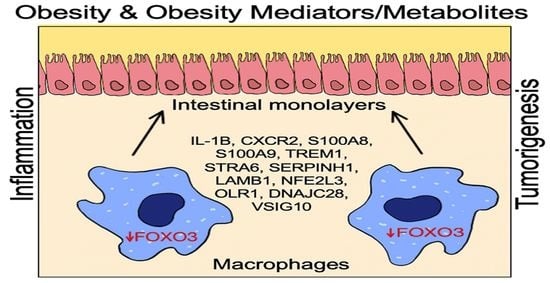
FOXO3 Expression in Macrophages Is Lowered by a High-Fat Diet and Regulates Colonic Inflammation and Tumorigenesis
Abstract
:1. Introduction
2. Results
2.1. HFD Obesity in Mice Mediates Increased Presence of Macrophages and Loss of FOXO3 in Colon
2.2. FOXO3 Deficiency in Macrophages Is Associated with Colonic Inflammation and Cancer
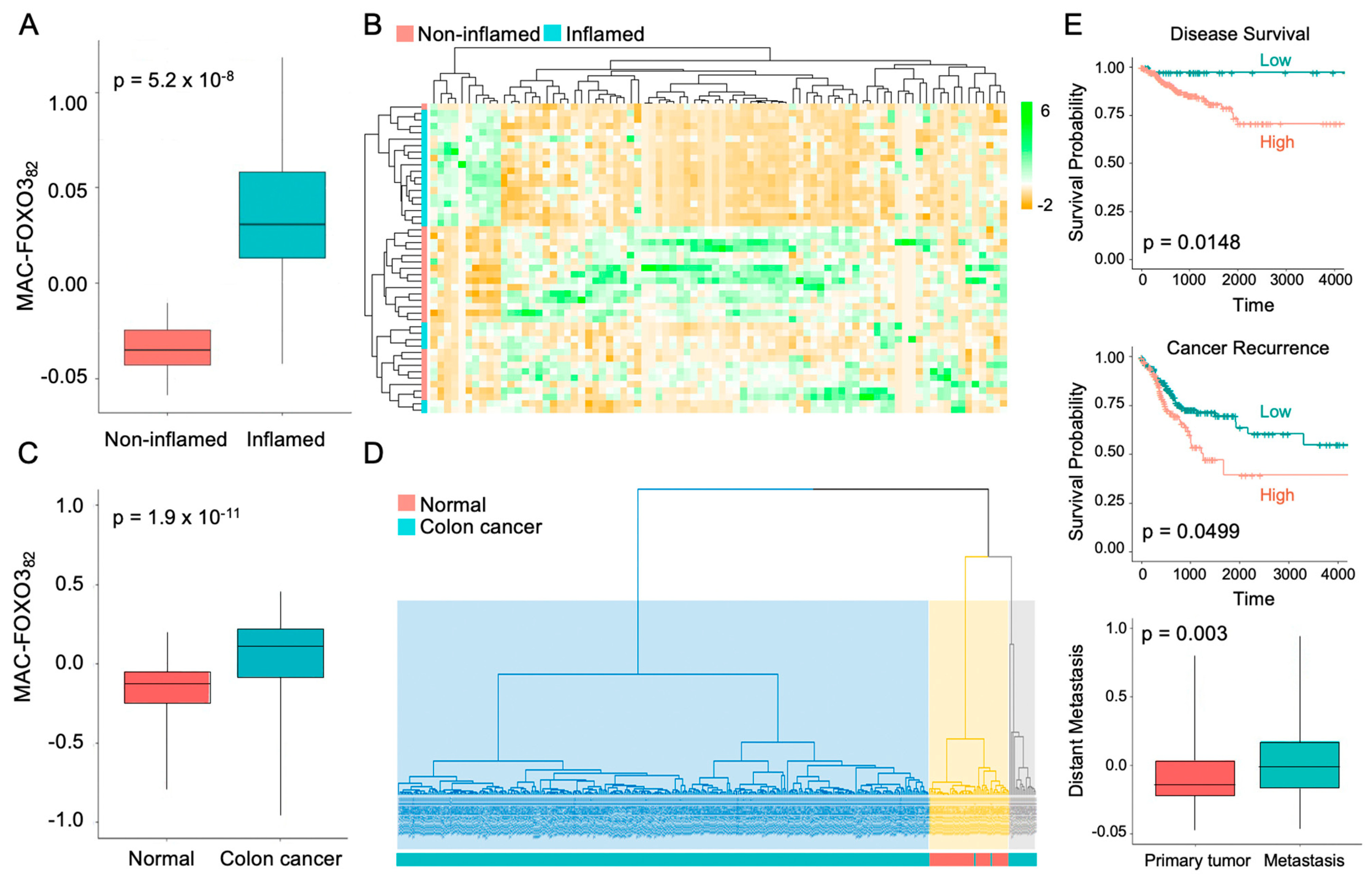
2.3. FOXO3-Deficient Macrophage Signature Is Prevalent in Human Colonic Inflammation and Cancer
2.4. Expression of Select FOXO3-Dependent Genes in Peritoneal Macrophages
3. Discussion
4. Materials and Methods
4.1. Human IBD and Colon Cancer Samples
4.2. Mice
4.3. Mouse Peritoneal Macrophage
4.4. RNA Isolation and cDNA Synthesis
4.5. qPCR
4.6. Protein Extraction and Immunoblot
4.7. RNA Sequencing and Differential Expression Testing
4.8. Transcriptome and Pathway Analysis
4.9. Hierarchical Clustering
4.10. Principal Component Analysis and Transcriptional Signature score
4.11. CIBERSORT
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, G.A. The underlying basis for obesity: Relationship to cancer. J. Nutr. 2002, 132, 3451S–3455S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demark-Wahnefried, W.; Platz, E.A.; Ligibel, J.A.; Blair, C.K.; Courneya, K.S.; Meyerhardt, J.A.; Ganz, P.A.; Rock, C.L.; Schmitz, K.H.; Wadden, T.; et al. The role of obesity in cancer survival and recurrence. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1244–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Jung, H.S.; Bae, S.W.; Kim, J.H.; Park, B.L.; Choi, Y.H.; Cho, H.Y.; Cheong, H.S.; Shin, H.D. Polymorphisms in FOXO gene family and association analysis with BMI. Obesity 2006, 14, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.L. Foxo in the immune system. Oncogene 2008, 27, 2337–2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senokuchi, T.; Liang, C.P.; Seimon, T.A.; Han, S.; Matsumoto, M.; Banks, A.S.; Paik, J.H.; DePinho, R.A.; Accili, D.; Tabas, I.; et al. Forkhead transcription factors (FoxOs) promote apoptosis of insulin-resistant macrophages during cholesterol-induced endoplasmic reticulum stress. Diabetes 2008, 57, 2967–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na, Y.R.; Stakenborg, M.; Seok, S.H.; Matteoli, G. Macrophages in intestinal inflammation and resolution: A potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Hamidzadeh, K.; Goncalves, R. Macrophages and the maintenance of homeostasis. Cell. Mol. Immunol. 2021, 18, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Munari, F.; Sanchez-Rodriguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Yang, M.; Ericsson, A.C. Function of Macrophages in Disease: Current Understanding on Molecular Mechanisms. Front. Immunol. 2021, 12, 620510. [Google Scholar] [CrossRef]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavez-Galan, L.; Olleros, M.L.; Vesin, D.; Garcia, I. Much More than M1 and M2 Macrophages, There are also CD169(+) and TCR(+) Macrophages. Front. Immunol. 2015, 6, 263. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.L.; Singer, K.; Lumeng, C.N. Adipose tissue macrophages: Phenotypic plasticity and diversity in lean and obese states. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 341–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isidro, R.A.; Appleyard, C.B. Colonic macrophage polarization in homeostasis, inflammation, and cancer. Am. J. Physiol.-Gastrointest. Liver Physiol. 2016, 311, G59–G73. [Google Scholar] [CrossRef] [PubMed]
