Phenolic Profile, Antioxidant and Enzyme Inhibition Properties of the Chilean Endemic Plant Ovidia pillopillo (Gay) Meissner (Thymelaeaceae)
Abstract
:1. Introduction
2. Results
2.1. UHPLC-OT-MS Analysis of Lloime (O. pillopillo)
2.1.1. Coumarins
2.1.2. Phenolic Acids
2.1.3. Flavonoids
2.1.4. Fatty Acids
2.1.5. Other Compounds
2.2. Antioxidant and Enzyme Inhibitory Capacities of Lloime
2.3. Docking Assays of Most Abundant Compounds
2.3.1. Acetylcholinesterase (TcAChE) Docking Results
2.3.2. Butyrylcholinesterase (hBuChE) Docking Results
2.3.3. Tyrosinase Docking Results
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Plant Material
4.3. Preparation of the Ethanolic Extract of O. pillopillo
4.4. Ultra-High Resolution Liquid Chromatography Orbitrap MS Analysis (UHPLC–PDA–OT-MS/MS) and Analysis Conditions
4.5. Determination of Total Phenolics and Flavonoids Content
4.6. Antioxidant Capacities of Lloime
4.6.1. ABTS Assay
4.6.2. DPPH Assay
4.6.3. Ferric Reducing/Antioxidant Power the FRAP Assay
4.6.4. ORAC Assay
4.7. Cholinesterase Enzymes (ChE and BuCHe) Inhibition
4.8. Docking Simulations
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morales, E. Vegetales venenosos que pueden provocar intoxicacion en el niño. Rev. Chil. Pediatr. 1945, 16, 307–349. [Google Scholar] [CrossRef]
- Espinoza, E. Plantas medicinales de Chile, 4th ed.; Gay, E., Vazquez, M., Murillo, J., Gajardo, A., Eds.; Mprenta Barcelona: Santiago, Chile, 1897. [Google Scholar]
- Tito Mansilla, J.; Tarcaya, V.P.; Cufre, I.M.; Fabrizio, M.C.; Wright, E.R.; Broussalis, A.M.; Rivera, M.C. Control of Rhizoctonia solani with extracts from Ovidia andina. Rev. Fac. Cienc. Agrar. 2018, 50, 2. [Google Scholar]
- Nanzhen, K.; Jieying, W.; Wenwei, Z.; Xiaoping, Z.; Yingyuan, F. Toxicological studies of daphnetin. Pharmacogn. Mag. 2018, 14, 561. [Google Scholar] [CrossRef]
- Cottiglia, F.; Loy, G.; Garau, D.; Floris, C.; Caus, M.; Pompei, R.; Bonsignore, L. Antimicrobial evaluation of coumarins and flavonoids from the stems of Daphne gnidium L. Phytomedicine 2001, 8, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wei, F.; Liu, S.; Zhao, S.; Liu, L.; Zhao, H.; Li, Y.; Zhang, T.; Wei, F.; Liu, S.; et al. Identification and Quantification of Chemical Constituents in Daphne altaica and their Antioxidant and Cytotoxic Activities. Int. J. Agric. Biol. 2019. [Google Scholar] [CrossRef]
- Deiana, M.; Rosa, A.; Casu, V.; Cottiglia, F.; Bonsignore, L.; Dessì, M.A. Chemical composition and antioxidant activity of extracts from Daphne gnidium L. JAOCS J. Am. Oil Chem. Soc. 2003, 80, 65–70. [Google Scholar] [CrossRef]
