; Cobb, J.E.; Groop, P.H.; Ferrannini, E. Metabolomic Profile Predicts Development of Microalbuminuria in Individuals with Type 1 Diabetes. Sci. Rep. 2018, 8, 13853. [Google Scholar] [CrossRef]
- Arneth, B.; Arneth, R.; Shams, M. Metabolomics of Type 1 and Type 2 Diabetes. Int. J. Mol. Sci. 2019, 20, 2467. [Google Scholar] [CrossRef] [Green Version]
- Vijan, S.; Sussman, J.B.; Yudkin, J.S.; Hayward, R.A. Effect of patients’ risks and preferences on health gains with plasma glucose level lowering in type 2 diabetes mellitus. JAMA Intern. Med. 2014, 174, 1227–1234. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K. Metabolic profiling in diabetes. J. Endocrinol. 2014, 221, R75–R85. [Google Scholar] [CrossRef] [Green Version]
- Gudmundsdottir, V.; Zaghlool, S.B.; Emilsson, V.; Aspelund, T.; Ilkov, M.; Gudmundsson, E.F.; Jonsson, S.M.; Zilhao, N.R.; Lamb, J.R.; Suhre, K.; et al. Circulating Protein Signatures and Causal Candidates for Type 2 Diabetes. Diabetes 2020, 69, 1843–1853. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, S.J.; Levin, D.; Looker, H.C.; Lindsay, R.S.; Wild, S.H.; Joss, N.; Leese, G.; Leslie, P.; McCrimmon, R.J.; Metcalfe, W.; et al. Estimated life expectancy in a Scottish cohort with type 1 diabetes, 2008–2010. JAMA 2015, 313, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Floegel, A.; von Ruesten, A.; Drogan, D.; Schulze, M.B.; Prehn, C.; Adamski, J.; Pischon, T.; Boeing, H. Variation of serum metabolites related to habitual diet: A targeted metabolomic approach in EPIC-Potsdam. Eur. J. Clin. Nutr. 2013, 67, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Pallares-Mendez, R.; Aguilar-Salinas, C.A.; Cruz-Bautista, I.; Del Bosque-Plata, L. Metabolomics in diabetes, a review. Ann. Med. 2016, 48, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Regan, J.A.; Shah, S.H. Obesity Genomics and Metabolomics: A Nexus of Cardiometabolic Risk. Curr. Cardiol. Rep. 2020, 22, 174. [Google Scholar] [CrossRef] [PubMed]
- McGarrah, R.W.; Crown, S.B.; Zhang, G.F.; Shah, S.H.; Newgard, C.B. Cardiovascular Metabolomics. Circ. Res. 2018, 122, 1238–1258. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.E.; Muoio, D.M.; Stevens, R.; Craig, D.; Bain, J.R.; Grass, E.; Haynes, C.; Kwee, L.; Qin, X.; Slentz, D.H.; et al. Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis. PLoS Genet. 2015, 11, e1005553. [Google Scholar] [CrossRef] [Green Version]
- Mayr, M.; Madhu, B.; Xu, Q. Proteomics and metabolomics combined in cardiovascular research. Trends Cardiovasc. Med. 2007, 17, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Mayr, M.; Chung, Y.L.; Mayr, U.; McGregor, E.; Troy, H.; Baier, G.; Leitges, M.; Dunn, M.J.; Griffiths, J.R.; Xu, Q. Loss of PKC-delta alters cardiac metabolism. Am. J. Physiol. Heart Circ. Physiol. 2004, 287, H937–H945. [Google Scholar] [CrossRef] [PubMed]
- Mayr, M.; Liem, D.; Zhang, J.; Li, X.; Avliyakulov, N.K.; Yang, J.I.; Young, G.; Vondriska, T.M.; Ladroue, C.; Madhu, B.; et al. Proteomic and metabolomic analysis of cardioprotection: Interplay between protein kinase C epsilon and delta in regulating glucose metabolism of murine hearts. J. Mol. Cell Cardiol. 2009, 46, 268–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayr, M.; Metzler, B.; Chung, Y.L.; McGregor, E.; Mayr, U.; Troy, H.; Hu, Y.; Leitges, M.; Pachinger, O.; Griffiths, J.R.; et al. Ischemic preconditioning exaggerates cardiac damage in PKC-delta null mice. Am. J. Physiol. Heart Circ. Physiol. 2004, 287, H946–H956. [Google Scholar] [CrossRef] [Green Version]
- Mayr, M.; Chung, Y.L.; Mayr, U.; Yin, X.; Ly, L.; Troy, H.; Fredericks, S.; Hu, Y.; Griffiths, J.R.; Xu, Q. Simultaneous in vivo assessment of cardiacfrom apolipoprotein E-deficient mice reveal alterations in inflammation, oxidative stress, and energy metabolism. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 2135–2142. [Google Scholar] [CrossRef] [Green Version]
- Le Page, L.M.; Ball, D.R.; Ball, V.; Dodd, M.S.; Miller, J.J.; Heather, L.C.; Tyler, D.J. Simultaneous in vivo assessment of cardiac and hepatic metabolism in the diabetic rat using hyperpolarized MRS. NMR Biomed. 2016, 29, 1759–1767. [Google Scholar] [CrossRef] [PubMed]
- Bernini, P.; Bertini, I.; Luchinat, C.; Tenori, L.; Tognaccini, A. The cardiovascular risk of healthy individuals studied by NMR metabonomics of plasma samples. J. Proteome Res. 2011, 10, 4983–4992. [Google Scholar] [CrossRef] [PubMed]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.e22. [Google Scholar] [CrossRef] [PubMed]
