Margaritaria nobilis L.F. (Phyllanthaceae): Ethnopharmacology and Application of Computational Tools in the Annotation of Bioactive Molecules
Abstract
1. Introduction
2. Results
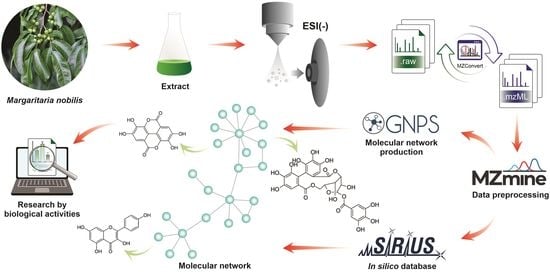
2.1. Characterization of Detectable Components in the EtOH Extract of Margaritaria nobilis
2.1.1. Phenolic Acids Derivatives
2.1.2. Flavonoids and O-Glycosylated Derivatives
2.1.3. Hydrolysable Tannins: Gallotannins and Ellagitannins
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Botanical Collection and Identification
4.3. Obtaining the Ethanol Extract
4.4. Sample Preparation for Analysis via UHPLC-MS/MS
4.5. Analysis via UHPLC-ESI-QToF-MS/MS
4.6. Processing of UHPLC-MS/MS Data
4.7. Resource-Based Molecular Network Creation
4.8. Putative Identification of Compounds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mao, X.; Wu, L.-F.; Guo, H.-L.; Chen, W.-J.; Cui, Y.-P.; Qi, Q.; Li, S.; Liang, W.-Y.; Yang, G.-H.; Shao, Y.-Y.; et al. The Genus Phyllanthus: An Ethnopharmacological, Phytochemical, and Pharmacological Review. Evid.-Based Complement. Altern. Med. 2016, 2016, 1–36. [Google Scholar] [CrossRef]
- Hoffmann, P.; Kathriarachchi, H.; Wurdack, K.J. A Phylogenetic Classification of Phyllanthaceae (Malpighiales; Euphorbiaceae Sensu Lato). Bulletin 2006, 61, 37–53. [Google Scholar]
- Agustini, M.B.; Wendt, L.; de Matos Malavasi, M.; de Bortolli, L.; Sabii, C.; Battistus, A.G.; Ricardo, P.; Correia, L. Superação de Dormência Fisiológica em Sementes de Margaritaria nobilis (Linnaeus). Innov. Sci. Technol. J. 2016, 2, 14–19. [Google Scholar]
- Moraes, L.S.; Donza, M.R.H.; Rodrigues, A.P.D.; Silva, B.J.M.; Brasil, D.S.B.; Zoghbi, M.D.G.B.; Andrade, E.H.A.; Guilhon, G.M.S.P.; Silva, E.O.; Schmidt, T.J. Leishmanicidal Activity of (+)-Phyllanthidine and the Phytochemical Profile of Margaritaria nobilis (Phyllanthaceae). Molecules 2015, 20, 22157–22169. [Google Scholar] [CrossRef]
- Sanz-Biset, J.; Campos-de-la-Cruz, J.; Epiquién-Rivera, M.A.; Cañigueral, S. A First Survey on the Medicinal Plants of the Chazuta Valley (Peruvian Amazon). J. Ethnopharmacol. 2009, 122, 333–362. [Google Scholar] [CrossRef]
- Caamal-Fuentes, E.; Torres-Tapia, L.W.; Simá-Polanco, P.; Peraza-Sánchez, S.R.; Moo-Puc, R. Screening of Plants Used in Mayan Traditional Medicine to Treat Cancer-like Symptoms. J. Ethnopharmacol. 2011, 135, 719–724. [Google Scholar] [CrossRef]
- Ekuadzi, E.; Dickson, R.; Fleischer, T.; Annan, K.; Pistorius, D.; Oberer, L.; Gibbons, S. Flavonoid Glycosides from the Stem Bark of Margaritaria discoidea Demonstrate Antibacterial and Free Radical Scavenging Activities. Phytother. Res. 2014, 28, 784–787. [Google Scholar] [CrossRef]
- Johnson-Ajinwo, O.R.; Richardson, A.; Li, W.-W. Cytotoxic Effects of Stem Bark Extracts and Pure Compounds from Margaritaria discoidea on Human Ovarian Cancer Cell Lines. Phytomedicine 2015, 22, 1–4. [Google Scholar] [CrossRef]
- Arbain, D.; Byrne, L.; Cannon, J.; Engelhardt, L.; White, A. The Alkaloids of Margaritaria indica (Euphorbiaceae). The Crystal Structure and Absolute Configuration of the Hydrobromide of (+)-15α-Methoxy-14,15-Dihydrophyllochrysine. Aust. J. Chem. 1990, 43, 439. [Google Scholar] [CrossRef]