- Fernández-Galleguillos, C.; Quesada-Romero, L.; Puerta, A.; Padrón, J.M.; Souza, E.; Romero-Parra, J.; Simirgiotis, M.J. Uhplc-ms chemical fingerprinting and antioxidant, antiproliferative, and enzyme inhibition potential of gaultheria pumila berries. Metabolites 2021, 11, 523. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, R.E.; Simirgiotis, M.J.; Palacios, J.; Paredes, A.; Bórquez, J.; Bravo, A.; Cifuentes, F. Chemical fingerprinting, isolation and characterization of polyphenol compounds from Heliotropium taltalense (Phil.) I.M. Johnst and its endothelium-dependent vascular relaxation effect in rat aorta. Molecules 2020, 25, 3105. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, F.; Palacios, J.; Bórquez, J.; Paredes, A.; Parra, C.; Bravo, A.; Simirgiotis, M.J. Fast Isolation of Flavonoids from the Endemic Species Nolana ramosissima I.M. Johnst and Its Endothelium-Independent Relaxation Effect in Rat Aorta. Molecules 2020, 25, 520. [Google Scholar] [CrossRef] [Green Version]
- Gómez, J.; Simirgiotis, M.J.; Manrique, S.; Lima, B.; Bórquez, J.; Feresin, G.E.; Tapia, A. UHPLC-HESI-OT-MS-MS Biomolecules Profiling, Antioxidant and Antibacterial Activity of the “Orange-Yellow Resin” from Zuccagnia punctata Cav. Antioxidants 2020, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Barrientos, R.E.; Ahmed, S.; Cortés, C.; Fernández-Galleguillos, C.; Romero-Parra, J.; Simirgiotis, M.J.; Echeverría, J. Chemical Fingerprinting and Biological Evaluation of the Endemic Chilean Fruit Greigia sphacelata (Ruiz and Pav.) Regel (Bromeliaceae) by UHPLC-PDA-Orbitrap-Mass Spectrometry. Molecules 2020, 25, 3750. [Google Scholar] [CrossRef]
- Nawwar, M.A.; Hashem, A.N.; Hussein, S.A.; Swilam, N.F.; Becker, A.; Haertel, B.; Lindequist, U.; El-Khatib, A.; Linscheid, M.W. Phenolic profiling of an extract from Eugenia jambos L. (Alston)—The structure of three flavonoid glycosides—Antioxidant and cytotoxic activities. Pharmazie 2016, 71, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lim, M.S.; Kim, S.I.; Lee, H.J.; Kim, S.S.; Kwon, Y.S.; Chun, W. Quercetin-3-O-β-D-glucuronide suppresses lipopolysaccharide-induced JNK and ERK phosphorylation in LPS-challenged RAW264.7 cells. Biomol. Ther. 2016, 24, 610–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.J.; Yokozawa, T.; Rhyu, D.Y.; Kim, H.Y.; Shibahara, N.; Park, J.C. The inhibitory effects of 12 medicinal plants and their component compounds on lipid peroxidation. Am. J. Chin. Med. 2003, 31, 907–917. [Google Scholar] [CrossRef] [Green Version]
- Del Carmen Juárez-Vázquez, M.; Josabad Alonso-Castro, A.; García-Carrancá, A. Kaempferitrin induces immunostimulatory effects in vitro. J. Ethnopharmacol. 2013, 148, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.C.; Park, J.; Cho, S. Anti-Inflammatory and Anti-Oxidative Effects of luteolin-7-O-glucuronide in LPS-Stimulated Murine Macrophages through TAK1 Inhibition and Nrf2 Activation. Int. J. Mol. Sci. 2020, 21, 2007. [Google Scholar] [CrossRef] [Green Version]
- Simirgiotis, M.J.; Ramirez, J.E.; Schmeda Hirschmann, G.; Kennelly, E.J. Bioactive coumarins and HPLC-PDA-ESI-ToF-MS metabolic profiling of edible queule fruits (Gomortega keule), an endangered endemic Chilean species. Food Res. Int. 2013, 54, 532–543. [Google Scholar] [CrossRef]
