Dysregulated Metabolism in EGFR-TKI Drug Resistant Non-Small-Cell Lung Cancer: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
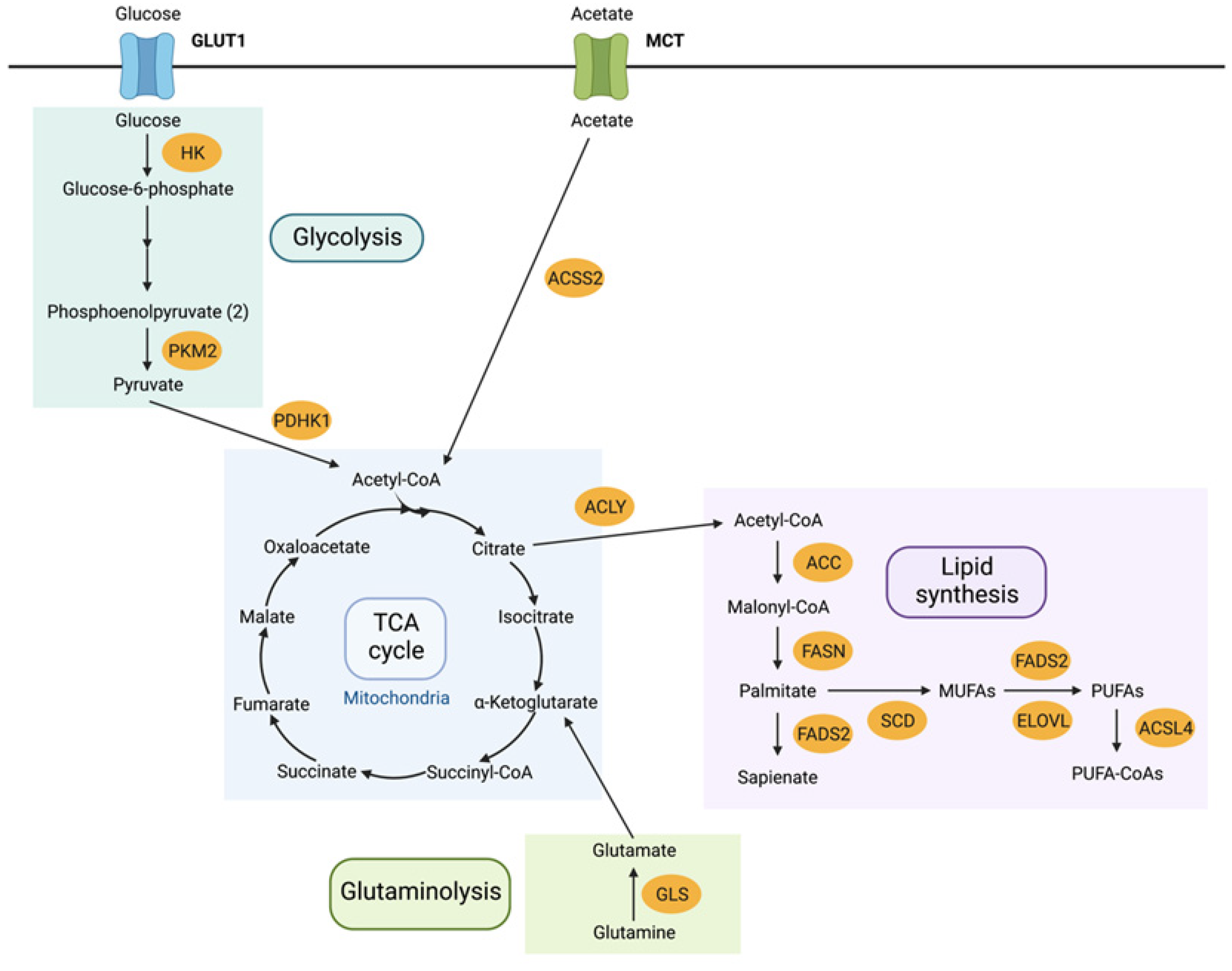
3.1. Metabolome Rewiring
3.2. Reversal of Drug Resistance by Metabolic Intervention
3.2.1. Protein Targets
3.2.2. Alternative Signalling Pathways
3.2.3. Transporters
3.2.4. Molecular Targets
3.3. Alterations in Dysregulated Lipid Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dyrstad, S.E.; Lotsberg, M.L.; Tan, T.Z.; Pettersen, I.K.N.; Hjellbrekke, S.; Tusubira, D.; Engelsen, A.S.T.; Daubon, T.; Mourier, A.; Thiery, J.P.; et al. Blocking Aerobic Glycolysis by Targeting Pyruvate Dehydrogenase Kinase in Combination with EGFR TKI and Ionizing Radiation Increases Therapeutic Effect in Non-Small Cell Lung Cancer Cells. Cancers 2021, 13, 941. [Google Scholar] [CrossRef] [PubMed]
- Gelatti, A.C.Z.; Drilon, A.; Santini, F.C. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer 2019, 137, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Stokes, W.; Chater, E.; Roy, R.; de Bruin, E.; Hu, Y.; Liu, Z.; Smit, E.F.; Heynen, G.J.; Downward, J.; et al. Decreased glutathione biosynthesis contributes to EGFR T790M-driven erlotinib resistance in non-small cell lung cancer. Cell Discov. 2016, 2, 16031. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Wang, J.; Ren, Y.; Meng, F.; Zeng, L. Pathological Mechanistic Studies of Osimertinib Resistance in Non-Small-Cell Lung Cancer Cells Using an Integrative Metabolomics-Proteomics Analysis. J. Oncol. 2020, 2020, 6249829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, W.; Huang, Y.; Gu, W.; Gan, J.; Wang, W.; Zhang, S.; Wang, K.; Zhan, J.; Yang, Y.; Huang, Y.; et al. PI3K-AKT-mTOR pathway alterations in advanced NSCLC patients after progression on EGFR-TKI and clinical response to EGFR-TKI plus everolimus combination therapy. Transl. Lung Cancer Res. 2020, 9, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Eberlein, C.A.; Stetson, D.; Markovets, A.A.; Al-Kadhimi, K.J.; Lai, Z.; Fisher, P.R.; Meador, C.B.; Spitzler, P.; Ichihara, E.; Ross, S.J.; et