- Dickson, R.; Fleischer, T.; Ekuadzi, E.; Mensah, A.; Annan, K.; Woode, E. Antibacterial, Antioxidant and Anti-Inflammatory Properties of Margaritaria discoidea, a Wound Healing Remedy from Ghana. Pharmacogn. J. 2010, 2, 32–39. [Google Scholar] [CrossRef]
- Adedapo, A.A.; Sofidiya, M.O.; Afolayan, A.J. Anti-Inflammatory and Analgesic Activities of the Aqueous Extracts of Margaritaria discoidea (Euphorbiaceae) Stem Bark in Experimental Animal Models. Rev. De Biol. Trop. 2008, 57. [Google Scholar] [CrossRef]
- Diallo, M.S.T.; Baldé, M.A.; Camara, A.; Traoré, M.S.; Bah, M.L.; Diallo, A.S.; Camara, A.K.; Laurent, S.; Roch, A.; Muller, R.N.; et al. Ethnomedical, Phytochemical and Biological Investigations of Margaritaria discoidea (Baill.) Webster, a Plant Species Widely Used in Guinean Traditional Medicine. J. Plant Sci. Spec. Issue Ethnopharmacol. Investig. Med. Plants 2015, 3, 40–46. [Google Scholar] [CrossRef]
- Suleimen, Y.M.; Jose, R.A.; Suleimen, R.N.; Arenz, C.; Ishmuratova, M.; Toppet, S.; Dehaen, W.; Alsfouk, A.A.; Elkaeed, E.B.; Eissa, I.H.; et al. Isolation and In Silico Anti-SARS-CoV-2 Papain-Like Protease Potentialities of Two Rare 2-Phenoxychromone Derivatives from Artemisia Spp. Molecules 2022, 27, 1216. [Google Scholar] [CrossRef] [PubMed]
- Edison, A.S.; Colonna, M.; Gouveia, G.J.; Holderman, N.R.; Judge, M.T.; Shen, X.; Zhang, S. NMR: Unique Strengths That Enhance Modern Metabolomics Research. Anal. Chem. 2021, 93, 478–499. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.A.; Nothias, L.-F.; Ludwig, M.; Fleischauer, M.; Gentry, E.C.; Witting, M.; Dorrestein, P.C.; Dührkop, K.; Böcker, S. High-Confidence Structural Annotation of Metabolites Absent from Spectral Libraries. Nat. Biotechnol. 2022, 40, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.; Quirós-Guerrero, L.; Silva, C.; Pamplona, S.; Boutin, J.A.; Eberlin, M.; Wolfender, J.-L.; Silva, M. Feature-Based Molecular Network-Guided Dereplication of Natural Bioactive Products from Leaves of Stryphnodendron pulcherrimum (Willd.) Hochr. Metabolites 2021, 11, 281. [Google Scholar] [CrossRef] [PubMed]
- Demarque, D.P.; Dusi, R.G.; de Sousa, F.D.M.; Grossi, S.M.; Silvério, M.R.S.; Lopes, N.P.; Espindola, L.S. Mass Spectrometry-Based Metabolomics Approach in the Isolation of Bioactive Natural Products. Sci. Rep. 2020, 10, 1051. [Google Scholar] [CrossRef]
- Buedenbender, L.; Astone, F.A.; Tasdemir, D. Bioactive Molecular Networking for Mapping the Antimicrobial Constituents of the Baltic Brown Alga Fucus vesiculosus. Mar. Drugs 2020, 18, 311. [Google Scholar] [CrossRef]
- Suntivich, R.; Songjang, W.; Jiraviriyakul, A.; Ruchirawat, S.; Chatwichien, J. LC-MS/MS Metabolomics-Facilitated Identification of the Active Compounds Responsible for Anti-Allergic Activity of the Ethanol Extract of Xenostegia tridentata. PLoS ONE 2022, 17, e0265505. [Google Scholar] [CrossRef]
