Infection Biomarkers Based on Metabolomics
Abstract
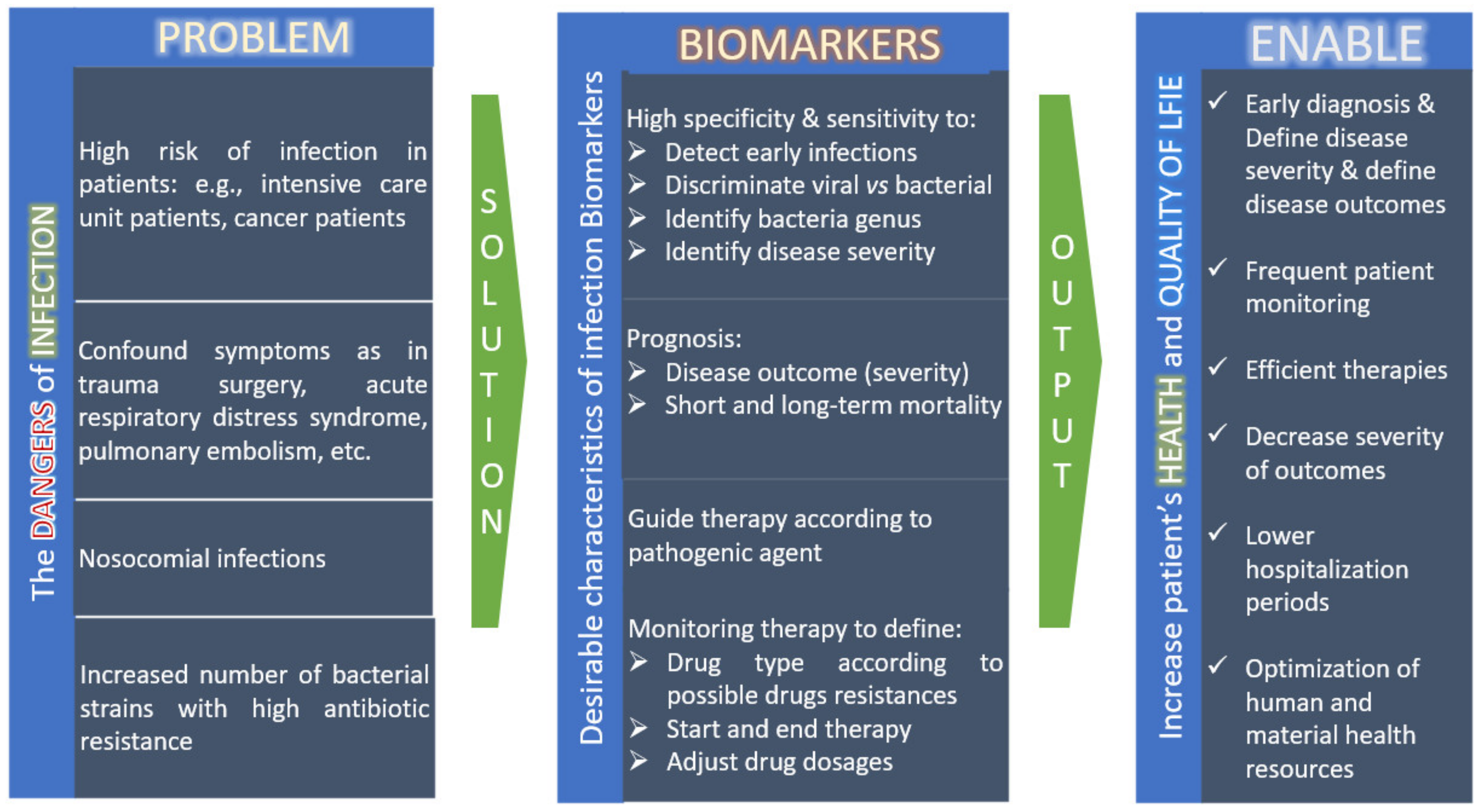
:1. Introduction
2. Metabolomics Overview
2.1. Major Analytical Techniques
2.2. Untargeted versus Targeted Analysis
2.3. Biomarkers
3. Metabolomics to Discover Biomarkers of Infection
3.1. Predict Infection, the Causative Agent, Diseases Severity and Disease Outcome
3.2. Metabolic Information
4. Metabolomics Integration with Other Omics
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Kinloch, N.K.; Ritchie, G.; Brumme, C.J.; Dong, W.; Dong, W.; Lawson, T.; Jones, R.B.; Montaner, J.S.; Leung, V.; Romney, M.G.; et al. Suboptimal Biological Sampling as a Probable Cause of False-Negative COVID-19 Diagnostic Test Results. J. Infect. Dis. 2020, 222, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Tsalik, E.L.; Jones, D.; Nicholson, B.; Waring, L.; Liesenfeld, O.; Park, L.P.; Glickman, S.W.; Caram, L.B.; Langley, R.J.; van Velkinburgh, J.C.; et al. Multiplex PCR to diagnose bloodstream infections in patients admitted from the emergency department with sepsis. J. Clin. Microbiol. 2010, 48, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Koenig, S.M.; Truwit, J.D. Ventilator-Associated Pneumonia: Diagnosis, Treatment, and Prevention. Clin. Microbiol. Rev. 2006, 19, 637–657. [Google Scholar] [CrossRef] [Green Version]
- Cong, S.; Ma, T.; Di, X.; Tiang, C.; Zhao, M.; Wang, K. Diagnostic value of neutrophil CD64, procalcitonin, and interleukin-6 in sepsis: A meta-analysis. BMC Infect. Dis. 2021, 21, 384. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.; Lai, Y.; Wang, H.; Peng, L.; Zhou, N.; Tian, Y.; Jiang, Y.; Gong, G. Diagnostic Accuracy of Procalcitonin Compared to C-Reactive Protein and Interleukin 6 in Recognizing Gram-Negative Bloodstream Infection: A Meta-Analytic Study. Dis. Markers 2020, 2020, 4873074. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.M.P.; Eslick, G.D.; Graig, J.C.; McLean, A.S. Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: Systematic review and meta-analysis. Lancet Infect. Dis. 2007, 7, 210–217. [Google Scholar] [CrossRef]
- Hoeboer, S.H.; van der Geest, P.J.; Nieber, D.; Groeneveld, A.B.J. The diagnostic accuracy of procalcitonin for bacteraemia: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2015, 21, 474–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, L.; Rabello, L.; Salluh, J.; Martin-Loeches, I.; Rodriguez, A.; Nseir, S.; Gomes, J.A.; Povoa, P.; TAVeM Study Group. C-reactive protein and procalcitonin profile in ventilator-associated lower respiratory infections. J. Crit. Care 2018, 48, 385–389. [Google Scholar] [CrossRef]