- Karlstaedt, A.; Zhang, X.; Vitrac, H.; Harmancey, R.; Vasquez, H.; Wang, J.H.; Goodell, M.A.; Taegtmeyer, H. Oncometabolite d-2-hydroxyglutarate impairs alpha-ketoglutarate dehydrogenase and contractile function in rodent heart. Proc. Natl. Acad. Sci. USA 2016, 113, 10436–10441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGranaghan, P.; Saxena, A.; Rubens, M.; Radenkovic, J.; Bach, D.; Schleussner, L.; Pieske, B.; Edelmann, F.; Trippel, T.D. Predictive value of metabolomic biomarkers for cardiovascular disease risk: A systematic review and meta-analysis. Biomarkers 2020, 25, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Pascale, R.M.; Calvisi, D.F.; Simile, M.M.; Feo, C.F.; Feo, F. The Warburg Effect 97 Years after Its Discovery. Cancers 2020, 12, 2819. [Google Scholar] [CrossRef]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phan, L.M.; Yeung, S.C.; Lee, M.H. Cancer metabolic reprogramming: Importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [Green Version]
- El Sayed, R.; Haibe, Y.; Amhaz, G.; Bouferraa, Y.; Shamseddine, A. Metabolic Factors Affecting Tumor Immunogenicity: What Is Happening at the Cellular Level? Int. J. Mol. Sci. 2021, 22, 2142. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Huang, X.; Zhang, G.; Hong, Z.; Bai, X.; Liang, T. Advantages of targeting the tumor immune microenvironment over blocking immune checkpoint in cancer immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 72. [Google Scholar] [CrossRef] [PubMed]
- Ananieva, E. Targeting amino acid metabolism in cancer growth and anti-tumor immune response. World J. Biol. Chem. 2015, 6, 281–289. [Google Scholar] [CrossRef]
- Bleve, A.; Durante, B.; Sica, A.; Consonni, F.M. Lipid Metabolism and Cancer Immunotherapy: Immunosuppressive Myeloid Cells at the Crossroad. Int. J. Mol. Sci. 2020, 21, 5845. [Google Scholar] [CrossRef] [PubMed]
- Ciocan-Cartita, C.A.; Jurj, A.; Buse, M.; Gulei, D.; Braicu, C.; Raduly, L.; Cojocneanu, R.; Pruteanu, L.L.; Iuga, C.A.; Coza, O.; et al. The Relevance of Mass Spectrometry Analysis for Personalized Medicine through Its Successful Application in Cancer “Omics”. Int. J. Mol. Sci. 2019, 20, 2576. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Song, L.; Liu, N.; He, C.; Li, Z. Decreased serum levels of free fatty acids are associated with breast cancer. Clin. Chim. Acta 2014, 437, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yu, D. Metabolomics study of oral cancers. Metabolomics 2019, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, F.J.; Mrdjen, D.; McCaffrey, E.; Glass, D.R.; Greenwald, N.F.; Bharadwaj, A.; Khair, Z.; Verberk, S.G.S.; Baranski, A.; Baskar, R.; et al. Single-Cell Metab.olic profiling of human cytotoxic T cells. Nat. Biotechnol. 2021, 39, 186–197. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacKinnon, N.; Khan, A.P.; Chinnaiyan, A.M.; Rajendiran, T.M.; Ramamoorthy, A. Androgen receptor activation results in metabolite signatures of an aggressive prostate cancer phenotype: An NMR-based metabonomics study. Metabolomics 2012, 8, 1026–1036. [Google Scholar] [CrossRef]
- Struck, W.; Siluk, D.; Yumba-Mpanga, A.; Markuszewski, M.; Kaliszan, R.; Markuszewski, M.J. Liquid chromatography tandem mass spectrometry study of urinary nucleosides as potential cancer markers. J. Chromatogr. A 2013, 1283, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Naseri, M.T.; Babri, M. Quantification of candidate prostate cancer metabolite biomarkers in urine using dispersive derivatization liquid-liquid microextraction followed by gas and liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2013, 81-82, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Soliman, L.C.; Hui, Y.; Hewavitharana, A.K.; Chen, D.D. Monitoring potential prostate cancer biomarkers in urine by capillary electrophoresis-tandem mass spectrometry. J. Chromatogr. A 2012, 1267, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Alberice, J.V.; Amaral, A.F.; Armitage, E.G.; Lorente, J.A.; Algaba, F.; Carrilho, E.; Marquez, M.; Garcia, A.; Malats, N.; Barbas, C. Searching for urine biomarkers of bladder cancer recurrence using a liquid chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry metabolomics approach. J. Chromatogr. A 2013, 1318, 163–170. [Google Scholar] [CrossRef]