- Ramabulana, A.-T.; Petras, D.; Madala, N.E.; Tugizimana, F. Metabolomics and Molecular Networking to Characterize the Chemical Space of Four Momordica Plant Species. Metabolites 2021, 11, 763. [Google Scholar] [CrossRef]
- da Silva Antonio, A.; Oliveira, D.S.; Cardoso dos Santos, G.R.; Pereira, H.M.G.; Wiedemann, L.S.M.; Veiga-Junior, V.F. da UHPLC-HRMS/MS on Untargeted Metabolomics: A Case Study with Copaifera (Fabaceae). RSC Adv. 2021, 11, 25096–25103. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lv, S.; Peng, Y.; Zhu, C.; Pan, S. Characterization of Phenolics and Antioxidant Abilities of Red Navel Orange “Cara Cara” Harvested from Five Regions of China. Int. J. Food Prop. 2018, 21, 1107–1116. [Google Scholar] [CrossRef]
- Pfundstein, B.; el Desouky, S.K.; Hull, W.E.; Haubner, R.; Erben, G.; Owen, R.W. Polyphenolic Compounds in the Fruits of Egyptian Medicinal Plants (Terminalia bellerica, Terminalia chebula and Terminalia horrida): Characterization, Quantitation and Determination of Antioxidant Capacities. Phytochemistry 2010, 71, 1132–1148. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Chen, J. Screening and Identification of Acetylcholinesterase Inhibitors from Terminalia chebula Fruits Based on Ultrafiltration and Ultra-Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry. Microchem. J. 2021, 168. [Google Scholar] [CrossRef]
- da Fontoura Sprenger, R.; Cass, Q.B. Characterization of Four Phyllanthus Species Using Liquid Chromatography Coupled to Tandem Mass Spectrometry. J. Chromatogr. A 2013, 1291, 97–103. [Google Scholar] [CrossRef]
- de Andrade Neves, N.; Stringheta, P.C.; Gómez-Alonso, S.; Hermosín-Gutiérrez, I. Flavonols and Ellagic Acid Derivatives in Peels of Different Species of Jabuticaba (Plinia Spp.) Identified by HPLC-DAD-ESI/MSn. Food Chem. 2018, 252, 61–71. [Google Scholar] [CrossRef]
- Okba, M.M.; El-Shiekh, R.A.; Abu-Elghait, M.; Sobeh, M.; Ashour, R.M.S. HPLC-PDA-ESI-MS/MS Profiling and Anti-Biofilm Potential of Eucalyptus sideroxylon Flowers. Antibiotics 2021, 10, 761. [Google Scholar] [CrossRef]
- Szparaga, A.; Kocira, S.; Findura, P.; Kapusta, I.; Zaguła, G.; Świeca, M. Uncovering the Multi-Level Response of Glycine max L. to the Application of Allelopathic Biostimulant from Levisticum officinale Koch. Sci. Rep. 2021, 11, 15360. [Google Scholar] [CrossRef]
- Silva, V.; Falco, V.; Dias, M.I.; Barros, L.; Silva, A.; Capita, R.; Alonso-Calleja, C.; Amaral, J.S.; Igrejas, G.; Ferreira, I.C.F.R.; et al. Evaluation of the Phenolic Profile of Castanea sativa Mill. By-Products and Their Antioxidant and Antimicrobial Activity against Multiresistant Bacteria. Antioxidants 2020, 9, 87. [Google Scholar] [CrossRef]
- Sinosaki, N.B.M.; Tonin, A.P.P.; Ribeiro, M.A.S.; Poliseli, C.B.; Roberto, S.B.; da Silveira, R.; Visentainer, J.V.; Santos, O.O.; Meurer, E.C. Structural Study of Phenolic Acids by Triple Quadrupole Mass Spectrometry with Electrospray Ionization in Negative Mode and H/D Isotopic Exchange. J. Braz. Chem. Soc. 2020, 31, 402–408. [Google Scholar] [CrossRef]