- Mohan, A.; Harikrishna, J. Biomarkers for the diagnosis of bacterial infections: In pursuit of the ‘Holy Grail’. Indian J. Med. Res. 2015, 141, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Assink-de Jong, E.; de Lange, D.W.; van Oers, J.A.; Njisten, M.W.; Twisk, J.W.; Beishuizen, A. Stop Antibiotics on guidance of Procalcitonin Study (SAPS): A randomised prospective multicenter investigator-initiated trial to analyse whether daily measurements of procalcitonin versus a standard-of-care approach can safely shorten antibiotic duration in intensive care unit patients-calculated sample size: 1816 patients. BMC Infect. Dis. 2013, 13, 178. [Google Scholar] [CrossRef] [Green Version]
- Arulkumaran, N.; Khpal, M.; Tam, K.; Baheerathan, A.; Corredor, C.; Singer, M. Effect of Antibiotic Discontinuation Strategies on Mortality and Infectious Complications in Critically Ill Septic Patients: A Meta-Analysis and Trial Sequential Analysis. Crit. Care Med. 2020, 48, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Chang, W.; Xie, J.-F.; Sun, Q.; Qiu, H.-B.; Yang, Y. Ineffectiveness of procalcitonin-guided antibiotic therapy in severely critically ill patients: A meta-analysis. Int. J. Infect. Dis. 2019, 85, 158–166. [Google Scholar] [CrossRef]
- Pepper, D.J.; Sun, J.; Rhee, C.; Welsh, J.; Powers, J.H.; Danner, R.L.; Kadri, S.S. Procalcitonin-Guided Antibiotic Discontinuation and Mortality in Critically Ill Adults: A Systematic Review and Meta-analysis. Chest 2019, 155, 1109–1118. [Google Scholar] [CrossRef]
- Meier, M.A.; Branche, A.; Neeser, O.L.; Wirz, Y.; Haubitz, S.; Bouadma, L.; Wolff, M.; Luyt, C.E.; Chastre, J.; Tubach, F.; et al. Procalcitonin-guided antibiotic treatment in patients with positive blood cultures: A patient-level meta-analysis of randomized trials. Clin. Infect. Dis. 2018, 69, 388–396. [Google Scholar] [CrossRef]
- Wirz, Y.; Meier, M.A.; Boudma, L.; Luyt, C.E.; Wolff, M.; Chastre, J.; Tubach, F.; Schroeder, S.; Nobre, V.; Annane, D.; et al. Effect of procalcitonin-guided antibiotic treatment on clinical outcomes in intensive care unit patients with infection and sepsis patients: A patient-level meta-analysis of randomized trials. Crit. Care 2018, 22, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, W.E.; Eickhoff, J.C.; Shukla, S.K.; Pantrangi, M.; Rooijakkers, S.; Cosgrove, S.E.; Nizet, V.; Sakoulas, G. Elevated Serum Interleukin-10 at Time of Hospital Admission Is Predictive of Mortality in Patients with Staphylococcus aureus Bacteremia. J. Infect. Dis. 2012, 206, 1604–1611. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, R.; Martin-Loeches, I.; Phillips, G.; Osborn, T.M.; Townsend, S.; Dellinger, R.P.; Artigas, A.; Schorr, C.; Levy, M.M. Empiric Antibiotic Treatment Reduces Mortality in Severe Sepsis and Septic Shock from the First Hour. Crit. Care Med. 2014, 42, 1749–1755. [Google Scholar] [CrossRef]