- Hamidzadeh, K.; Christensen, S.M.; Dalby, E.; Chandrasekaran, P.; Mosser, D.M. Macrophages and the Recovery from Acute and Chronic Inflammation. Annu. Rev. Physiol. 2017, 79, 567–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Dasgupta, S.; Fu, Y.; Bailey, B.; Roy, C.; Lightcap, E.; Faustin, B. Subsets of mononuclear phagocytes are enriched in the inflamed colons of patients with IBD. BMC Immunol. 2019, 20, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhl, A.A.; Erben, U.; Kredel, L.I.; Siegmund, B. Diversity of Intestinal Macrophages in Inflammatory Bowel Diseases. Front. Immunol. 2015, 6, 613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, S.R.; Schmid, M.C. Macrophages as Key Drivers of Cancer Progression and Metastasis. Mediat. Inflamm. 2017, 2017, 9624760. [Google Scholar] [CrossRef] [PubMed]
- Dandekar, R.C.; Kingaonkar, A.V.; Dhabekar, G.S. Role of macrophages in malignancy. Ann. Maxillofac. Surg. 2011, 1, 150–154. [Google Scholar] [CrossRef] [Green Version]
- Qiu, S.Q.; Waaijer, S.J.H.; Zwager, M.C.; de Vries, E.G.E.; van der Vegt, B.; Schroder, C.P. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat. Rev. 2018, 70, 178–189. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Liu, Q.; Liao, Q. Tumor-Associated Macrophages in Pancreatic Ductal Adenocarcinoma: Origin, Polarization, Function, and Reprogramming. Front. Cell Dev. Biol. 2020, 8, 607209. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chen, B.; Yang, Z. The Role of Tumor-Associated Macrophages in Colorectal Carcinoma Progression. Cell. Physiol. Biochem. 2018, 45, 356–365. [Google Scholar] [CrossRef]
- Edin, S.; Wikberg, M.L.; Dahlin, A.M.; Rutegard, J.; Oberg, A.; Oldenborg, P.A.; Palmqvist, R. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS ONE 2012, 7, e47045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefanetti, R.J.; Voisin, S.; Russell, A.; Lamon, S. Recent advances in understanding the role of FOXO3. F1000Research 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhu, X.; Sun, X.; Yang, X.; Chang, X.; Xia, M.; Lu, Y.; Xia, P.; Yan, H.; Bian, H.; et al. FoxO3 regulates hepatic triglyceride metabolism via modulation of the expression of sterol regulatory-element binding protein 1c. Lipids Health Dis. 2019, 18, 197. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Dong, H.H. FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 2017, 233, R67–R79. [Google Scholar] [CrossRef] [Green Version]
- Zemva, J.; Schilbach, K.; Stohr, O.; Moll, L.; Franko, A.; Krone, W.; Wiesner, R.J.; Schubert, M. Central FoxO3a and FoxO6 expression is down-regulated in obesity induced diabetes but not in aging. Exp. Clin. Endocrinol. Diabetes 2012, 120, 340–350. [Google Scholar] [CrossRef]
- Heller, S.; Cable, C.; Penrose, H.; Makboul, R.; Biswas, D.; Cabe, M.; Crawford, S.E.; Savkovic, S.D. Intestinal inflammation requires FOXO3 and Prostaglandin E2 dependent lipogenesis and elevated lipid droplets. Am. J. Physiol.-Gastrointest. Liver Physiol. 2016, 310, G844–G854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, W.; Fitchev, P.S.; Cornwell, M.L.; Greenberg, J.; Cabe, M.; Weber, C.R.; Roy, H.K.; Crawford, S.E.; Savkovic, S.D. FOXO3 growth inhibition of colonic cells is dependent on intraepithelial lipid droplet density. J. Biol. Chem. 2013, 288, 16274–16281. [Google Scholar] [CrossRef] [Green Version]