- Singh, A.; Bajpai, V.; Kumar, S.; Sharma, K.R.; Kumar, B. Profiling of Gallic and Ellagic Acid Derivatives in Different Plant Parts of Terminalia arjuna by HPLC-ESI-QTOF-MS/MS. Nat. Prod. Commun. 2016, 11, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cao, A.; Ouyang, C.; Li, Y.; Wei, Y. Rapid Screening and Identification of Non-Target Flavonoid Components in Invasive Weeds by LC/MS-IT-TOF. Anal. Methods 2015, 7, 10207–10216. [Google Scholar] [CrossRef]
- Pilon, A.C.; Gu, H.; Raftery, D.; Bolzani, V.D.S.; Lopes, N.P.; Castro-Gamboa, I.; Carnevale Neto, F. Mass Spectral Similarity Networking and Gas-Phase Fragmentation Reactions in the Structural Analysis of Flavonoid Glycoconjugates. Anal. Chem. 2019, 91, 10413–10423. [Google Scholar] [CrossRef] [PubMed]
- Arapitsas, P. Hydrolyzable Tannin Analysis in Food. Food Chem. 2012, 135, 1708–1717. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic Classification of Unknown Metabolites Using High-Resolution Fragmentation Mass Spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef]
- Chen Liu, K.C.S.; Lin, M.-T.; Lee, S.-S.; Chiou, J.-F.; Ren, S.; Lien, E.J. Antiviral Tannins from Two Phyllanthus Species. Planta Med. 1999, 65, 043–046. [Google Scholar] [CrossRef]
- Graham, J.G.; Pendland, S.L.; Prause, J.L.; Danzinger, L.H.; Schunke Vigo, J.; Cabieses, F.; Farnsworth, N.R. Antimycobacterial Evaluation of Peruvian Plants. Phytomedicine 2003, 10, 528–535. [Google Scholar] [CrossRef]
- Dias, Ê.R.; Freire Dias, T.D.L.M.; Alexandre-Moreira, M.S.; Branco, A. Flavonoid-Rich Fraction from Pleroma pereirae (Melastomataceae): Effects on Calcium Oxalate Crystallization, Antioxidant and Antinociceptive Activities. Eur. J. Integr. Med. 2020, 35, 101095. [Google Scholar] [CrossRef]
- Hernandez-Leon, A.; Fernández-Guasti, A.; González-Trujano, M.E. Rutin Antinociception Involves Opioidergic Mechanism and Descending Modulation of Ventrolateral Periaqueductal Grey Matter in Rats. Eur. J. Pain 2016, 20, 274–283. [Google Scholar] [CrossRef]
- Küpeli, E.; Tatli, I.I.; Akdemir, Z.S.; Yesilada, E. Estimation of Antinociceptive and Anti-Inflammatory Activity on Geranium pratense Subsp. finitimum and Its Phenolic Compounds. J. Ethnopharmacol. 2007, 114, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Devi, K.P.; Malar, D.S.; Nabavi, S.F.; Sureda, A.; Xiao, J.; Nabavi, S.M.; Daglia, M. Kaempferol and Inflammation: From Chemistry to Medicine. Pharmacol. Res. 2015, 99, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, C.; Zhang, H. Hepatoprotective Effects of Kaempferol 3-O-Rutinoside and Kaempferol 3-O-Glucoside from Carthamus tinctorius L. on CCl4-Induced Oxidative Liver Injury in Mice. J. Food Drug Anal. 2015, 23, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Campos-Vidal, Y.; Herrera-Ruiz, M.; Trejo-Tapia, G.; Gonzalez-Cortazar, M.; Aparicio, A.J.; Zamilpa, A. Gastroprotective Activity of Kaempferol Glycosides from Malvaviscus arboreus Cav. J. Ethnopharmacol. 2021, 268, 113633. [Google Scholar] [CrossRef] [PubMed]
- Aa, L.; Fei, F.; Qi, Q.; Sun, R.; Gu, S.; Di, Z.; Aa, J.; Wang, G.; Liu, C. Rebalancing of the Gut Flora and Microbial Metabolism Is Responsible for the Anti-Arthritis Effect of Kaempferol. Acta Pharmacol. Sin. 2020, 41, 73–81. [Google Scholar] [CrossRef]