- de Souza, L.G.; Rennã, M.N.; Figueroa-Villar, J.D. Coumarins as cholinesterase inhibitors: A review. Chem. Biol. Interact. 2016, 254, 11–23. [Google Scholar] [CrossRef]
- Stefanachi, A.; Leonetti, F.; Pisani, L.; Catto, M.; Carotti, A. Coumarin: A natural, privileged and versatile scaffold for bioactive compounds. Molecules 2018, 23, 250. [Google Scholar] [CrossRef] [Green Version]
- Akkol, E.K.; Genç, Y.; Karpuz, B.; Sobarzo-Sánchez, E.; Capasso, R. Coumarins and coumarin-related compounds in pharmacotherapy of cancer. Cancers 2020, 12, 1959. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, J.Y.; Park, P.S.; Bang, S.H.; Lee, K.M.; Lee, Y.R.; Jang, Y.H.; Kim, M.J.; Chun, W.; Heo, M.Y.; et al. Flavonoid Glycosides as Acetylcholinesterase Inhibitors from the Whole Plants of Persicaria thunbergii. Nat. Prod. Sci. 2014, 20, 191–195. [Google Scholar]
- Lee, S.G.; Karadeniz, F.; Seo, Y.; Kong, C.S. Anti-melanogenic effects of flavonoid glycosides from limonium tetragonum (thunb.) bullock via inhibition of tyrosinase and tyrosinase-related proteins. Molecules 2017, 22, 1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, J.; Simirgiotis, M.J.; Manrique, S.; Piñeiro, M.; Lima, B.; Bórquez, J.; Feresin, G.E.; Tapia, A. Uhplc-esi-ot-ms phenolics profiling, free radical scavenging, antibacterial and nematicidal activities of “yellow-brown resins” from Larrea spp. Antioxidants 2021, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, R.; Fernández-Galleguillos, C.; Pastene, E.; Simirgiotis, M.; Romero-Parra, J.; Ahmed, S.; Echeverría, J. Metabolomic Analysis, Fast Isolation of Phenolic Compounds, and Evaluation of Biological Activities of the Bark from Weinmannia trichosperma Cav. (Cunoniaceae). Front. Pharmacol. 2020, 11, 780. [Google Scholar] [CrossRef] [PubMed]
- Larrazábal-Fuentes, M.J.; Fernández-Galleguillos, C.; Palma-Ramírez, J.; Romero-Parra, J.; Sepúlveda, K.; Galetovic, A.; González, J.; Paredes, A.; Bórquez, J.; Simirgiotis, M.J.; et al. Chemical Profiling, Antioxidant, Anticholinesterase, and Antiprotozoal Potentials of Artemisia copa Phil. (Asteraceae). Front. Pharmacol. 2020, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model Seeking for parameter-free double-hybrid functionals: The PBE0-DH model Accurate excitation energies from time-dependent density functional theory: Assessing the PBE0 model toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 2889. [Google Scholar] [CrossRef]
- Petersson, G.A.; Bennett, A.; Tensfeldt, T.G.; Al-Laham, M.A.; Shirley, W.A.; Mantzaris, J. A complete basis set model chemistry. I. The total energies of closed-shell atoms and hydrides of the first-row elements. J. Chem. Phys. 1988, 89, 2193–2218. [Google Scholar] [CrossRef]
- Frisch, A. Gaussian 09W Reference; Gaussian: Wallingford, UK, 2009. [Google Scholar]
- Release, S. 2: Maestro; Version 11.8; Schrdinger, LLC: New York, NY, USA, 2018; Available online: https://scirp.org/reference/referencespapers.aspx?referenceid=2581072 (accessed on 4 December 2021).