- ECDC. Healthcare-Associated Infections Acquired in Intensive Care Units. 2016. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/AER-HCAI_ICU_3_0.pdf (accessed on 3 January 2021).
- Restrepo, M.I.; Faverio, P.; Anzueto, A. Long-term prognosis in community-acquired pneumonia. Curr. Opin. Infect. Dis. 2013, 26, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Paiva, J.; Rello, J. Assessing Severity of Patients with Community-Acquired Pneumonia. Semin. Respir. Crit. Care Med. 2012, 33, 272–283. [Google Scholar] [CrossRef]
- Kim, M.W.; Lim, J.Y.; Oh, S.H. Mortality prediction using serum biomarkers and various clinical risk scales in community-acquired pneumonia. Scand. J. Clin. Lab. Investig. 2017, 77, 486–492. [Google Scholar] [CrossRef]
- Banoei, M.M.; Vogel, H.J.; Weljie, A.M.; Yende, S.; Angus, D.C.; Winston, B.W. Plasma lipid profiling for the prognosis of 90-day mortality, in-hospital mortality, ICU admission, and severity in bacterial community-acquired pneumonia (CAP). Crit. Care 2020, 24, 461. [Google Scholar] [CrossRef]
- Breitling, R. What is systems biology? Front. Physiol. 2010, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated omics: Tools, advances and future approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef] [Green Version]
- Tebani, A.; Afonso, C.; Bekri, S. Advances in metabolome information retrieval: Turning chemistry into biology. Part II: Biological information recovery. J. Inherit. Metab. Dis. 2018, 41, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Sun, H.; Wang, X. Serum metabolomics as a novel diagnostic approach for disease: A systematic review. Anal. Bioanal. Chem. 2012, 404, 1239–1245. [Google Scholar] [CrossRef]
- Vinayavekhin, N.; Homan, E.A.; Saghatelian, A. Exploring Disease through Metabolomics. ACS Chem. Biol. 2010, 5, 91–103. [Google Scholar] [CrossRef]
- Vargas, A.J.; Harris, C.C. Biomarker development in the precision medicine era: Lung cancer as a case study. Nat. Rev. Cancer 2016, 16, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banoei, M.M.; Vogel, H.J.; Weljie, A.M.; Kumar, A.; Yende, S.; Angus, D.C.; Winston, B.W. Plasma metabolomics for the diagnosis and prognosis of H1N1 influenza pneumonia. Crit. Care 2017, 21, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, E.P.; Gerszten, R.E. Metabolomics and Cardiovascular Biomarker Discovery. Clin. Chem. 2012, 58, 139–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WGBSE. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Cunha, B.R.; Aleixo, S.M.; Fonseca, L.P.; Calado, C.R.C. Fast identification of off-target liabilities in early antibiotic discovery with Fourier-transform infrared spectroscopy. Biotechnol. Bioeng. 2021, 118, 4465–4476. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Jiang, D.; Feng, S.; Ren, C.; Guo, J. µ-NMR at the point of care testing. Electrophoresis 2020, 41, 319–327. [Google Scholar] [CrossRef]