- Felice, M.R.; Maugeri, A.; de Sarro, G.; Navarra, M.; Barreca, D. Molecular Pathways Involved in the Anti-Cancer Activity of Flavonols: A Focus on Myricetin and Kaempferol. Int. J. Mol. Sci. 2022, 23, 4411. [Google Scholar] [CrossRef]
- Jantas, D.; Malarz, J.; Le, T.N.; Stojakowska, A. Neuroprotective Properties of Kempferol Derivatives from Maesa Membranacea against Oxidative Stress-Induced Cell Damage: An Association with Cathepsin D Inhibition and PI3K/Akt Activation. Int. J. Mol. Sci. 2021, 22, 363. [Google Scholar] [CrossRef]
- García-Niño, W.R.; Zazueta, C. Ellagic Acid: Pharmacological Activities and Molecular Mechanisms Involved in Liver Protection. Pharmacol. Res. 2015, 97, 84–103. [Google Scholar] [CrossRef]
- Ríos, J.-L.; Giner, R.; Marín, M.; Recio, M. A Pharmacological Update of Ellagic Acid. Planta Med. 2018, 84, 1068–1093. [Google Scholar] [CrossRef]
- Farha, A.K.; Yang, Q.-Q.; Kim, G.; Li, H.-B.; Zhu, F.; Liu, H.-Y.; Gan, R.-Y.; Corke, H. Tannins as an Alternative to Antibiotics. Food Biosci. 2020, 38, 100751. [Google Scholar] [CrossRef]
- Pizzi, A. Tannins Medical/Pharmacological and Related Applications: A Critical Review. Sustain. Chem. Pharm. 2021, 22, 100481. [Google Scholar] [CrossRef]
- Li, X.; Deng, Y.; Zheng, Z.; Huang, W.; Chen, L.; Tong, Q.; Ming, Y. Corilagin, a Promising Medicinal Herbal Agent. Biomed. Pharmacother. 2018, 99, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yi, D.; Lei, X.; Zhao, J.; Zhang, Y.; Cui, X.; Xiao, X.; Jiao, T.; Dong, X.; Zhao, X.; et al. Corilagin Inhibits SARS-CoV-2 Replication by Targeting Viral RNA-Dependent RNA Polymerase. Acta Pharm. Sin. B 2021, 11, 1555–1567. [Google Scholar] [CrossRef]
- Kim, Y.S.; Chung, H.-S.; Noh, S.G.; Lee, B.; Chung, H.Y.; Choi, J.-G. Geraniin Inhibits the Entry of SARS-CoV-2 by Blocking the Interaction between Spike Protein RBD and Human ACE2 Receptor. Int. J. Mol. Sci. 2021, 22, 8604. [Google Scholar] [CrossRef]
- Liu, C.; Cai, D.; Zhang, L.; Tang, W.; Yan, R.; Guo, H.; Chen, X. Identification of Hydrolyzable Tannins (Punicalagin, Punicalin and Geraniin) as Novel Inhibitors of Hepatitis B Virus Covalently Closed Circular DNA. Antivir. Res. 2016, 134, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Perera, A.; Ton, S.H.; Palanisamy, U.D. Perspectives on Geraniin, a Multifunctional Natural Bioactive Compound. Trends Food Sci. Technol. 2015, 44, 243–257. [Google Scholar] [CrossRef]
- Shanmuganathan, S.; Angayarkanni, N. Chebulagic Acid Chebulinic Acid and Gallic Acid, the Active Principles of Triphala, Inhibit TNFα Induced pro-Angiogenic and pro-Inflammatory Activities in Retinal Capillary Endothelial Cells by Inhibiting P38, ERK and NFkB Phosphorylation. Vasc. Pharmacol. 2018, 108, 23–35. [Google Scholar] [CrossRef]