- Greenblatt, H.M.; Kryger, G.; Lewis, T.; Silman, I.; Sussman, J.L. Structure of acetylcholinesterase complexed with (-)-galanthamine at 2.3 A resolution. FEBS Lett. 1999, 463, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Nachon, F.; Carletti, E.; Ronco, C.; Trovaslet, M.; Nicolet, Y.; Jean, L.; Renard, P.Y. Crystal structures of human cholinesterases in complex with huprine W and tacrine: Elements of specificity for anti-Alzheimer’s drugs targeting acetyl- and butyryl-cholinesterase. Biochem. J. 2013, 453, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Sussman, J.L.; Harel, M.; Frolow, F.; Oefner, C.; Goldman, A.; Toker, L.; Silman, I. Atomic structure of acetylcholinesterase from Torpedo californica: A prototypic acetylcholine-binding protein. Science 1991, 253, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Silman, I.; Harel, M.; Axelsen, P.; Raves, M.; Sussman, J.L. Three-dimensional structures of acetylcholinesterase and of its complexes with anticholinesterase agents. Biochem. Soc. Trans. 1994, 22, 745–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolet, Y.; Lockridge, O.; Masson, P.; Fontecilla-Camps, J.C.; Nachon, F. Crystal Structure of Human Butyrylcholinesterase and of Its Complexes with Substrate and Products. J. Biol. Chem. 2003, 278, 41141–41147. [Google Scholar] [CrossRef] [Green Version]
- Tallini, L.R.; Bastida, J.; Cortes, N.; Osorio, E.H.; Theoduloz, C.; Schmeda-Hirschmann, G. Cholinesterase inhibition activity, alkaloid profiling and molecular docking of chilean Rhodophiala (Amaryllidaceae). Molecules 2018, 23, 1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, A.P.; Silva, N.; de Silva, N.F.; Andrade, E.H.A.; Gratieri, T.; Setzer, W.N.; Maia, J.G.S.; da Silva, J.K.R. Tyrosinase inhibitory activity, molecular docking studies and antioxidant potential of chemotypes of Lippia origanoides (Verbenaceae) essential oils. PLoS ONE 2017, 12, e0175598. [Google Scholar] [CrossRef]
- Chen, J.; Ye, Y.; Ran, M.; Li, Q.; Ruan, Z.; Jin, N. Inhibition of Tyrosinase by Mercury Chloride: Spectroscopic and Docking Studies. Front. Pharmacol. 2020, 11, 81. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [Green Version]
- PyMOL. pymol.org. Available online: https://pymol.org/2/ (accessed on 6 July 2021).