- Slupsky, C.M. Nuclear magnetic resonance-based analysis of urine for the rapid etiological diagnosis of pneumonia. Expert Opin. Med. Diagn. 2011, 5, 63–73. [Google Scholar] [CrossRef]
- Lau, S.K.; Lee, K.-C.; Curreem, S.O.; Chow, W.-N.; To, K.K.W.; Hung, I.F.N.; Ho, D.T.; Sridhar, S.; Li, I.W.; Ding, V.S.; et al. Metabolomic Profiling of Plasma from Patients with Tuberculosis by Use of Untargeted Mass Spectrometry Reveals Novel Biomarkers for Diagnosis. J. Clin. Microbiol. 2015, 53, 3750–3759. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.; Lee, K.-C.; Lo, G.; Ding, V.; Chow, W.-N.; Ke, T.; Curreem, S.O.; To, K.K.; Ho, D.T.; Sridhar, S.; et al. Metabolomic Profiling of Plasma from Melioidosis Patients Using UHPLC-QTOF MS Reveals Novel Biomarkers for Diagnosis. Int. J. Mol. Sci. 2016, 17, 307. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.-H.; Fan, J.; Zhu, J.; Zhao, Y.-S.; Wang, C.-J.; Zhang, M.; Xu, F. Exploring plasma metabolomic changes in sepsis: A clinical matching study based on gas chromatography–mass spectrometry. Ann. Transl. Med. 2020, 8, 1568. [Google Scholar] [CrossRef]
- Adamko, D.J.; Saude, E.; Bear, M.; Regush, S.; Robinson, J.L. Urine metabolomic profiling of children with respiratory tract infections in the emergency department: A pilot study. BMC Infect. Dis. 2016, 16, 439. [Google Scholar] [CrossRef] [Green Version]
- Kelly, R.S.; Lasky-Su, J.; Yeung, S.-C.J.; Stone, R.M.; Caterino, J.M.; Hagan, S.C.; Lyman, G.H.; Baden, L.R.; Glotzbecker, B.E.; Coyne, C.J.; et al. Integrative omics to detect bacteremia in patients with febrile neutropenia. PLoS ONE 2018, 13, e0197049. [Google Scholar] [CrossRef]
- Slupsky, C.M.; Rankin, K.N.; Fu, H.; Chang, D.; Rowe, B.H.; Charles, P.G.P.; McGeer, A.; Low, D.; Long, R.; Kunimoto, D.; et al. Pneumococcal Pneumonia: Potential for Diagnosis through a Urinary Metabolic Profile. J. Proteome Res. 2009, 8, 5550–5558. [Google Scholar] [CrossRef]
- Sarafidis, K.; Chatziioannou, A.; Thomaidou, A.; Gika, H.; Mikros, E.; Benaki, D.; Diamanti, E.; Agakidis, C.; Raikos, N.; Drossou, V.; et al. Urine metabolomics in neonates with late-onset sepsis in a case-control study. Sci. Rep. 2017, 7, 45506. [Google Scholar] [CrossRef] [PubMed]
- Mickiewicz, B.; Vogel, H.J.; Wong, H.R.; Winston, B.W. Metabolomics as a novel approach for early diagnosis of pediatric septic shock and its mortality. Am. J. Respir. Crit. Care Med. 2013, 187, 967–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, P.; Zheng, Y.; Luo, Q.; Liu, X.; Kang, Y.; Zhang, Y.; Zhang, R.; Xu, Y.; Yang, D.; Xi, W.; et al. Metabolic profiles in community-acquired pneumonia: Developing assessment tools for disease severity. Crit. Care. 2018, 22, 130. [Google Scholar] [CrossRef] [Green Version]