- Lam, C.W.; Law, C.Y.; To, K.K.; Cheung, S.K.; Lee, K.C.; Sze, K.H.; Leung, K.F.; Yuen, K.Y. NMR-based metabolomic urinalysis: A rapid screening test for urinary tract infection. Clin. Chim. Acta 2014, 436, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Davis, V.W.; Schiller, D.E.; Eurich, D.; Bathe, O.F.; Sawyer, M.B. Pancreatic ductal adenocarcinoma is associated with a distinct urinary metabolomic signature. Ann. Surg. Oncol. 2013, 20 (Suppl. 3), S415–S423. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [Green Version]
- Bonanomi, M.; Salmistraro, N.; Fiscon, G.; Conte, F.; Paci, P.; Bravata, V.; Forte, G.I.; Volpari, T.; Scorza, M.; Mastroianni, F.; et al. Transcriptomics and Metabolomics Integration Reveals Redox-Dependent Metabolic Rewiring in Breast Cancer Cells. Cancers 2021, 13, 5058. [Google Scholar] [CrossRef] [PubMed]
- LeWitt, P.A.; Li, J.; Lu, M.; Guo, L.; Auinger, P.; Parkinson Study Group, D.I. Metabolomic biomarkers as strong correlates of Parkinson disease progression. Neurology 2017, 88, 862–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varma, V.R.; Oommen, A.M.; Varma, S.; Casanova, R.; An, Y.; Andrews, R.M.; O’Brien, R.; Pletnikova, O.; Troncoso, J.C.; Toledo, J.; et al. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: A targeted metabolomics study. PLoS Med. 2018, 15, e1002482. [Google Scholar] [CrossRef]
- MahmoudianDehkordi, S.; Arnold, M.; Nho, K.; Ahmad, S.; Jia, W.; Xie, G.; Louie, G.; Kueider-Paisley, A.; Moseley, M.A.; Thompson, J.W.; et al. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimers Dement. 2019, 15, 76–92. [Google Scholar] [CrossRef]
- Cirstea, M.S.; Yu, A.C.; Golz, E.; Sundvick, K.; Kliger, D.; Radisavljevic, N.; Foulger, L.H.; Mackenzie, M.; Huan, T.; Finlay, B.B.; et al. Microbiota Composition and Metabolism Are Associated With Gut Function in Parkinson’s Disease. Mov. Disord. 2020, 35, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Karlíková, R.; Mičová, K.; Najdekr, L.; Gardlo, A.; Adam, T.; Majerová, P.; Friedecký, D.; Kováč, A. Metabolic status of CSF distinguishes rats with tauopathy from controls. Alzheimers Res. Ther. 2017, 9, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamid, Z.; Basit, A.; Pontis, S.; Piras, F.; Assogna, F.; Bossù, P.; Pontieri, F.E.; Stefani, A.; Spalletta, G.; Franceschi, P.; et al. Gender specific decrease of a set of circulating N-acylphosphatidyl ethanolamines (NAPEs) in the plasma of Parkinson’s disease patients. Metabolomics 2019, 15, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virel, A.; Dudka, I.; Laterveer, R.; Bjerkén, S.A. 1H NMR profiling of the 6-OHDA parkinsonian rat brain reveals metabolic alterations and signs of recovery after N-acetylcysteine treatment. Mol. Cell. Neurosci. 2019, 98, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Riano, C.; Saiz, J.; Barbas, C.; Bergareche, A.; Huerta, J.M.; Ardanaz, E.; Konjevod, M.; Mondragon, E.; Erro, M.E.; Chirlaque, M.D.; et al. Prognostic biomarkers of Parkinson’s disease in the Spanish EPIC cohort: A multiplatform metabolomics approach. NPJ Parkinsons Dis. 2021, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, E.; Trivedi, D.K.; Sarkar, D.; Walton-Doyle, C.; Milne, J.; Kunath, T.; Rijs, A.M.; de Bie, R.M.A.; Goodacre, R.; Silverdale, M.; et al. Metabolomics of sebum reveals lipid dysregulation in Parkinson’s disease. Nat. Commun. 2021, 12, 1592. [Google Scholar] [CrossRef]
- Teruya, T.; Chen, Y.J.; Kondoh, H.; Fukuji, Y.; Yanagida, M. Whole-blood metabolomics of dementia patients reveal classes of disease-linked metabolites. Proc. Natl. Acad. Sci. USA 2021, 118, e2022857118. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, S.M. Implications of the exposome for exposure science. J. Expo. Sci. Environ. Epidemiol. 2011, 21, 5–9. [Google Scholar] [CrossRef]
- Saracci, R.; Vineis, P. Disease proportions attributable to environment. Environ. Health 2007, 6, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willett, W.; Manson, J.; Liu, S. Glycemic index, glycemic load, and risk of type 2 diabetes. Am. J. Clin. Nutr. 2002, 76, 274S–280S. [Google Scholar] [CrossRef]
- Cohn, B.A.; Cirillo, P.M.; Terry, M.B. DDT and Breast Cancer: Prospective Study of Induction Time and Susceptibility Windows. J. Natl. Cancer Inst. 2019, 111, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Cohn, B.A.; La Merrill, M.; Krigbaum, N.Y.; Yeh, G.; Park, J.S.; Zimmermann, L.; Cirillo, P.M. DDT Exposure in Utero and Breast Cancer. J. Clin. Endocrinol. Metab. 2015, 100, 2865–2872. [Google Scholar] [CrossRef] [PubMed]