| Peak # | Retention Time | UV Max | Tentative Identification | Molecular Formula [M-H] | Measured Mass (m/z) | Theoretical Mass (m/z) | Accuracy (ppm) | Ions MSn |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.45 | 227–272 | Quinic acid | C7H11O6− | 191.05610 | 191.05610 | 1.385 | 175.06064 |
| 2 | 1.79 | 226 | Isocitric acid | C6H7O7− | 191.02007 | 191.01917 | −1.441 | 175.02426 |
| 3 | 8.90 | 227 | Vanillic acid * | C8H8O4− | 168.04117 | 168.04225 | −0.540 | 151.03951 |
| 4 | 8.95 | 227–290sh–325 | Daphnin 8-O-methyl ether, (daphnetin 7 O-glucose, 8-methyl ether) | C16H17O9− | 353.08923 | 353.08779 | 2.521 | 191.05622 323.11307 |
| 5 | 9.26 | 228–260 | Sinapoyl glucose | C17H21O10− | 385.11575 | 385.35066 | 2.287 | 341.06777, 223.05322 |
| 6 | 10.45 | 245–295sh–324 | Daphnin 8-O-methyl ether-6″-O-methyl-glucose, (daphnetin 7 O-(6″-O-methyl-glucose, 8-O-methyl ether) | C17H19O9− | 367.10498 | 367.10345 | 2.621 | 323.11307, 191.05556, 175.03951 |
| 7 | 10.81 | 245–295sh 325 | 8-O-methyl daphnin, (daphnetin 7 O-(5″-O-methyl-glucose, 8-O-methyl ether) | C17H19O9− | 367.10492 | 367.10492 | 2.561 | 191.05620, 135.04460 |
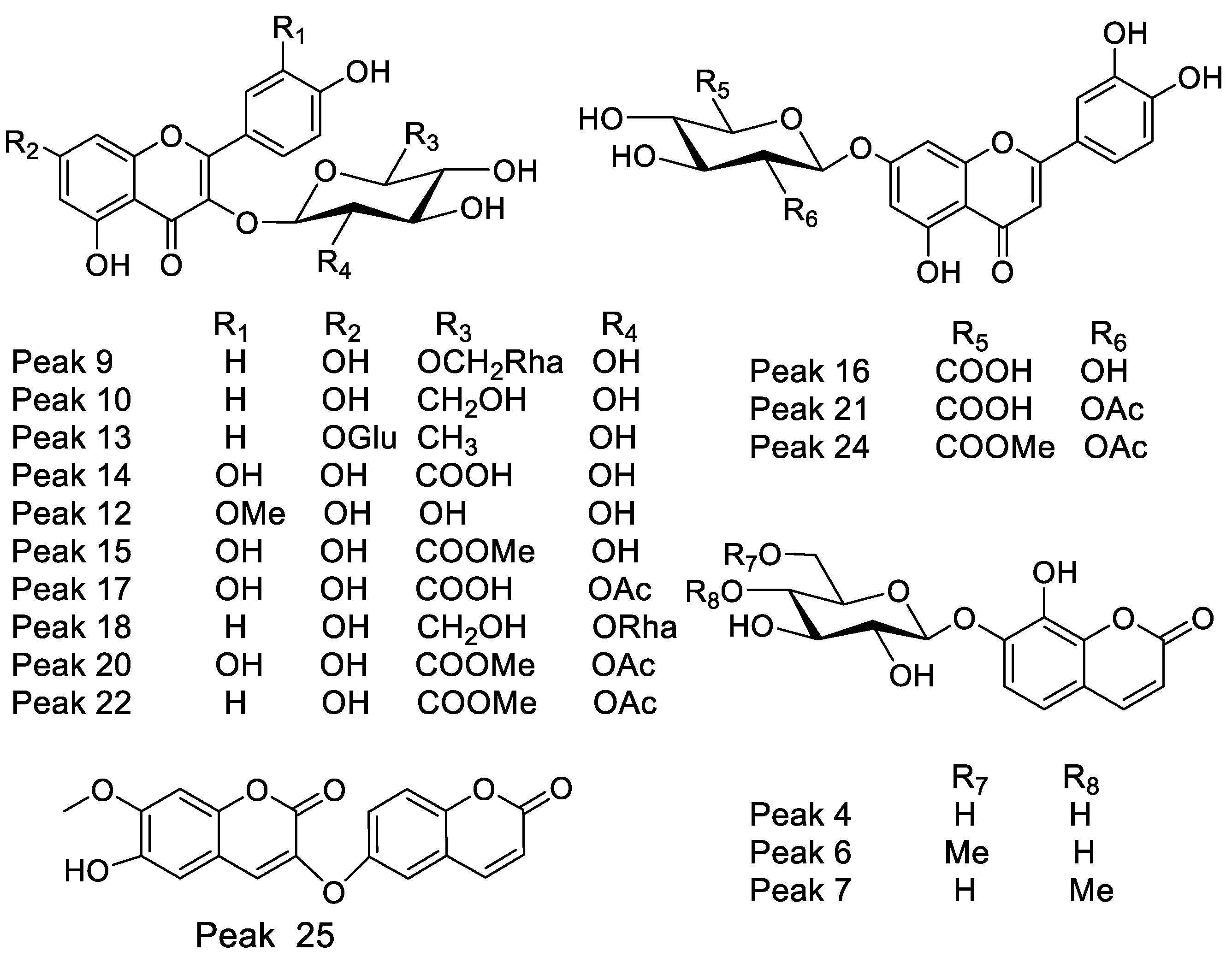
| 8 | 10.87 | 255–355 | Quercetin 3-O-xylosyl-(1-2)-rhamnoside | C26H27O15− | 579.13782 | 579.12446 | 3.374 | 301.03524 |
| 9 | 11.11 | 264–365 | Kaempferol 3-O-rutinose | C27H29O15− | 593.15356 | 593.15356 | 5.83 | 449.10838, 285.03991 |
| 10 | 11.21 | 264–365 | Kaempferol 3-O-β-D-glucose | C21H19O11− | 447.09503 | 447.09603 | 2.294 | 285.03991, 325.05595 |
| 11 | 11.40 | 277 | Proanthocyanidin Geranin B | C30H23O11− | 559.12659 | 559.13186 | 5.548 | 255.06628, 541.11402 |
| 12 | 11.49 | 240–340 | Isoscoparine | C22H21O11− | 461.10895 | 461.10894 | 1.112 | 299.05556, 283.02426 |
| 13 | 11.65 | 264–365 | Kaempferitrin | C27H29O14− | 577.15786 | 577.15845 | 5.66 | 431.09837, 325.03537 |
| 14 | 11.88 | 254–354 | Quercetin 3-O-(β-D-glucuronide) * | C21H18O13− | 477.06943 | 477.06691 | 3.063 | 301.03482, 433.07708 |
| 15 | 12.46 | 254–354 | Quercetin 3-O-β-D-glucuronide- methyl ester | C21H17O12− | 491.08499 | 491.08470 | 6.054 | 301.03537 |
| 16 | 12.51 | 270–338 | Luteolin-7-O-glucuronide | C21H17O12− | 461.07443 | 461.07254 | 5.269 | 285.03991, 267.02934 |
| 17 | 12.83 | 254–354 | Quercetin-3-O-β-D(2″-acetyl glucuronide | C23H19O14− | 519.07990 | 519.07961 | 2.968 | 301.03529 |
| 18 | 13.45 | 265–365 | Kaempferol-3-O-neohesperidose | C27H25O5− | 593.15596 | 593.15344 | 3.423 | 363.0770, 285.04843, 247.37323, 163.03951 |