- Snoeks, L.; Weber, C.R.; Turner, J.R.; Bhattacharyya, M.; Wasland, K.; Savkovic, S.D. Tumor suppressor Foxo3a is involved in the regulation of lipopolysaccharide-induced interleukin-8 in intestinal HT-29 cells. Infect. Immun. 2008, 76, 4677–4685. [Google Scholar] [CrossRef] [Green Version]
- Snoeks, L.; Weber, C.R.; Wasland, K.; Turner, J.R.; Vainder, C.; Qi, W.; Savkovic, S.D. Tumor suppressor FOXO3 participates in the regulation of intestinal inflammation. Lab. Investig. J. Tech. Methods Pathol. 2009, 89, 1053–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.C.; Espeli, M.; Anderson, C.A.; Linterman, M.A.; Pocock, J.M.; Williams, N.J.; Roberts, R.; Viatte, S.; Fu, B.; Peshu, N.; et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 2013, 155, 57–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Huang, Y.; Zhao, Y.; Zheng, J. Transcription factor FOXO3a mediates apoptosis in HIV-1-infected macrophages. J. Immunol. 2008, 180, 898–906. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.L. Forkhead transcription factors in chronic inflammation. Int. J. Biochem. Cell Biol. 2010, 42, 482–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Ao, X.; Ding, W.; Ponnusamy, M.; Wu, W.; Hao, X.; Yu, W.; Wang, Y.; Li, P.; Wang, J. Critical role of FOXO3a in carcinogenesis. Mol. Cancer 2018, 17, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullock, M.D.; Bruce, A.; Sreekumar, R.; Curtis, n.; Cheung, T.; Reading, I.; Primrose, J.N.; Ottensmeier, C.; Packham, G.K.; Thomas, G.; et al. FOXO3 expression during colorectal cancer progression: Biomarker potential reflects a tumour suppressor role. Br. J. Cancer 2013, 109, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Weber, C.R.; Wasland, K.; Roy, H.; Wali, R.; Joshi, S.; Savkovic, S.D. Tumor suppressor FOXO3 mediates signals from the EGF receptor to regulate proliferation of colonic cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 2011, 300, G264–G272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penrose, H.M.; Cable, C.; Heller, S.; Ungerleider, n.; Nakhoul, H.; Baddoo, M.; Hartono, A.B.; Lee, S.B.; Burow, M.E.; Flemington, E.F.; et al. Loss of Forkhead Box O3 Facilitates Inflammatory Colon Cancer: Transcriptome Profiling of the Immune Landscape and Novel Targets. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 391–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castoldi, A.; Naffah de Souza, C.; Camara, N.O.; Moraes-Vieira, P.M. The Macrophage Switch in Obesity Development. Front. Immunol. 2015, 6, 637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, Y.; Nakae, J.; Watanabe, N.; Kikuchi, T.; Tateya, S.; Tamori, Y.; Kaneko, M.; Abe, T.; Onodera, M.; Itoh, H. Colonic Pro-inflammatory Macrophages Cause Insulin Resistance in an Intestinal Ccl2/Ccr2-Dependent Manner. Cell Metab. 2016, 24, 295–310. [Google Scholar] [CrossRef] [Green Version]
- Penrose, H.M.; Heller, S.; Cable, C.; Nakhoul, H.; Baddoo, M.; Flemington, E.; Crawford, S.E.; Savkovic, S.D. High-fat diet induced leptin and Wnt expression: RNA-sequencing and pathway analysis of mouse colonic tissue and tumors. Carcinogenesis 2017, 38, 302–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, L.C.; Rosas, M.; Jenkins, S.J.; Liao, C.T.; Scurr, M.J.; Brombacher, F.; Fraser, D.J.; Allen, J.E.; Jones, S.A.; Taylor, P.R. Distinct bone marrow-derived and tissue-resident macrophage lineages proliferate at key stages during inflammation. Nat. Commun. 