- Ferrarini, A.; Righetti, L.; Martínez, M.P.; Fernández-López, M.; Mastrangelo, A.; Horcajada, J.P.; Betbesé, A.; Esteban, A.; Ordóñez, J.; Gea, J.; et al. Discriminant biomarkers of acute respiratory distress syndrome associated to H1N1 influenza identified by metabolomics HPLC-QTOF-MS/MS platform. Electrophoresis 2017, 38, 2341–2348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wozniak, J.M.; Mills, R.H.; Olson, J.; Caldera, J.R.; Sepich-Poore, G.D.; Carrillo-Terrazas, M.; Tsai, C.M.; Vargas, F.; Knight, R.; Dorrestein, P.C.; et al. Mortality Risk Profiling of Staphylococcus aureus Bacteremia by Multi-omic Serum Analysis Reveals Early Predictive and Pathogenic Signatures. Cell 2020, 182, 1311–1327.e14. [Google Scholar] [CrossRef]
- Rogers, A.J.; McGeachie, M.; Baron, R.M.; Gazourian, L.; Haspel, J.A.; Nakahira, K.; Fredenburgh, L.E.; Hunninghake, G.M.; Raby, B.A.; Matthay, M.A.; et al. Metabolomic Derangements Are Associated with Mortality in Critically Ill Adult Patients. PLoS ONE 2014, 9, e87538. [Google Scholar] [CrossRef] [Green Version]
- Seymour, C.W.; Yende, S.; Scott, M.J.; Pribis, J.; Mohney, R.P.; Bell, L.N.; Chen, Y.F.; Zuckerbraun, B.S.; Bigbee, W.L.; Yealy, D.M.; et al. Metabolomics in pneumonia and sepsis: An analysis of the GenIMS cohort study. Intensive Care Med. 2013, 39, 1423–1434. [Google Scholar] [CrossRef]
- Roberts, I.; Muelas, M.W.; Taylor, J.M.; Davison, A.S.; Xu, Y.; Grixti, J.M.; Gotts, N.; Sorokin, A.; Goodacre, R.; Kell, D.B. Untargeted metabolomics of COVID-19 patient serum reveals potential prognostic markers of both severity and outcome. Metabbolomics 2021, 18, 6. [Google Scholar] [CrossRef]
- Wu, D.; Shu, T.; Yang, X.; Song, J.X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Delafiori, J.; Navarro, L.C.; Siciliano, R.F.; De Melo, G.C.; Busanello, E.N.B.; Nicolau, J.C.; Sales, G.M.; De Oliveira, A.N.; Val, F.F.A.; De Oliveira, D.N.; et al. Covid-19 Automated Diagnosis and Risk Assessment through Metabolomics and Machine Learning. Anal. Chem. 2021, 93, 2471–2479. [Google Scholar] [CrossRef]
- Langley, R.J.; Tsalik, E.L.; Velkinburgh, J.C.; Glickman, S.W.; Rice, B.J.; Wang, C.; Chen, B.; Carin, L.; Suarez, A.; Mohney, R.P.; et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci. Transl. Med. 2013, 5, 195ra95. [Google Scholar] [CrossRef] [Green Version]