al. Acquired Resistance to the Mutant-Selective EGFR Inhibitor AZD9291 Is Associated with Increased Dependence on RAS Signaling in Preclinical Models. Cancer Res. 2015, 75, 2489–2500. [Google Scholar] [CrossRef] [Green Version]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [Green Version]
- De Rosa, V.; Iommelli, F.; Monti, M.; Fonti, R.; Votta, G.; Stoppelli, M.P.; Del Vecchio, S. Reversal of Warburg Effect and Reactivation of Oxidative Phosphorylation by Differential Inhibition of EGFR Signaling Pathways in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2015, 21, 5110–5120. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.J.; Eberlein, C.; Taylor, M.; Ashton, S.; Robinson, D.; Cross, D. Inhibition of oxidative phosphorylation suppresses the development of osimertinib resistance in a preclinical model of EGFR-driven lung adenocarcinoma. Oncotarget 2016, 7, 86313–86325. [Google Scholar] [CrossRef] [Green Version]
- Pham-Danis, C.; Gehrke, S.; Danis, E.; Rozhok, A.I.; Daniels, M.W.; Gao, D.; Collins, C.; Paola, J.T.D.; D’Alessandro, A.; DeGregori, J. Urea Cycle Sustains Cellular Energetics upon EGFR Inhibition in EGFR-Mutant NSCLC. Mol. Cancer Res. 2019, 17, 1351–1364. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Nam, B.; Choi, Y.J.; Kim, S.Y.; Lee, J.-E.; Sung, K.J.; Kim, W.S.; Choi, C.-M.; Chang, E.-J.; Koh, J.S.; et al. Enhanced Glycolysis Supports Cell Survival in EGFR-Mutant Lung Adenocarcinoma by Inhibiting Autophagy-Mediated EGFR Degradation. Cancer Res. 2018, 78, 4482–4496. [Google Scholar] [CrossRef] [Green Version]
- Röhrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turkoglu, O.; Zeb, A.; Graham, S.; Szyperski, T.; Szender, J.B.; Odunsi, K.; Bahado-Singh, R. Metabolomics of biomarker discovery in ovarian cancer: A systematic review of the current literature. Metabolomics 2016, 12, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Diao, X.-Y.; Zhang, X.; Shao, Q.; Feng, Y.