2013, 4, 1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Layoun, A.; Samba, M.; Santos, M.M. Isolation of murine peritoneal macrophages to carry out gene expression analysis upon Toll-like receptors stimulation. J. Vis. Exp. 2015, e52749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misharin, A.V.; Saber, R.; Perlman, H. Eosinophil contamination of thioglycollate-elicited peritoneal macrophage cultures skews the functional readouts of in vitro assays. J. Leukoc. Biol. 2012, 92, 325–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Ding, S.; Jiang, H.; Liu, G. Roles of Macrophages in the Development and Treatment of Gut Inflammation. Front. Cell Dev. Biol. 2021, 9, 625423. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic reprogramming in macrophage responses. Biomark. Res. 2021, 9, 1. [Google Scholar] [CrossRef]
- Franze, E.; Laudisi, F.; Di Grazia, A.; Maronek, M.; Bellato, V.; Sica, G.; Monteleone, G. Macrophages produce and functionally respond to interleukin-34 in colon cancer. Cell Death Discov. 2020, 6, 117. [Google Scholar] [CrossRef]
- Wu, X.; Fan, Z.; Chen, M.; Chen, Y.; Rong, D.; Cui, Z.; Yuan, Y.; Zhuo, L.; Xu, Y. Forkhead transcription factor FOXO3a mediates interferon-gamma-induced MHC II transcription in macrophages. Immunology 2019, 158, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Braster, R.; Bogels, M.; Beelen, R.H.; van Egmond, M. The delicate balance of macrophages in colorectal cancer; their role in tumour development and therapeutic potential. Immunobiology 2017, 222, 21–30. [Google Scholar] [CrossRef]
- Mao, L.; Kitani, A.; Strober, W.; Fuss, I.J. The Role of NLRP3 and IL-1beta in the Pathogenesis of Inflammatory Bowel Disease. Front. Immunol. 2018, 9, 2566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, L.; Pappan, L.; Galliher-Beckley, A.; Shi, J. IL-1beta promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Mol. Cancer 2012, 11, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akasaki, Y.; Hasegawa, A.; Saito, M.; Asahara, H.; Iwamoto, Y.; Lotz, M.K. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthr. Cartil. 2014, 22, 162–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, A.; Park, L.; Shah, M.; Drotar, M.; Calaminus, S.; Hopcroft, L.E.; Kinstrie, R.; Guitart, A.V.; Dunn, K.; Abraham, S.A.; et al. CXCR2 and CXCL4 regulate survival and self-renewal of hematopoietic stem/progenitor cells. Blood 2016, 128, 371–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; He, H.; Fan, L.; Ma, C.; Xu, Z.; Xue, Y.; Wang, Y.; Zhang, C.; Zhou, G. Blockade of CXCR2 suppresses proinflammatory activities of neutrophils in ulcerative colitis. Am. J. Transl. Res. 2020, 12, 5237–5251. [Google Scholar] [PubMed]
- Desurmont, T.; Skrypek, n.; Duhamel, A.; Jonckheere, n.; Millet, G.; Leteurtre, E.; Gosset, P.; Duchene, B.; Ramdane, n.; Hebbar, M.; et al. Overexpression of chemokine receptor CXCR2 and ligand CXCL7 in liver metastases from colon cancer is correlated to shorter disease-free and overall survival. Cancer Sci. 2015, 106, 262–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in Inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Williams, R.; Wang, L.; Vogl, T.; Srikrishna, G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol. Cancer Res. 2011, 9, 133–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, K.; Okabe, M.; Kimura, Y.; Itoh, H.; Ikemoto, M. Serum S100A8/A9 as a Potentially Sensitive Biomarker for Inflammatory Bowel Disease. Lab. Med. 2019, 50, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Caer, C.; Gorreja, F.; Forsskahl, S.K.; Brynjolfsson, S.F.; Szeponik, L.; Magnusson, M.K.; Borjesson, L.G.; Block, M.; Bexe-Lindskog, E.; Wick, M.J. TREM-1+ Macrophages Define a Pathogenic Cell Subset in the Intestine of Crohn’s Disease Patients. J. Crohns Colitis 2021, 15, 1346–1361. [Google Scholar] [CrossRef]