- Cohn, B.A.; Terry, M.B.; Plumb, M.; Cirillo, P.M. Exposure to polychlorinated biphenyl (PCB) congeners measured shortly after giving birth and subsequent risk of maternal breast cancer before age 50. Breast Cancer Res. Treat. 2012, 136, 267–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohn, B.A.; Wolff, M.S.; Cirillo, P.M.; Sholtz, R.I. DDT and breast cancer in young women: New data on the significance of age at exposure. Environ. Health Perspect. 2007, 115, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cirillo, P.; Hu, X.; Tran, V.; Krigbaum, N.; Yu, S.; Jones, D.P.; Cohn, B. Understanding mixed environmental exposures using metabolomics via a hierarchical community network model in a cohort of California women in 1960’s. Reprod. Toxicol. 2020, 92, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, J.H.; Fredrik Jarskog, L.; Vadlamudi, S.; Lauder, J.M. Prenatal infection and risk for schizophrenia: IL-1beta, IL-6, and TNFalpha inhibit cortical neuron dendrite development. Neuropsychopharmacology 2004, 29, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Thacher, J.D.; Gruzieva, O.; Pershagen, G.; Neuman, A.; Wickman, M.; Kull, I.; Melen, E.; Bergstrom, A. Pre- and postnatal exposure to parental smoking and allergic disease through adolescence. Pediatrics 2014, 134, 428–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, D.I.; Valvi, D.; Rothman, N.; Lan, Q.; Miller, G.W.; Jones, D.P. The metabolome: A key measure for exposome research in epidemiology. Curr. Epidemiol. Rep. 2019, 6, 93–103. [Google Scholar] [PubMed]
- Vermeulen, R.; Schymanski, E.L.; Barabasi, A.L.; Miller, G.W. The exposome and health: Where chemistry meets biology. Science 2020, 367, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.W.; Jones, D.P. The nature of nurture: Refining the definition of the exposome. Toxicol. Sci. 2014, 137, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Andres Lacueva, C.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F.; et al. Nutrimetabolomics: An Integrative Action for Metabolomic Analyses in Human Nutritional Studies. Mol. Nutr. Food Res. 2019, 63, e1800384. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.I.; Perry-Walker, K.; Finnell, R.H.; Pennell, K.D.; Tran, V.; May, R.C.; McElrath, T.F.; Meador, K.J.; Pennell, P.B.; Jones, D.P. Metabolome-wide association study of anti-epileptic drug treatment during pregnancy. Toxicol. Appl. Pharmacol. 2019, 363, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Carlsten, C.; Chaleckis, R.; Hanhineva, K.; Huang, M.; Isobe, T.; Koistinen, V.M.; Meister, I.; Papazian, S.; Sdougkou, K.; et al. Defining the Scope of Exposome Studies and Research Needs from a Multidisciplinary Perspective. Environ. Sci. Technol. Lett. 2021, 8, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Dominguez, R.; Jauregui, O.; Queipo-Ortuno, M.I.; Andres-Lacueva, C. Characterization of the Human Exposome by a Comprehensive and Quantitative Large-Scale Multianalyte Metabolomics Platform. Anal. Chem. 2020, 92, 13767–13775. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, J.; Pomeranz, B.H.; Corey, P.N. Incidence of adverse drug reactions in hospitalized patients: A meta-analysis of prospective studies. JAMA 1998, 279, 1200–1205. [Google Scholar] [CrossRef]
- Pirmohamed, M.; James, S.; Meakin, S.; Green, C.; Scott, A.K.; Walley, T.J.; Farrar, K.; Park, B.K.; iménez-García, R. Trends of adverse drug reactions 18 820 patients. BMJ 2004, 329, 15–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrasco-Garrido, P.; De Andrés, L.A.; Barrera, V.H.; De Miguel, G.Á.; Jiménez-García, R. Trends of adverse drug reactions related-hospitalizations in Spain (2001–2006). BMC Health Serv. Res. 2010, 10, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giardina, C.; Cutroneo, P.M.; Mocciaro, E.; Russo, G.T.; Mandraffino, G.; Basile, G.; Rapisarda, F.; Ferrara, R.; Spina, E.; Arcoraci, V. Adverse drug reactions in hospitalized patients: Results of the FORWARD (facilitation of reporting in hospital ward) study. Front. Pharmacol. 2018, 9, 350. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.M.; Mendes, M.A.S.; Ma, J.D. Thiopurine methyltransferase (TPMT) genotyping to predict myelosuppression risk. PLoS Curr. 2011, 3, RRN1236. [Google Scholar] [CrossRef] [PubMed]