-F.; An, X.; Wang, H.-Y. Identification of genetic alterations associated with primary resistance to EGFR-TKIs in advanced non-small-cell lung cancer patients with EGFR sensitive mutations. Cancer Commun. 2019, 39, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiying, C.; Zeyun, L.; Yongliang, Y.; Zijia, Z.; Ji, Z.; Xin, T.; Xiaojian, Z. A comprehensive analysis of metabolomics and transcriptomics in non-small cell lung cancer. PLoS ONE 2020, 15, e0232272. [Google Scholar] [CrossRef] [PubMed]
- Noreldeen, H.A.A.; Liu, X.; Xu, G. Metabolomics of lung cancer: Analytical platforms and their applications. J. Sep. Sci. 2020, 43, 120–133. [Google Scholar] [CrossRef]
- Dieterle, F.; Riefke, B.; Schlotterbeck, G.; Ross, A.; Senn, H.; Amberg, A. NMR and MS methods for metabonomics. Methods Mol. Biol. 2011, 691, 385–415. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, L.; Wang, D.; Jiang, S.; Cao, D.; Zhao, Z.; Huang, M.; Jin, J. Lipidomics reveals that sustained SREBP-1-dependent lipogenesis is a key mediator of gefitinib-acquired resistance in EGFR-mutant lung cancer. Cell Death Discov. 2021, 7, 353. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, D.; Owonikoko, T.K.; Ramalingam, S.S.; Sun, S.-Y. Induction of SREBP1 degradation coupled with suppression of SREBP1-mediated lipogenesis impacts the response of EGFR mutant NSCLC cells to osimertinib. Oncogene 2021, 40, 6653–6665. [Google Scholar] [CrossRef]
- Guo, S.; Wang, Y.; Zhou, D.; Li, Z. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci. Rep. 2015, 4, 5959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhu, X.; Wang, C.; Zhang, H.; Cai, Z. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget 2016, 7, 63437–63448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. CMB 2012, 1, 92–107. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 10, 89. [Google Scholar] [CrossRef]
- Covidence—Better Systematic Review Management. Available online: https://www.covidence.org/ (accessed on 15 February 2022).