- Ho, C.C.; Liao, W.Y.; Wang, C.Y.; Lu, Y.H.; Huang, H.Y.; Chen, H.Y.; Chan, W.K.; Chen, H.W.; Yang, P.C. TREM-1 expression in tumor-associated macrophages and clinical outcome in lung cancer. Am. J. Respir. Crit. Care Med. 2008, 177, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Saurer, L.; Zysset, D.; Rihs, S.; Mager, L.; Gusberti, M.; Simillion, C.; Lugli, A.; Zlobec, I.; Krebs, P.; Mueller, C. TREM-1 promotes intestinal tumorigenesis. Sci. Rep. 2017, 7, 14870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, M.; von Lintig, J. STRA6: Role in cellular retinol uptake and efflux. Hepatobiliary Surg. Nutr. 2015, 4, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Gokulakrishnan, K.; Pandey, G.K.; Sathishkumar, C.; Sundararajan, S.; Durairaj, P.; Manickam, N.; Mohan, V.; Balasubramanyam, M. Augmentation of RBP4/STRA6 signaling leads to insulin resistance and inflammation and the plausible therapeutic role of vildagliptin and metformin. Mol. Biol. Rep. 2021, 48, 4093–4106. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.M.; Teixeira, F.M.E.; Sato, M.N. Impact of Retinoic Acid on Immune Cells and Inflammatory Diseases. Mediat. Inflamm. 2018, 2018, 3067126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muniz-Hernandez, S.; Velazquez-Fernandez, J.B.; Diaz-Chavez, J.; Mondragon-Fonseca, O.; Mayen-Lobo, Y.; Ortega, A.; Lopez-Lopez, M.; Arrieta, O. STRA6 Polymorphisms Are Associated with EGFR Mutations in Locally-Advanced and Metastatic Non-Small Cell Lung Cancer Patients. Front. Oncol. 2020, 10, 579561. [Google Scholar] [CrossRef] [PubMed]
- Karunanithi, S.; Levi, L.; DeVecchio, J.; Karagkounis, G.; Reizes, O.; Lathia, J.D.; Kalady, M.F.; Noy, N. RBP4-STRA6 Pathway Drives Cancer Stem Cell Maintenance and Mediates High-Fat Diet-Induced Colon Carcinogenesis. Stem Cell Rep. 2017, 9, 438–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widmer, C.; Gebauer, J.M.; Brunstein, E.; Rosenbaum, S.; Zaucke, F.; Drogemuller, C.; Leeb, T.; Baumann, U. Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc. Natl. Acad. Sci. USA 2012, 109, 13243–13247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi-Galibov, O.; Lavon, H.; Wassermann-Dozorets, R.; Pevsner-Fischer, M.; Mayer, S.; Wershof, E.; Stein, Y.; Brown, L.E.; Zhang, W.; Friedman, G.; et al. Heat Shock Factor 1-dependent extracellular matrix remodeling mediates the transition from chronic intestinal inflammation to colon cancer. Nat. Commun. 2020, 11, 6245. [Google Scholar] [CrossRef] [PubMed]
- Spenle, C.; Lefebvre, O.; Lacroute, J.; Mechine-Neuville, A.; Barreau, F.; Blottiere, H.M.; Duclos, B.; Arnold, C.; Hussenet, T.; Hemmerle, J.; et al. The laminin response in inflammatory bowel disease: Protection or malignancy? PLoS ONE 2014, 9, e111336. [Google Scholar] [CrossRef] [Green Version]
- Consortium, U.I.G.; Barrett, J.C.; Lee, J.C.; Lees, C.W.; Prescott, N.J.; Anderson, C.A.; Phillips, A.; Wesley, E.; Parnell, K.; Zhang, H.; et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009, 41, 1330–1334. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.I.; Lees, C.W. Genetics of ulcerative colitis. Inflamm. Bowel Dis. 2011, 17, 831–848. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Lim, H.S.; Lin, H.L.; Tan, H.T.; Lim, T.K.; Cheong, W.K.; Cheah, P.Y.; Tang, C.L.; Chow, P.K.; Chung, M.C. Analysis of colorectal cancer glyco-secretome identifies laminin beta-1 (LAMB1) as a potential serological biomarker for colorectal cancer. Proteomics 2015, 15, 3905–3920. [Google Scholar] [CrossRef] [PubMed]