- Langley, R.J.; Tipper, J.L.; Bruse, S.; Baron, R.M.; Tsalik, E.L.; Huntley, J.; Rogers, A.J.; Jaramillo, R.J.; O’Donnell, D.; Mega, W.M.; et al. Integrative “Omic” Analysis of Experimental Bacteremia Identifies a Metabolic Signature That Distinguishes Human Sepsis from Systemic Inflammatory Response Syndromes. Am. J. Respir. Crit. Care Med. 2014, 190, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Bernatchez, J.A.; McCall, L.-I. Insights gained into respiratory infection pathogenesis using lung tissue metabolomics. PLoS Pathog. 2020, 16, e1008662. [Google Scholar] [CrossRef] [PubMed]
- Nickler, M.; Ottiger, M.; Steuer, C.; Huber, A.; Anderson, J.B.; Müller, B.; Schuetz, P. Systematic review regarding metabolic profiling for improved pathophysiological understanding of disease and outcome prediction in respiratory infections. Respir. Res. 2015, 16, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurfluh, S.; Baumgartner, T.; Meier, M.A.; Ottiger, M.; Voegeli, A.; Bernasconi, L.; Neyer, P.; Mueller, B.; Schuetz, P. The role of metabolomic markers for patients with infectious diseases: Implications for risk stratification and therapeutic modulation. Expert Rev. Anti-Infect. Ther. 2018, 16, 133–142. [Google Scholar] [CrossRef]
- Izquierdo-Garcia, J.L.; Nin, N.; Jimenez-Clemente, J.; Horcajada, J.P.; del Mar Arenas-Miras, M.; Gea, J.; Esteban, A.; Ruiz-Cabello, J.; Lorente, J.A. Metabolomic Profile of ARDS by Nuclear Magnetic Resonance Spectroscopy in Patients With H1N1 Influenza Virus Pneumonia. Shock 2018, 50, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Et Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Lorente, J.A.; Nin, N.; Villa, P.; Vasco, D.; Miguel-Coello, A.B.; Rodriguez, I.; Herrero, R.; Peñuelas, O.; Ruiz-Cabello, J.; Izquierdo-Garcia, J.L. Metabolomic diferences between COVID-19 and H1N1 influenza induced ARDS. Crit. Care 2021, 25, 390. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Doğan, H.O.; Şenol, O.; Bolat, S.; Yıldız, Ş.N.; Büyüktuna, S.A.; Sarıismailoğlu, R.; Doğan, K.; Hasbek, M.; Hekim, S.N. Understanding the pathophysiological changes via untargeted metabolomics in COVID-19 patients. J. Med. Virol. 2021, 93, 2340–2349. [Google Scholar] [CrossRef] [PubMed]
- Páez-Franco, J.C.; Torres-Ruiz, J.; Sosa-Hernández, V.A.; Cervantes-Díaz, R.; Romero-Ramírez, S.; Pérez-Fragoso, A.; Meza-Sánchez, D.E.; Germán-Acacio, J.M.; Maravillas-Montero, J.L.; Mejía-Domínguez, N.R.; et al. Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e20. [Google Scholar] [CrossRef]
- Robles, A.I.; Harris, C.C. Integration of multiple “OMIC” biomarkers: A precision medicine strategy for lung cancer. Lung Cancer 2017, 107, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Bartel, J.; Krumsiek, J.; Schramm, K.; Adamski, J.; Gieger, C.; Herder, C.; Carstensen, M.; Peters, A.; Rathmann, W.; Roden, M.; et al. The Human Blood Metabolome-Transcriptome Interface. PLoS Genet. 2015, 11, e1005274. [Google Scholar] [CrossRef]