- Cristoni, S.; Bernardi, L.R.; Malvandi, A.M.; Larini, M.; Longhi, E.; Sortino, F.; Conti, M.; Pantano, N.; Puccio, G. A case of personalized and precision medicine: Pharmacometabolomic applications to rare cancer, microbiological investigation, and therapy. Rapid Commun. Mass Spectrom. 2020, 35, e8976. [Google Scholar] [CrossRef]
- Ellero-Simatos, S.; Lewis, J.P.; Georgiades, A.; Yerges-Armstrong, L.M.; Beitelshees, A.L.; Horenstein, R.B.; Dane, A.; Harms, A.C.; Ramaker, R.; Vreeken, R.J.; et al. Pharmacometabolomics reveals that serotonin is implicated in aspirin response variability. CPT Pharmacomet. Syst. Pharmacol. 2014, 3, e125. [Google Scholar] [CrossRef]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.C.; He, X.; Kamp, M.A.; Sabel, M.; Barker, R.A.; Steiger, H.J.; et al. A comparative pharmaco-metabolomic study of glutaminase inhibitors in glioma stem-like cells confirms biological effectiveness but reveals differences in target-specificity. Cell Death Discov. 2020, 6, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haraszti, R.A.; Didiot, M.C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef] [PubMed]
- Puhka, M.; Takatalo, M.; Nordberg, M.E.; Valkonen, S.; Nandania, J.; Aatonen, M.; Yliperttula, M.; Laitinen, S.; Velagapudi, V.; Mirtti, T.; et al. Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer-related changes. Theranostics 2017, 7, 3824–3841. [Google Scholar] [CrossRef]
- Luo, P.; Mao, K.; Xu, J.; Wu, F.; Wang, X.; Wang, S.; Zhou, M.; Duan, L.; Tan, Q.; Ma, G.; et al. Metabolic characteristics of large and small extracellular vesicles from pleural effusion reveal biomarker candidates for the diagnosis of tuberculosis and malignancy. J. Extracell. Vesicles 2020, 9, 1790158. [Google Scholar] [CrossRef]
- Royo, F.; Gil-Carton, D.; Gonzalez, E.; Mleczko, J.; Palomo, L.; Perez-Cormenzana, M.; Mayo, R.; Alonso, C.; Falcon-Perez, J.M. Differences in the metabolite composition and mechanical properties of extracellular vesicles secreted by hepatic cellular models. J. Extracell. Vesicles 2019, 8, 1575678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Datta-Chaudhuri, A.; Deme, P.; Dickens, A.; Dastgheyb, R.; Bhargava, P.; Bi, H.; Haughey, N.J. Lipidomic characterization of extracellular vesicles in human serum. J. Circ. Biomark. 2019, 8, 1849454419879848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhto, N.; Vinaiphat, A.; Thongboonkerd, V. Discrimination of urinary exosomes from microvesicles by lipidomics using thin layer liquid chromatography (TLC) coupled with MALDI-TOF mass spectrometry. Sci. Rep. 2019, 9, 13834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinzman, C.P.; Baulch, J.E.; Mehta, K.Y.; Girgis, M.; Bansal, S.; Gill, K.; Li, Y.; Limoli, C.L.; Cheema, A.K. Plasma-derived extracellular vesicles yield predictive markers of cranial irradiation exposure in mice. Sci. Rep. 2019, 9, 9460. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, F.; Yuste, J.E.; Teruel-Montoya, R.; Luengo-Gil, G.; Bohdan, N.; Espín, S.; García-Barberá, N.; Martínez, C.; Vicente, V.; Espín, J.C.; et al. First exploratory study on the metabolome from plasma exosomes in patients with paroxysmal nocturnal hemoglobinuria. Thromb. Res. 2019, 183, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Bestard-escalas, J.; Maimó-barceló, A.; Lopez, D.H.; Reigada, R.; Guardiola-serrano, F.; Ramos-vivas, J.; Hornemann, T.; Okazaki, T.; Barceló-coblijn, G. Common and differential traits of the membrane lipidome of colon cancer cell lines and their secreted vesicles: Impact on studies using cell lines. Cancers 2020, 12, 1293. [Google Scholar] [CrossRef] [PubMed]
- Čuperlović-Culf, M.; Khieu, N.H.; Surendra, A.; Hewitt, M.; Charlebois, C.; Sandhu, J.K. Analysis and simulation of glioblastoma cell lines-derived extracellular vesicles metabolome. Metabolites 2020, 10, 88. [Google Scholar] [CrossRef]