- Chevillard, G.; Blank, V. NFE2L3 (NRF3): The Cinderella of the Cap‘n’Collar transcription factors. Cell. Mol. Life Sci. 2011, 68, 3337–3348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Sun, T. Transcription Factors That Regulate the Pathogenesis of Ulcerative Colitis. BioMed Res. Int. 2020, 2020, 7402657. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, A.; Katoh, H.; Hatanaka, A.; Iwanari, H.; Nakamura, N.; Hamakubo, T.; Natsume, T.; Waku, T.; Kobayashi, A. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci. Rep. 2017, 7, 12494. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, n.; Richard, S.; Silvy, F.; Verine, A.; Lombardo, D. Lectin-like Ox-LDL receptor is expressed in human INT-407 intestinal cells: Involvement in the transcytosis of pancreatic bile salt-dependent lipase. Mol. Biol. Cell 2003, 14, 2861–2875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Roso, M.B.; Gil-Zamorano, J.; Lopez de Las Hazas, M.C.; Tome-Carneiro, J.; Crespo, M.C.; Latasa, M.J.; Briand, O.; Sanchez-Lopez, D.; Ortiz, A.I.; Visioli, F.; et al. Intestinal Lipid Metabolism Genes Regulated by miRNAs. Front. Genet. 2020, 11, 707. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Xiong, G.; Feng, M.; Zhao, F.; Qiu, J.; Liu, Y.; Cao, Z.; Wang, H.; Yang, J.; You, L.; et al. OLR1 Promotes Pancreatic Cancer Metastasis via Increased c-Myc Expression and Transcription of HMGA2. Mol. Cancer Res. 2020, 18, 685–697. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, H.A.; Iliopoulos, D.; Joshi, A.; Zhang, Y.; Jaeger, S.A.; Bulyk, M.; Tsichlis, P.N.; Shirley Liu, X.; Struhl, K. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell 2010, 17, 348–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murdocca, M.; Capuano, R.; Pucci, S.; Cicconi, R.; Polidoro, C.; Catini, A.; Martinelli, E.; Paolesse, R.; Orlandi, A.; Mango, R.; et al. Targeting LOX-1 Inhibits Colorectal Cancer Metastasis in an Animal Model. Front. Oncol. 2019, 9, 927. [Google Scholar] [CrossRef] [PubMed]
- Rand, T.A.; Sutou, K.; Tanabe, K.; Jeong, D.; Nomura, M.; Kitaoka, F.; Tomoda, E.; Narita, M.; Nakamura, M.; Nakamura, M.; et al. MYC Releases Early Reprogrammed Human Cells from Proliferation Pause via Retinoblastoma Protein Inhibition. Cell Rep. 2018, 23, 361–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarouchlioti, C.; Parfitt, D.A.; Li, W.; Gittings, L.M.; Cheetham, M.E. DNAJ Proteins in neurodegeneration: Essential and protective factors. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daemen, S.; Schilling, J.D. The Interplay Between Tissue Niche and Macrophage Cellular Metabolism in Obesity. Front. Immunol. 2019, 10, 3133. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.P.; Han, S.; Senokuchi, T.; Tall, A.R. The macrophage at the crossroads of insulin resistance and atherosclerosis. Circ. Res. 2007, 100, 1546–1555. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Hron, J.D.; Peng, S.L. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity 2004, 21, 203–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Goncalves, R.; Mosser, D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008, Chapter 14, Unit 14 11. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [Green Version]





| Gene | Gene Name | FC | p-Value | |
|---|---|---|---|---|
| 1 | Stfa1 | Stefin-A 1 | 225.6 | 4.8 × 10−8 |