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; the PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serizawa, M.; Kusuhara, M.; Zangiacomi, V.; Urakami, K.; Watanabe, M.; Takahashi, T.; Yamaguchi, K.; Yamamoto, N.; Koh, Y. Identification of metabolic signatures associated with erlotinib resistance of non-small cell lung cancer cells. Anticancer Res. 2014, 34, 2779–2787. [Google Scholar]
- Thiagarajan, P.S.; Wu, X.; Zhang, W.; Shi, I.; Bagai, R.; Leahy, P.; Feng, Y.; Veigl, M.; Lindner, D.; Danielpour, D.; et al. Transcriptomic-metabolomic reprogramming in EGFR-mutant NSCLC early adaptive drug escape linking TGFβ2-bioenergetics-mitochondrial priming. Oncotarget 2016, 7, 82013–82027. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.C.; Wells, J.M.; Chow, K.-H.; Huang, H.; Yuan, M.; Saxena, T.; Melnick, M.A.; Politi, K.; Asara, J.M.; Costa, D.B.; et al. miR-147b-mediated TCA cycle dysfunction and pseudohypoxia initiate drug tolerance to EGFR inhibitors in lung adenocarcinoma. Nat. Metab. 2019, 1, 460–474. [Google Scholar] [CrossRef]
- Suda, K.; Mitsudomi, T. Drug Tolerance to EGFR Tyrosine Kinase Inhibitors in Lung Cancers with EGFR Mutations. Cells 2021, 10, 1590. [Google Scholar] [CrossRef]
- Zhang, Z.; Deng, X.; Liu, Y.; Liu, Y.; Sun, L.; Chen, F. PKM2, function and expression and regulation. Cell Biosci. 2019, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Xie, C.; Fan, X.-X.; Jiang, Z.-B.; Wong, V.K.-W.; Xu, J.-H.; Yao, X.-J.; Liu, L.; Leung, E.L.-H. Novel direct AMPK activator suppresses non-small cell lung cancer through inhibition of lipid metabolism. Oncotarget 2017, 8, 96089–96102. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Yun, M.R.; Hong, Y.K.; Solca, F.; Kim, J.-H.; Kim, H.-J.; Cho, B.C. Glycolysis inhibition sensitizes non-small cell lung cancer with T790M mutation to irreversible EGFR inhibitors via translational suppression of Mcl-1 by AMPK activation. Mol. Cancer Ther. 2013, 12, 2145–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Jin, J.; Bao, X.; Zhan, W.-H.; Han, T.-Y.; Gan, M.; Zhang, C.; Wang, J. Inhibition of mitochondrial glutaminase activity reverses acquired erlotinib resistance in non-small cell lung cancer. Oncotarget 2016, 7, 610–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Zhang, S.-L.; Hu, X.; Tam, K.Y. Inhibition of pyruvate dehydrogenase kinase 1 enhances the anti-cancer effect of EGFR tyrosine kinase inhibitors in non-small cell lung cancer. Eur. J. Pharmacol. 2018, 838, 41–52. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [Green Version]
- Ye, M.; Wang, S.; Wan, T.; Jiang, R.; Qiu, Y.; Pei, L.; Pang, N.; Huang, Y.; Huang, Y.; Zhang, Z.; et al. Combined Inhibitions of Glycolysis and AKT/autophagy Can Overcome Resistance to EGFR-targeted Therapy of Lung Cancer. J. Cancer 2017, 8, 3774–3784. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Mei, H.; Gao, Y.; Xie, X.; Nie, H.; Li, T.; Zhang, H.; Jia, L. Co-delivery of oxygen and erlotinib by aptamer-modified liposomal complexes to reverse hypoxia-induced drug resistance in lung cancer. Biomaterials 2017, 145, 56–71. [Google Scholar] [CrossRef]
- Ye, M.; Pang, N.; Wan, T.; Huang, Y.; Wei, T.; Jiang, X.; Zhou, Y.; Huang, Y.; Yang, H.; Zhang, Z.; et al. Oxidized Vitamin C (DHA) Overcomes Resistance to EGFR-targeted Therapy of Lung Cancer through Disturbing Energy Homeostasis. J. Cancer 2019, 10, 757–764. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Okada, M.; Takeda, H.; Kuramoto, K.; Sanomachi, T.; Togashi, K.; Seino, S.; Yamamoto, M.; Yoshioka, T.; Kitanaka, C. Involvement of GLUT1-mediated glucose transport and metabolism in gefitinib resistance of non-small-cell lung cancer cells. Oncotarget 2018, 9, 32667–32679. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-Y.; Hsu, L.-H.; Chen, C.-Y.; Chang, G.-C.; Chang, H.-W.; Hung, Y.-M.; Liu, K.-J.; Kao, S.-H. Inhibition of Alternative Cancer Cell Metabolism of EGFR Mutated Non-Small Cell Lung Cancer Serves as a Potential Therapeutic Strategy. Cancers 2020, 12, 181. [Google Scholar] [CrossRef] [Green Version]