- Nishida-Aoki, N.; Izumi, Y.; Takeda, H.; Takahashi, M.; Ochiya, T.; Bamba, T. Lipidomic analysis of cells and extracellular vesicles from high-and low-metastatic triple-negative breast cancer. Metabolites 2020, 10, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzozowski, J.S.; Jankowski, H.; Bond, D.R.; McCague, S.B.; Munro, B.R.; Predebon, M.J.; Scarlett, C.J.; Skelding, K.A.; Weidenhofer, J. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018, 17, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreri, C.; Sansone, A.; Buratta, S.; Urbanelli, L.; Costanzi, E.; Emiliani, C.; Chatgilialoglu, C. The n-10 fatty acids family in the lipidome of human prostatic adenocarcinoma cell membranes and extracellular vesicles. Cancers 2020, 12, 900. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.H.; Lee, M.Y.; Choi, D.Y.; Lee, J.W.; You, S.; Lee, K.Y.; Kim, J.; Kim, K.P. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics 2015, 15, 824–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Royo, F.; Palomo, L.; Mleczko, J.; Gonzalez, E.; Alonso, C.; Martínez, I.; Pérez-Cormenzana, M.; Castro, A.; Falcon-Perez, J.M. Metabolically active extracellular vesicles released from hepatocytes under drug-induced liver-damaging conditions modify serum metabolome and might affect different pathophysiological processes. Eur. J. Pharm. Sci. 2017, 98, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Moreno, L.; Mleczko, J.; Palomo, L.; Gonzalez, E.; Cabrera, D.; Cogolludo, A.; Vizcaino, F.P.; Van-Liempd, S.; Falcon-Perez, J.M. Hepatocyte-secreted extracellular vesicles modify blood metabolome and endothelial function by an arginase-dependent mechanism. Sci. Rep. 2017, 7, 42798. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saari, H.; Kärkkäinen, O.; Pekkinen, J.; Auriola, S.; Yliperttula, M.; Puhka, M.; Hanhineva, K.; Siljander, P.R.M. Metabolic signature of extracellular vesicles depends on the cell culture conditions. J. Extracell. Vesicles 2019, 8, 1596669. [Google Scholar] [CrossRef] [Green Version]
- Hough, K.P.; Wilson, L.S.; Trevor, J.L.; Strenkowski, J.G.; Maina, N.; Kim, Y.I.; Spell, M.L.; Wang, Y.; Chanda, D.; Dager, J.R.; et al. Unique Lipid Signatures of Extracellular Vesicles from the Airways of Asthmatics. Sci. Rep. 2018, 8, 10340. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Ren, Y.; Zhao, Q.; Ma, Z.; Wang, X.; Liang, M.; Wang, W.; Su, F.; Yang, H. Lipidomic biomarkers of extracellular vesicles for the prediction of preterm birth in the early second trimester. J. Proteome Res. 2020, 19, 4104–4113. [Google Scholar] [CrossRef]
- Guan, F.; Xiang, X.; Xie, Y.; Li, H.; Zhang, W.; Shu, Y.; Wang, J.; Qin, W. Simultaneous metabolomics and proteomics analysis of plasma-derived extracellular vesicles. Anal. Methods 2021, 13, 1930–1938. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S. Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics. Metabolites 2019, 9, 301. [Google Scholar] [CrossRef] [Green Version]
- Sebastiani, P.; Solovieff, N.; DeWan, A.T.; Walsh, K.M.; Puca, A.; Hartley, S.W.; Melista, E.; Andersen, S.; Dworkis, D.A.; Wilk, J.B.; et al. Genetic signatures of exceptional longevity in humans. PLoS ONE 2012, 7, e29848. [Google Scholar] [CrossRef] [Green Version]
- Giblin, W.; Skinner, M.E.; Lombard, D.B. Sirtuins: Guardians of mammalian healthspan. Trends Genet. 2014, 30, 271–286. [Google Scholar] [CrossRef] [Green Version]
- Uno, M.; Nishida, E. Lifespan-regulating genes in c. Elegans. NPJ Aging Mech. Dis. 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Ruby, J.G.; Wright, K.M.; Rand, K.A.; Kermany, A.; Noto, K.; Curtis, D.; Varner, N.; Garrigan, D.; Slinkov, D.; Dorfman, I.; et al. Estimates of the Heritability of Human Longevity Are Substantially Inflated due to Assortative Mating. Genetics 2018, 210, 1109–1124. [Google Scholar] [CrossRef] [Green Version]
- Mattison, J.A.; Roth, G.S.; Beasley, T.M.; Tilmont, E.M.; Handy, A.M.; Herbert, R.L.; Longo, D.L.; Allison, D.B.; Young, J.E.; Bryant, M.; et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 2012, 489, 318–321. [Google Scholar] [CrossRef] [Green Version]
- Parkhitko, A.A.; Filine, E.; Mohr, S.E.; Moskalev, A.; Perrimon, N. Targeting metabolic pathways for extension of lifespan and healthspan across multiple species. Ageing Res. Rev. 2020, 64, 101188. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.M.; Ross, C.; Tran, V.L.; Promislow, D.E.L.; Tardif, S.; Jones, D.P. The metabolome as a biomarker of mortality risk in the common marmoset. Am. J. Primatol. 2019, 81, e22944. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.L.; Shi, X.; Liu, J.; Ding, A.J.; Pu, Y.Z.; Li, Z.; Wu, G.S.; Luo, H.R. Metabolomic signature associated with reproduction-regulated aging in Caenorhabditis elegans. Aging 2017, 9, 447–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viltard, M.; Durand, S.; Pérez-Lanzón, M.; Aprahamian, F.; Lefevre, D.; Leroy, C.; Madeo, F.; Kroemer, G.; Friedlander, G. The metabolomic signature of extreme longevity: Naked mole rats versus mice. Aging 2019, 11, 4783–4800. [Google Scholar] [CrossRef]