| 2 | Entpd3 | Ectonucleoside Triphosphate Diphosphohydrolase 3 | 113.6 | 3.0 × 10−9 |
| 3 | Amer2 | APC Membrane Recruitment Protein 2 | 67.7 | 7.6 × 10−10 |
| 4 | S100a9 | S100 Calcium Binding Protein A9 | 66.7 | 1.1 × 10−4 |
| 5 | Mrgpra2b | MAS Related GPR Family Member X2 | 35.3 | 2.4 × 10−4 |
| 6 | Asprv1 | Aspartic Peptidase Retroviral-like 1 | 33.9 | 7.5 × 10−18 |
| 7 | Sycp2 | Synaptonemal Complex Protein 2 | 30.2 | 1.6 × 10−13 |
| 8 | Chil1 | Chitinase 3-like 1 | 29.5 | 1.4 × 10−6 |
| 9 | Olfm4 | Olfactomedin 4 | 24.3 | 6.2 × 10−8 |
| 10 | Cxcr2 | C-X-C Motif Chemokine Receptor 2 | 21.5 | 8.4 × 10−35 |
| 11 | Catspere2 | Catsper Channel Auxiliary Subunit Epsilon | 18.6 | 9.7 × 10−5 |
| 12 | Il1f9 | Interleukin 1 Family, Member 9 | 13.6 | 5.9 × 10−13 |
| 13 | Amd2 | Adenosylmethionine Decarboxylase 1 Pseudogene 2 | 11.8 | 2.6 × 10−34 |
| 14 | Alas2 | 5′-Aminolevulinate Synthase 2 | 10.1 | 1.9 × 10−5 |
| 15 | S100a8 | S100 Calcium-binding Protein A8 | 9.9 | 4.9 × 10−7 |
| 16 | Steap4 | Six-Transmembrane Epithelial Antigen Of Prostate 4 | 8.0 | 2.1 × 10−12 |
| 17 | Il1r2 | Interleukin 1 Receptor Type 2 | 7.9 | 6.6 × 10−8 |
| 18 | Hba-a2 | Hemoglobin Subunit Alpha 2 | 7.7 | 3.1 × 10−6 |
| 19 | Slc38a4 | Solute Carrier Family 38 Member 4 | 7.7 | 3.9 × 10−8 |
| 20 | Stra6 | Stimulated By Retinoic Acid 6 | 7.2 | 3.7 × 10−5 |
| 21 | Col24a1 | Collagen Type XXIV Alpha 1 Chain | 6.7 | 9.3 × 10−6 |
| 22 | Hba-a1 | Hemoglobin Subunit Alpha 1 | 5.9 | 1.7 × 10−7 |
| 23 | Trem1 | Triggering Receptor Expressed On Myeloid Cells 1 | 5.7 | 1.2 × 10−15 |
| 24 | Lin28a | Lin-28 Homolog A | 5.6 | 1.5 × 10−4 |
| 25 | Il1b | Interleukin 1 Beta | 5.4 | 1.5 × 10−9 |
| 26 | Ambp | Alpha-1-Microglobulin/Bikunin Precursor | 5.1 | 2.3 × 10−4 |
| 27 | Ifitm1 | Interferon-induced Transmembrane Protein 1 | 5.1 | 5.8 × 10−5 |
| 28 | Nfe2l3 | Nuclear Factor, Erythroid 2-like 3 | 4.9 | 1.9 × 10−5 |
| 29 | Kirrel | Kin Of Irregular Chiasm-like Protein 1 | 4.9 | 9.2 × 10−5 |
| 30 | Lamb1 | Laminin Subunit Beta 1 | 4.3 | 6.0 × 10−7 |
| 31 | Serpinh1 | Serpin Peptidase Inhibitor, Clade H, Member 1, | 4.0 | 1.7 × 10−6 |
| 32 | Adamts1 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 1 | 3.9 | 1.8 × 10−4 |
| 33 | Pkhd1l1 | Polycystic Kidney and Hepatic Disease 1-like 1 | 3.8 | 1.8 × 10−4 |
| 34 | Fads2 | Fatty Acid Desaturase 2 | 3.7 | 6.2 × 10−6 |
| 35 | Col1a1 | Collagen Type I Alpha 1 Chain | 3.6 | 2.5 × 10−4 |
| 36 | Dmkn | Dermokine | 3.6 | 4.0 × 10−5 |
| 37 | Rbp1 | Retinol-binding Protein 1 | 3.5 | 7.2 × 10−5 |
| 38 | Tcaf1 | TRPM8 Channel Associated Factor 1 | 3.4 | 9.0 × 10−8 |
| 39 | Hpgd | 15-Hydroxyprostaglandin Dehydrogenase | 3.4 | 2.1 × 10−7 |
| 40 | septin3 | Neuronal-specific Septin-3 | 3.4 | 6.2 × 10−5 |
| 41 | Nr4a3 | Nuclear Receptor Subfamily 4 Group A Member 3 | 3.3 | 5.8 × 10−7 |
| 42 | Ccl2 | C-C Motif Chemokine Ligand 2 | 3.3 | 3.1 × 10−5 |
| 43 | Ltbp2 | Latent Transforming Growth Factor Beta-binding Protein 2 | 3.3 | 1.6 × 10−4 |
| 44 | Dclk1 | Doublecortin-like Kinase 1 | 3.2 | 3.6 × 10−6 |
| 45 | Map1b | Microtubule Associated Protein 1B | 3.2 | 2.2 × 10−4 |
| 46 | Wt1 | Wilms Tumor 1 | 3.2 | 4.1 × 10−5 |
| 47 | Col1a2 | Collagen Type I Alpha 2 Chain | 3.1 | 1.9 × 10−4 |
| 48 | Ptprf | Protein Tyrosine Phosphatase Receptor Type F | 3.0 | 2.2 × 10−4 |
| 49 | Krt19 | Keratin 19 | 2.9 | 1.0 × 10−4 |
| 50 | Arhgef17 | Rho-specific Guanine-Nucleotide Exchange Factor | 2.8 | 1.5 × 10−4 |
| 51 | Il1r1 | Interleukin 1 Receptor Type 1 | 2.7 | 6.3 × 10−5 |