- Salazar, M.G.; Neugebauer, S.; Kacprowski, T.; Michalik, S.; Ahnert, P.; Creutz, P.; Rosolowski, M.; Löffler, M.; Bauer, M.; Suttorp, N.; et al. Association of proteome and metabolome signatures with severity in patients with community-acquired pneumonia. J. Proteom. 2020, 214, 103627. [Google Scholar] [CrossRef]
- Jefferies, K.; Drysdale, S.B.; Robinson, H.; Clutterbuck, E.A.; Blackwell, L.; McGinley, J.; Lin, G.L.; Galal, U.; Nair, H.; Aerssens, J.; et al. Presumed Risk Factors and Biomarkers for Severe Respiratory Syncytial Virus Disease and Related Sequelae: Protocol for an Observational Multicenter, Case-Control Study from the Respiratory Syncytial Virus Consortium in Europe (RESCEU). J. Infect. Dis. 2020, 222, S658–S665. [Google Scholar] [CrossRef]
- Öner, D.; Drysdale, S.B.; McPherson, C.; Lin, G.-L.; Janet, S.; Broad, J.; Pollard, A.J.; Aerssens, J. Biomarkers for Disease Severity in Children Infected with Respiratory Syncytial Virus: A Systematic Literature Review. J. Infect. Dis. 2020, 222, S648–S657. [Google Scholar] [CrossRef] [PubMed]

| Biomarker Type, Models Predictability | Biofluid/Analytical Technique | Patients with a Defined Condition, Population Dimension | Ref. |
|---|---|---|---|
| Infection vs. non-infected and mechanical ventilated patients, Q2 (NMRS) = 0.789 & Q2 (GC-MS) = 0.971 Discriminate viral and bacterial pneumonia, Q2 (NMRS) = 0.789 & Q2 (GC-MS) = 0.971 90 days mortality, Q2 (NMRS) = 0.597 & Q2 (GC-MS) = 0.829 | Plasma NMR & GC-MS | H1N1 viral pneumonia, n = 42 Non-infected patients, n = 31 With CAP, n = 30 Disease progression among patients with viral pneumonia, n = 21 | [31] |
| Discriminate active from non-active tuberculosis, AUC = 0.914 or from CAP, AUC = 0.894 | Plasma LC-MS | Active tuberculosis, n = 46 Non active tuberculosis infection, n = 30 With CAP, n = 30 | [37] |
| Identify melioidosis, AUC = 1.0 Discriminate from bacteremia, AUC = 0.998 | Plasma LC-MS | With Burkholderia pseudomallei, n = 22 Non-infected patients, n = 30 With bacteremia, n = 24 | [38] |
| Sepsis, AUC = 0.719 | Plasma GC-MS | Sepsis, n = 31 Healthy subjects, n = 23 | [39] |
| Discriminate children with respiratory syncytial virus from healthy ones, Q2 = 0.76 Disease severity, Q2 = 0.81 | Urine NMR | Children with respiratory syncytial virus, n = 55 Healthy children, n = 37 | [40] |
| Infection in cancer patients with chemotherapy-associated neutropenia, AUC = 0.991 | Plasma LC-MS | With infection, n = 14 Without infection, n = 25 | [41] |
| Infection vs. healthy, Q2 = 0.820 Infection among other diseases states, Q2 = 0.828 Discriminate S. pneumoniae pneumonia from viral pneumonia, Q2 = 0.665 Discriminate from other bacterial pneumonia, Q2 = 0.680 | Urine NMR | With S. pneumococcal pneumonia, n = 62 Healthy subjects, n = 115 Non-infectious metabolic stress, n = 55 With viral infection, n = 57 With pneumonia from other bacteria, n = 80 | [42] |