- Orwoll, E.S.; Wiedrick, J.; Nielson, C.M.; Jacobs, J.; Baker, E.S.; Piehowski, P.; Petyuk, V.; Gao, Y.; Shi, T.; Smith, R.D.; et al. Proteomic assessment of serum biomarkers of longevity in older men. Aging Cell 2020, 19, e13253. [Google Scholar] [CrossRef]
- Dall’Olio, F.; Vanhooren, V.; Chen, C.C.; Slagboom, P.E.; Wuhrer, M.; Franceschi, C. N-glycomic biomarkers of biological aging and longevity: A link with inflammaging. Ageing Res. Rev. 2013, 12, 685–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, K.; Kettunen, J.; Wurtz, P.; Haller, T.; Havulinna, A.S.; Kangas, A.J.; Soininen, P.; Esko, T.; Tammesoo, M.L.; Magi, R.; et al. Biomarker profiling by nuclear magnetic resonance spectroscopy for the prediction of all-cause mortality: An observational study of 17,345 persons. PLoS Med. 2014, 11, e1001606. [Google Scholar] [CrossRef]
- Cheng, S.; Larson, M.G.; McCabe, E.L.; Murabito, J.M.; Rhee, E.P.; Ho, J.E.; Jacques, P.F.; Ghorbani, A.; Magnusson, M.; Souza, A.L.; et al. Distinct metabolomic signatures are associated with longevity in humans. Nat. Commun. 2015, 6, 6791. [Google Scholar] [CrossRef] [PubMed]
- Després, J.P. Predicting longevity using metabolomics: A novel tool for precision lifestyle medicine? Nat. Rev. Cardiol. 2020, 17, 67–68. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De San-Martin, B.S.; Ferreira, V.G.; Bitencourt, M.R.; Pereira, P.C.G.; Carrilho, E.; de Assuncao, N.A.; de Carvalho, L.R.S. Metabolomics as a potential tool for the diagnosis of growth hormone deficiency (GHD): A review. Arch. Endocrinol. Metab. 2020, 64, 654–663. [Google Scholar] [CrossRef]
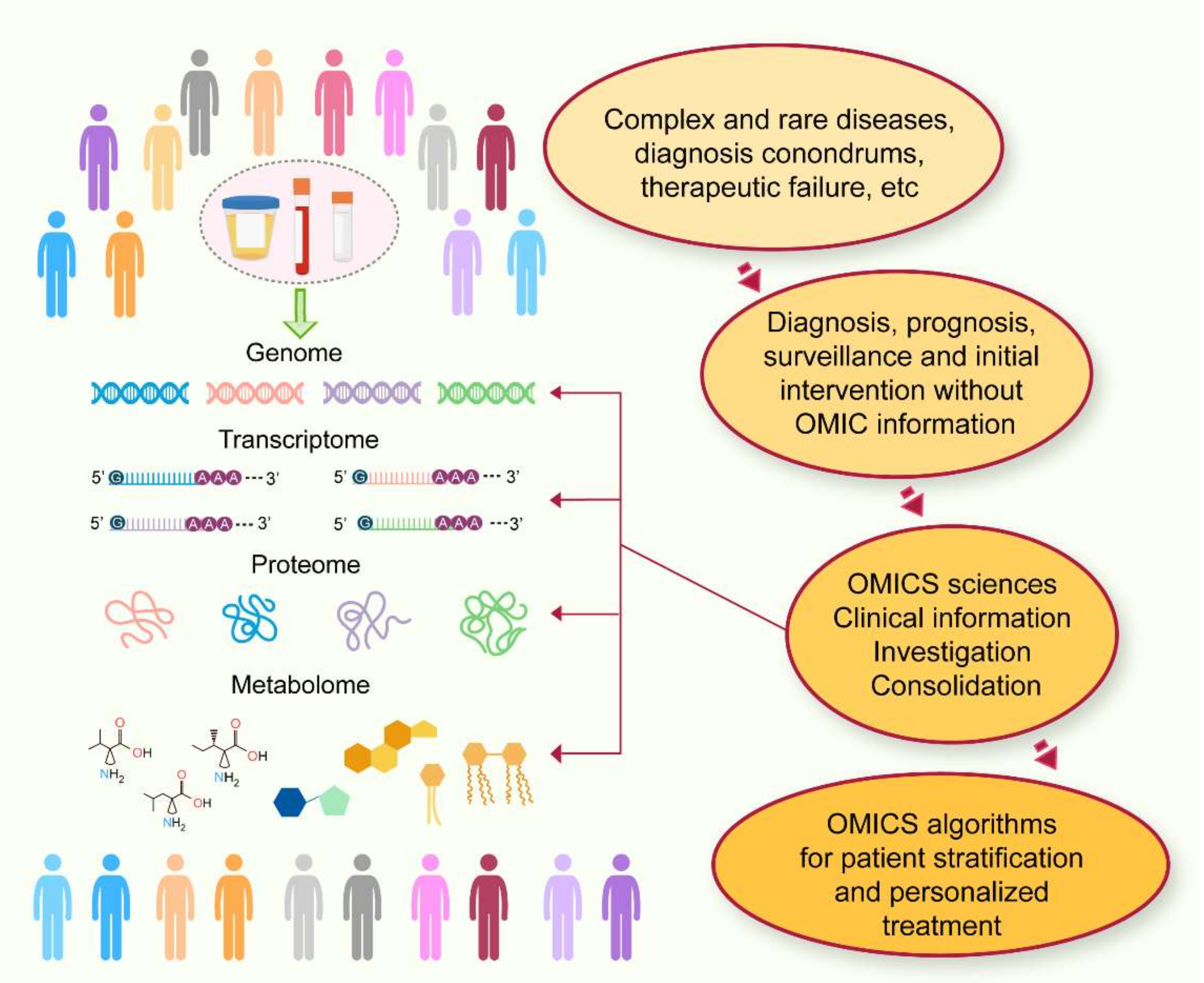
- Krassowski, M.; Das, V.; Sahu, S.K.; Misra, B.B. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front. Genet. 2020, 11, 610798. [Google Scholar] [CrossRef] [PubMed]
- Lotta, L.A.; Scott, R.A.; Sharp, S.J.; Burgess, S.; Luan, J.; Tillin, T.; Schmidt, A.F.; Imamura, F.; Stewart, I.D.; Perry, J.R.; et al. Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis. PLoS Med. 2016, 13, e1002179. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Kopczynski, D.; Coman, C.; Zahedi, R.P.; Lorenz, K.; Sickmann, A.; Ahrends, R. Multi-OMICS: A critical technical perspective on integrative lipidomics approaches. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 808–811. [Google Scholar] [CrossRef]
- Li, Y.; Bouza, M.; Wu, C.; Guo, H.; Huang, D.; Doron, G.; Temenoff, J.S.; Stecenko, A.A.; Wang, Z.L.; Fernandez, F.M. Sub-nanoliter metabolomics via mass spectrometry to characterize volume-limited samples. Nat. Commun. 2020, 11, 5625. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, M.; da Veiga Leprevost, F.; Squizzato, S.; Park, Y.M.; Haug, K.; Carroll, A.J.; Spalding, D.; Paschall, J.; Wang, M.; et al. Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 2017, 35, 406–409. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.Y.; White, K.; Loscalzo, J. Deciphering the molecular basis of human cardiovascular disease through network biology. Curr. Opin. Cardiol. 2012, 27, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, D.J.; Nielsen, J. Genome-scale metabolic models applied to human health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9, e1393. [Google Scholar] [CrossRef] [PubMed]
- Crowther, L.M.; Poms, M.; Plecko, B. Multiomics tools for the diagnosis and treatment of rare neurological disease. J. Inherit. Metab. Dis. 2018, 41, 425–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Species | Metabolomic Platform | Trait | Fluid/Tissue | Refs. |
|---|---|---|---|---|
| Diabetes | ||||
| Phe, Gly, diacyl-phosphatidylcholines SM(C16:1), acyl-alkyl-PC, etc. | Metabolomics, LC-MS | Predictive of T2D | serum | [50] |
| PC (34:2), PC (36:2), TG (52:1), long chain PUFA, total TG, ceramide (22:0) | Lipidomics, LC-MS/MS | Associated with T2D | plasma | [51] |
| Ile, Phe, Ser, Tyr, Gly, palmitoyl SM stearoylcarnitine, etc. | Metabolomics, GC/MS LC/MS/MS, fatty acids | Predictive of T2D | plasma | [52] |
| Leu, Ile, Val, γ-glutamyl-derivates, PC aa (OH, COOH) C28:4, etc. | Metabolomics, NMR, GC- MS, FIA-MS, LC/MS | Associated with T2D | plasma | [53] |