- Bach, D.-H.; Luu, T.-T.-T.; Kim, D.; An, Y.J.; Park, S.; Park, H.J.; Lee, S.K. BMP4 Upregulation Is Associated with Acquired Drug Resistance and Fatty Acid Metabolism in EGFR-Mutant Non-Small-Cell Lung Cancer Cells. Mol. Ther. Nucleic Acids 2018, 12, 817–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, C.-T.; Demetriou, A.N.; Ung, N.; Choudhury, N.; Ghaffarian, K.; Ruderman, D.L.; Mumenthaler, S.M. mTORC2 contributes to the metabolic reprogramming in EGFR tyrosine-kinase inhibitor resistant cells in non-small cell lung cancer. Cancer Lett. 2018, 434, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, K.; Zhang, X. Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: Mini review. Oncotarget 2017, 8, 115774–115786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Levantini, E.; Teo, J.T.; Goggi, J.; Clohessy, J.G.; Wu, C.S.; Chen, L.; Yang, H.; Krishnan, I.; Kocher, O.; et al. Fatty acid synthase mediates EGFR palmitoylation in EGFR mutated non-small cell lung cancer. EMBO Mol. Med. 2018, 10, e8313. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-A.; Xiong, X.; Zaytseva, Y.Y.; Napier, D.L.; Vallee, E.; Li, A.T.; Wang, C.; Weiss, H.L.; Evers, B.M.; Gao, T. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018, 9, 265. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yan, H.; Zhao, L.; Jia, W.; Yang, H.; Liu, L.; Zhou, X.; Miao, P.; Sun, X.; Song, S.; et al. Inhibition of SREBP increases gefitinib sensitivity in non-small cell lung cancer cells. Oncotarget 2016, 7, 52392–52403. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Pan, Z.; Zhao, M.; Wang, Q.; Qiao, C.; Miao, L.; Ding, X. High cholesterol in lipid rafts reduces the sensitivity to EGFR-TKI therapy in non-small cell lung cancer. J. Cell Physiol. 2018, 233, 6722–6732. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, Y.; Peng, P.; Zhan, J.; Wang, Z.; Zhu, Z.; Zhang, Z.; Liu, L.; Fang, W.; Zhang, L. Cholesterol synthesis disruption combined with a molecule-targeted drug is a promising metabolic therapy for EGFR mutant non-small cell lung cancer. Transl. Lung Cancer Res. 2021, 10, 128–142. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, Q.; Li, D.; Wei, X.; Jia, Y.; Zhang, Z.; Ai, B.; Cao, X.; Guo, T.; Liao, Y. Co-administration of 20(S)-protopanaxatriol (g-PPT) and EGFR-TKI overcomes EGFR-TKI resistance by decreasing SCD1 induced lipid accumulation in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2019, 38, 1–20. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, M.Y.; Choi, D.-Y.; Lee, J.W.; You, S.; Lee, K.Y.; Kim, J.; Kim, K.P. Phospholipids of tumor extracellular vesicles stratify gefitinib-resistant nonsmall cell lung cancer cells from gefitinib-sensitive cells. Proteomics 2015, 15, 824–835. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babuta, J.; Hall, Z.; Athersuch, T. Dysregulated Metabolism in EGFR-TKI Drug Resistant Non-Small-Cell Lung Cancer: A Systematic Review. Metabolites 2022, 12, 644. https://doi.org/10.3390/metabo12070644
Babuta J, Hall Z, Athersuch T. Dysregulated Metabolism in EGFR-TKI Drug Resistant Non-Small-Cell Lung Cancer: A Systematic Review. Metabolites. 2022; 12(7):644. https://doi.org/10.3390/metabo12070644
Chicago/Turabian StyleBabuta, Julia, Zoe Hall, and Toby Athersuch. 2022. "Dysregulated Metabolism in EGFR-TKI Drug Resistant Non-Small-Cell Lung Cancer: A Systematic Review" Metabolites 12, no. 7: 644. https://doi.org/10.3390/metabo12070644