| Sepsis (NMR), AUC = 0.94 Sepsis (LC-MS/MS), AUC = 0.97 | Urine NMR & LC-MS/MS | Neonates with sepsis, n = 16 Healthy neonates, n = 16 | [43] |
| Septic shock vs. healthy, AUC= 0.98 SIRS vs. healthy, AUC= 0.95 Septic shock vs. SIRS, AUC = 0.82 | Serum NMR | Pediatric (neonates to 11 years old) Septic shock, n = 60 SIRS, n = 40 Healthy, n = 40 | [44] |
| CAP severity, AUC = 0.911 | Serum LC-MS/MS | Discovery cohort (n = 102); Validated cohort (n = 73) | [45] |
| Progression to ARDS, specificity = 1, sensitivity =1 | Serum LC-MS/MS | With H1N1 virus infection, n = 25 that developed to ARDS, n = 17 | [46] |
| Mortality, AUC = 0.75 | Serum LC-MS/MS | S. aureus bacteremia, n = 200 | [47] |
| 90 days mortality, AUC = 0.91, sensitivity 0.82, specificity 0.91 | Plasma DI-MS/MS | With CAP, n = 150 | [23] |
| 28 days mortality ROCI, AUC = 0.91 CAPSOD, AUC = 0.74 | Plasma GC-MS& LC-MS | ROCI (patients with SIRS, sepsis, sepsis-induced ARDS), n = 90; CAPSOD, n = 149 (validation) | [48] |
| 90 days mortality, AUC = 0.67 | Plasma GC-MS& LC-MS | CAP patients that died, n = 15 that survived, n = 15 | [49] |
| COVID-19 severity, AUC= 0.83 COVID-19 mortality, AUC = 0.760 | Serum LC-MS/MS | COVID-19 patients, n = 120 for validation, n = 90 | [50] |
| COVID-19 infection, AUC = 1.00 Mild vs. severe COVID-19, AUC = 0.708 Death from severe group, AUC = 0.737 Death from mild severe group, AUC = 0.865 | Plasma LC-MS | Non infected, n = 10; Mild (n = 14) and severe (n = 11) COVID-19; Fatal COVID-19, n = 9 | [51] |
| COVID-19 infection, specificity >0.96, sensitivity >0.83 Disease severity, specificity >0.80 and sensitivity >0.85 | Plasma LC-MS/MS | With COVID-19, n = 442 Non-COVID-19, n = 350 | [52] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo, R.; Bento, L.F.N.; Fonseca, T.A.H.; Von Rekowski, C.P.; da Cunha, B.R.; Calado, C.R.C. Infection Biomarkers Based on Metabolomics. Metabolites 2022, 12, 92. https://doi.org/10.3390/metabo12020092
Araújo R, Bento LFN, Fonseca TAH, Von Rekowski CP, da Cunha BR, Calado CRC. Infection Biomarkers Based on Metabolomics. Metabolites. 2022; 12(2):92. https://doi.org/10.3390/metabo12020092
Chicago/Turabian StyleAraújo, Rúben, Luís F. N. Bento, Tiago A. H. Fonseca, Cristiana P. Von Rekowski, Bernardo Ribeiro da Cunha, and Cecília R. C. Calado. 2022. "Infection Biomarkers Based on Metabolomics" Metabolites 12, no. 2: 92. https://doi.org/10.3390/metabo12020092
APA StyleAraújo, R., Bento, L. F. N., Fonseca, T. A. H., Von Rekowski, C. P., da Cunha, B. R., & Calado, C. R. C. (2022). Infection Biomarkers Based on Metabolomics. Metabolites, 12(2), 92. https://doi.org/10.3390/metabo12020092