| 52 | Treml4 | Triggering Receptor Expressed On Myeloid Cells-like 4 | 2.7 | 1.9 × 10−4 |
| 53 | Vcan | Versican | 2.5 | 1.3 × 10−5 |
| 54 | Klrb1b | Killer Cell Lectin-like Receptor B1 | 2.4 | 2.9 × 10−5 |
| 55 | Olr1 | Oxidized Low Density Lipoprotein Receptor 1 | 2.1 | 2.8 × 10−4 |
| 56 | Pram1 | PML-RARA Regulated Adaptor Molecule 1 | 2.1 | 2.2 × 10−5 |
| 57 | Ccnb1 | Cyclin B1 | 2.1 | 1.9 × 10−4 |
| 58 | Npl | N-Acetylneuraminate Pyruvate Lyase | 2.0 | 6.2 × 10−5 |
| 59 | Cdk1 | Cyclin-dependent Kinase 1 | 1.9 | 1.5 × 10−4 |
| 60 | Tbc1d16 | TBC1 Domain Family Member 16 | 1.8 | 7.4 × 10−5 |
| 61 | Dab2 | DAB Adaptor Protein 2 | 1.8 | 3.1 × 10−7 |
| 62 | Nfil3 | Nuclear Factor, Interleukin 3 Regulated | 1.8 | 2.9 × 10−5 |
| 63 | Tbc1d4 | TBC1 Domain Family Member 4 | 1.7 | 1.2 × 10−4 |
| 64 | Samsn1 | SAM Domain, SH3 Domain And Nuclear Localization Signals 1 | 1.5 | 2.6 × 10−5 |
| 65 | Kcnk13 | Potassium Two Pore Domain Channel Subfamily K Member 13 | 1.5 | 1.4 × 10−4 |
| 66 | Fmo5 | Flavin Containing Dimethylaniline Monoxygenase 5 | −1.6 | 1.2 × 10−4 |
| 67 | Ip6k1 | Inositol Hexakisphosphate Kinase 1 | −1.6 | 8.4 × 10−5 |
| 68 | Wdr45 | WD Repeat Domain 45 | −1.7 | 2.2 × 10−5 |
| 69 | Serinc5 | Serine Incorporator 5 | −1.7 | 3.2 × 10−5 |
| 70 | Mrgprx2 | MAS-Related GPR Family Member X2 | −1.9 | 3.7 × 10−1 |
| 71 | Maml2 | Mastermind-like Transcriptional Coactivator 2 | −1.9 | 7.2 × 10−5 |
| 72 | Vsig10 | V-Set And Immunoglobulin Domain-containing 10 | −2.2 | 4.5 × 10−5 |
| 73 | Dnajc28 | DnaJ Heat Shock Protein Family (Hsp40) Member C28 | −2.2 | 7.9 × 10−7 |
| 74 | Rab6b | RAB6B, Member RAS Oncogene Family | −2.5 | 2.5 × 10−5 |
| 75 | Ypel3 | Yippee Like 3 | −2.9 | 4.2 × 10−19 |
| 76 | Armc2 | Armadillo Repeat-containing 2 | −3.1 | 2.8 × 10−6 |
| 77 | Cdkn2a | Cyclin-dependent Kinase Inhibitor 2A | −5.5 | 8.4 × 10−6 |
| 78 | Slc15a2 | Solute Carrier Family 15 Member 2 | −6.6 | 1.0 × 10−4 |
| 79 | Clec2g | C-Type Lectin Domain Family 2 Member D | −8.9 | 2.1 × 10−20 |
| 80 | Camk2b | Calcium/Calmodulin-dependent Protein Kinase II Beta | −9.4 | 3.2 × 10−11 |
| 81 | Slc25a27 | Solute Carrier Family 25 Member 27 | −9.4 | 4.3 × 10−6 |
| 82 | Foxo3a | Forkhead Box O3 | −22.0 | 8.1 × 10−43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iftikhar, R.; Penrose, H.M.; King, A.N.; Kim, Y.; Ruiz, E.; Kandil, E.; Machado, H.L.; Savkovic, S.D. FOXO3 Expression in Macrophages Is Lowered by a High-Fat Diet and Regulates Colonic Inflammation and Tumorigenesis. Metabolites 2022, 12, 250. https://doi.org/10.3390/metabo12030250
Iftikhar R, Penrose HM, King AN, Kim Y, Ruiz E, Kandil E, Machado HL, Savkovic SD. FOXO3 Expression in Macrophages Is Lowered by a High-Fat Diet and Regulates Colonic Inflammation and Tumorigenesis. Metabolites. 2022; 12(3):250. https://doi.org/10.3390/metabo12030250
Chicago/Turabian StyleIftikhar, Rida, Harrison M. Penrose, Angelle N. King, Yunah Kim, Emmanuelle Ruiz, Emad Kandil, Heather L. Machado, and Suzana D. Savkovic. 2022. "FOXO3 Expression in Macrophages Is Lowered by a High-Fat Diet and Regulates Colonic Inflammation and Tumorigenesis" Metabolites 12, no. 3: 250. https://doi.org/10.3390/metabo12030250
APA StyleIftikhar, R., Penrose, H. M., King, A. N., Kim, Y., Ruiz, E., Kandil, E., Machado, H. L., & Savkovic, S. D. (2022). FOXO3 Expression in Macrophages Is Lowered by a High-Fat Diet and Regulates Colonic Inflammation and Tumorigenesis. Metabolites, 12(3), 250. https://doi.org/10.3390/metabo12030250