| 19 | 13.75 | 280 | 3,8″-Binaringenin | C30H23O10− | 543.13165 | 543.13260 | 5.665 | 271.06119, 513.11910 |
| 20 | 13.91 | 254–354 | Quercetin 3-O-β-D-(2″-O-acetyl- glucuronide methyl ester | C24H21O14− | 533.09556 | 533.01351 | 3.058 | 489.08342, 301.03412, 285.04843 |
| 21 | 13.92 | 265–355 | Luteolin 7-O-(β-D-2′O-acetyl glucuronide | C23H19O13− | 503.08521 | 503.08521 | 3.058 | 491.08256, 315.05047, 285.03991 |
| 22 | 13.96 | 265–365 | Kaempferol 3-O-β-(2′-O-acetyl-β-D-glucuronide | C23H19O13− | 503.08511 | 503.08310 | 3.093 | 285.03991, 461.07200, 459.09273 |
| 23 | 14.41 | 265–365 | Kaempferol 3-O-glucuronide methyl ester | C22H19O12− | 475.09003 | 475.08856 | 2.928 | 285.03991, 429.08212 |
| 24 | 17.06 | 248–347 | Luteolin 7-O-(β-D-2″O-acetylglucuronide methyl ester | C24H21O13− | 517.10034 | 517.10046 | 2.85 | 285.03991, 461.07200, 355.04539 |
| 25 | 19.33 | 227–290sh-325 | Daphnoretin | C19H12O7− | 351.05252 | 351.05127 | 2.592 | 207.02995, 161.02441 |
| 26 | 19.33 | 265–365 | Kaempferide (methyl kaempferol) | C16H11O6− | 299.05740 | 299.05556 | 2.385 | 273.03991, 151.00313, 147.04460 |
| 27 | 20.12 | 270–338 | Quercetin * | C15H9O7− | 301.03513 | 301.03537 | 0.851 | 301.03427, 108.02057 |
| 28 | 20.46 | 267–335 | Acacetin * | C16H11O5− | 283.06250 | 283.06065 | 1.852 | 273.03991, 257.05599 |
| 29 | 20.61 | 253–343 | Hydroxyoctadecaenoic acid | C18H35O3− | 299.25907 | 299.25940 | 4.43 | 249.14966 |
| 30 | 20.63 | 253–343 | 3,5,7-Trihydroxyoleic acid | C18H33O5− | 329.23335 | 329.23346 | 3.12 | 119.04924 (C8H7O−) |
| 31 | 22.65 | 268–330 | Hexadecatrienoic acid | C16H25O2− | 249.17975 | 249.18600 | −5.157 | 233.0 |
| 32 | 24.11 | 220 | 2-Hydroxypalmitate | C16H31O3− | 271.22677 | 271.22911 | 8.90 | 253.225 |
| Sample | DPPH a | ABTS a | ORAC b | FRAP b | TPC c | TFC d | AChE e | BuChE e | Tyr e |
|---|---|---|---|---|---|---|---|---|---|
| Lloime ethanol extract | IC50 = 6.65 ± 0.5 | IC50 = 9.95 ± 0.05 | 25.33 ± 1.2 | 45.56 ± 1.32 | 57.33 ± 0.82 | 38.42 ± 1.32 | 1.94 ± 0.07 | 2.73 ± 0.05 | 9.92 ± 0.05 |
| Gallic acid | 14.32 ± 0.5 | 1.67 ± 0.25 | - | - | - | - | - | - | - |
| Kojic acid | - | - | - | - | - | - | - | - | 3.51 ± 0.02 |
| Galantamine | - | - | - | - | - | - | 0.26 ± 0.02 e | 3.82 ± 0.02 | - |