| Diabetes kidney disease | ||||
| C8:1, C10:1 | LC-MS | Increases prediction clinical models | blood/urine | [54] |
| C0, C10:2 and urinary C12:1 s | LC-MS | Albuminuria | urine | [54] |
| Gly, Phe, citrate, glycerol | NMD spectroscopy, amino acids, metabolites | Negatively associated with eGFR | urine | [55] |
| Ala, Val, pyruvate | NMD spectroscopy, metabolites | Positive association | serum | [55] |
| Cancer | ||||
| C16:1, C18:2, C20:4, and C22:6 | CBDI- nanoESI-FTICR MS, FFA | Colorectal cancer diagnosis. | serum | [56] |
| PC, Glu, Arg, hypoxanthine, α-glucose | Metabolomics, NMR, LC/MS | Prostate cancer | tissue | [57] |
| Obesity | ||||
| Arg, Leu/Ile, Tyr, Val, Pro | MS/MS | Childhood obesity and serum triglycerides | serum | [58] |
| Leu, Ile, Val, and Tyr | Metabolomics, NMR | Abdominally obese females | serum | [59] |
| Val, Phe, Tyr, and Gln | Metabolomics, NMR | Insulin resistance | serum | [59] |
| BCAA catabolites | Insulin resistance and abnormal brain function | serum | [47] | |
| Pharmacometabolomics | ||||
| ACs | Metabolomics, HILIC LC-MS/MS | Elevated in irinotecan exposure | plasma/serum | [60] |
| SM, dihydroceramide, PC, PS, PE, cys | Metabolomics/LC-MS/MS | Higher in lorlatinib treatment | plasma/serum | [31] |
| Palmitoleate (C16:1n-7), DHA; 22:6n-3 and EPA; 20:5n-3 | Lipidomics/LC-MS/MS | Associated with fish oil antiobesity effects | plasma/serum | [61] |
| Proteobacteria and Firmicutes | Microbiome/metagenomics | Associated to beta lactam antibiotic resistance | feces | [62] |
| Akkermansia muciniphila | Microbiome/metagenomics | Increased efficacy of programmed cell death 1 protein (PD-1) immunotherapy | plasma/feces | [63] |
| Escherichia coli | Microbiome/metagenomics | Associated to metformin efficacy and toxicity | plasma/feces | [62] |
| B. thetaiotaomicron | LC-MS/MS, microbiome analysis | Diltiazem and 46 different drugs | plasma/feces | [64] |
| Longevity | ||||
| PC (O-34:3, O-34:1, O-36:3), SM (d18:1/14:0), PE (38:6) | Lipidomics, LC-MS/MS, | Familial longevity, higher in females | plasma | [51] |
| Lipids in chylomicrons, VLDL HDL, VLDL size, PUFA, Val, histidine, Leu, and albumin | Metabolomics LC-MS/MS | Longevity, decrease mortality | plasma | [65] |
| Alzheimer’s disease | ||||
| Prostaglandin, diacylglycerols and oleamide | Lipidomics, LC-MS/MS | Altered NT systems & membrane integrity | serum | [66] |
| 3-hydroxyisovalerate | Metabolomics, NMR | Increased plasma levels; mitochondrial dysfunction | plasma | [67] |
| Biogenic amine, citrulline, Pro Arg, Ala, Thr, ACs | Metabolomics, LC-MS/MS | Nitric oxide pathway alterations; mitochondrial function | plasma | [68] |
| Bile acid metabolites, glycolithocholic acid taurolithocholic acid | Metabolomics, LC-MS/MS | Reduced glucose metabolism in the brain & structural atrophy; levels associated with Aβ1–42, p-tau181, t-tau | bile, serum | [69] |
| Gln, serotonin, and sphingomyelin C18:0 | Metabolomics, LC-MS/MS | Memory impairment | brain cortex | [70] |
| Parkinson’s disease | ||||
| Phe, Tyr, His, Gly, acetoacetate, taurine, TMAO, GABA, N-acetylglutamate, acetoin, acetate, Ala, Ile, Val, Cys, Pro, ornithine, fucose, propionate, and PE | Metabolomics; UPLC-MS, NMR | Disease onset | serum, saliva | [71,72] |
| Tricarboxylic acid cycle and purine pathway metabolites | Metabolomics, LC-Ms, GC-MS, UPLC-MS | Alteration of energy metabolism and neurotransmitter regulation | whole brain, striatum | [73,74,75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Covarrubias, V.; Martínez-Martínez, E.; del Bosque-Plata, L. The Potential of Metabolomics in Biomedical Applications. Metabolites 2022, 12, 194. https://doi.org/10.3390/metabo12020194
Gonzalez-Covarrubias V, Martínez-Martínez E, del Bosque-Plata L. The Potential of Metabolomics in Biomedical Applications. Metabolites. 2022; 12(2):194. https://doi.org/10.3390/metabo12020194
Chicago/Turabian StyleGonzalez-Covarrubias, Vanessa, Eduardo Martínez-Martínez, and Laura del Bosque-Plata. 2022. "The Potential of Metabolomics in Biomedical Applications" Metabolites 12, no. 2: 194. https://doi.org/10.3390/metabo12020194
APA StyleGonzalez-Covarrubias, V., Martínez-Martínez, E., & del Bosque-Plata, L. (2022). The Potential of Metabolomics in Biomedical Applications. Metabolites, 12(2), 194. https://doi.org/10.3390/metabo12020194