| Compound | Binding Energy (kcal/mol) Acetylcholinesterase | Binding Energy (kcal/mol) Butyrylcholinesterase | Binding Energy (kcal/mol) Tyrosinase |
|---|---|---|---|
| 5-hydroxy-7-methoxy-2-phenyl-4H-chromen-4-one | −9.154 | −7.987 | −5.908 |
| Luteolin 7,4′-dimethyl ether | −10.506 | −8.562 | −6.054 |
| Apigenin 5-glucoside | −12.798 | −10.378 | −9.018 |
| Quercetin 3-O-ß-D-2″-acetylglucuronide | −14.064 | −10.933 | −10.333 |
| Quercetin 3-O-ß-D-2″-acetylglucuronide methyl ester | −15.497 | −11.803 | −9.018 |
| Quercetin 3-O-(ß-D-glucuronide) | −14.144 | −12.888 | −10.038 |
| Quercetin 3-O-ß-D-(glucuronide methyl ester) | −14.518 | −12.169 | −9.169 |
| 8-O-methyl daphnin (daphnetin 7 O-(5″O-methyl-glucose, 8-methyl ether) | −12.651 | −10.812 | −8.456 |
| Galantamine | −12.989 | −7.125 | - |
| Kojic acid | - | - | −6.050 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortés, C.; González-Cabrera, D.A.; Barrientos, R.; Parra, C.; Romero-Parra, J.; Pertino, M.W.; Areche, C.; Sepúlveda, B.; Bórquez, J.; Torres-Benítez, A.; et al. Phenolic Profile, Antioxidant and Enzyme Inhibition Properties of the Chilean Endemic Plant Ovidia pillopillo (Gay) Meissner (Thymelaeaceae). Metabolites 2022, 12, 90. https://doi.org/10.3390/metabo12020090
Cortés C, González-Cabrera DA, Barrientos R, Parra C, Romero-Parra J, Pertino MW, Areche C, Sepúlveda B, Bórquez J, Torres-Benítez A, et al. Phenolic Profile, Antioxidant and Enzyme Inhibition Properties of the Chilean Endemic Plant Ovidia pillopillo (Gay) Meissner (Thymelaeaceae). Metabolites. 2022; 12(2):90. https://doi.org/10.3390/metabo12020090
Chicago/Turabian StyleCortés, Carmen, Diego A. González-Cabrera, Ruth Barrientos, Claudio Parra, Javier Romero-Parra, Mariano Walter Pertino, Carlos Areche, Beatriz Sepúlveda, Jorge Bórquez, Alfredo Torres-Benítez, and et al. 2022. "Phenolic Profile, Antioxidant and Enzyme Inhibition Properties of the Chilean Endemic Plant Ovidia pillopillo (Gay) Meissner (Thymelaeaceae)" Metabolites 12, no. 2: 90. https://doi.org/10.3390/metabo12020090
APA StyleCortés, C., González-Cabrera, D. A., Barrientos, R., Parra, C., Romero-Parra, J., Pertino, M. W., Areche, C., Sepúlveda, B., Bórquez, J., Torres-Benítez, A., & Simirgiotis, M. J. (2022). Phenolic Profile, Antioxidant and Enzyme Inhibition Properties of the Chilean Endemic Plant Ovidia pillopillo (Gay) Meissner (Thymelaeaceae). Metabolites, 12(2), 90. https://doi.org/10.3390/metabo12020090