- Yang, Y.; Xiu, J.; Liu, J.; Zhang, L.; Li, X.; Xu, Y.; Qin, C.; Zhang, L. Chebulagic Acid, a Hydrolyzable Tannin, Exhibited Antiviral Activity in Vitro and in Vivo against Human Enterovirus 71. Int. J. Mol. Sci. 2013, 14, 9618–9627. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Guo, J.; Shen, S.; Xing, S.; Yu, H.; Huan, T. ISFrag: De Novo Recognition of In-Source Fragments for Liquid Chromatography–Mass Spectrometry Data. Anal. Chem. 2021, 93, 10243–10250. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A Public Repository for Sharing Mass Spectral Data for Life Sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]






| Peak | R.T. (min) | [M−H]− Exp. (Error, ppm) | Molecular Formula | Characteristic Ions (MS2) | Putative Identification | Spectrum Reference |
|---|---|---|---|---|---|---|
| 1 | 1.84 | 169.0138 (0.6) | C7H6O5 | 125 | Gallic acid | a CCMSLIB00004691622 |
| 2 | 4.51 | 355.0661 (1.1) | C15H16O10 | 337, 313, 209, 191, 163, 147, 129 | O-Coumaroylgalactaric acid | a CCMSLIB00005745086 |
| 3 | 4.91 | 385.0766 (1.3) | C16H18O11 | 209, 191, 173, 147 | O-Feruloylgalactaric acid | [22] |
| 4 | 5.17 | 183.0285 (4.4) | C8H8O5 | 168, 124 | Methyl gallate | [23] |
| 5 | 5.92 | 951.0703 (3.9) | C41H28O27 | 933, 915, 763, 633, 463, 461, 443, 301, 275, 273, 169 | Galloyl-DHHDP-HHDP-glucose | [24] |
| 6 | 6.09 | 633.0710 (2.8) | C27H22O18 | 463, 301, 275, 249, 169 | Galloyl-HHDP-glucose | a CCMSLIB00000847042 |
| 7 | 6.56 | 953.0888 (0.8) | C41H30O27 | 935, 909, 801, 783, 765, 633, 481, 463, 337, 319, 301, 293, 275, 249, 169 | Galloyl-Che-HHDP-glucose Isomer I | a CCMSLIB00004692930 |
| 8 | 6.56 | 635.0866 (2.8) | C27H24O18 | 465, 313, 271, 221, 211, 193, 169, 125 | Trigalloyl-glucose | a CCMSLIB00000845184 |
| 9 | 6.92 | 163.0389 (3.7) | C9H8O3 | 119 | p-Coumaric acid | a CCMSLIB00005741418 |
| 10 | 6.98 | 625.1368 (5.9) | C27H30O17 | 301, 300, 271, 255, 243, 179, 151 | Quercetin 3-O-glucosyl-glucoside | a CCMSLIB00000847258 |
| 11 | 7.18 | 197.0445 (2.5) | C9H10O5 | 169, 168, 125, 124 | Ethyl gallate | a CCMSLIB00006691851 |
| 12 | 7.24 | 925.0983 (3.6) | C40H30O26 | 755, 615, 605, 453, 435, 309, 301, 275, 249, 247, 169 | Phyllanthusiin C Isomer | [25] |
| 13 | 7.55 | 433.0410 (0.7) | C19H14O12 | 301, 300 | Ellagic acid O-xyloside | [26] |
| 14 | 7.67 | 595.1321 (3.7) | C26H28O16 | 301, 300, 271, 255, 243, 179, 151 | Quercetin 3-O-xylosyl-glucoside | a CCMSLIB00004718534 |
| 15 | 7.84 | 609.1427 (4.8) | C27H30O16 | 301, 300, 271, 255, 243, 179, 151 | Quercetin 3-O-rhamnosyl-glucoside | a CCMSLIB00005778075 |
| 16 | 7.87 | 447.0585 (4.7) | C20H16O12 | 301, 300 | Ellagic acid O-rhamnoside | [27] |
| 17 | 7.96 | 953.0904 (0.8) | C41H30O27 | 935, 909, 801, 783, 765, 633, 481, 463, 337, 319, 301, 293, 275, 249, 169 | Galloyl-Che-HHDP-glucose Isomer II | a CCMSLIB00004692930 |
| 18 | 8.01 | 300.9972 (4.0) | C14H6O8 | 283, 245, 229, 201, 185, 173, 145 | Ellagic acid | a CCMSLIB00004694147 |
| 19 | 8.39 | 785.0847 (1.3) | C34H26O22 | 633, 615, 463, 301, 275, 249, 169 | Digalloyl-HHDP-glucose | [23] |
| 20 | 8.39 | 985.1155 (0.3) | C42H34O28 | 783, 633, 463, 351, 301, 169 | Methyl neochebulagate Isomer | [23] |
| 21 | 8.62 | 463.0890 (2.8) | C21H20O12 | 301, 300, 271, 255, 243, 179, 151 | Quercetin 3-O-glucoside Isomer I | a CCMSLIB00004684243 |
| 22 | 8.73 | 857.1077 (3.3) | C37H30O24 | 825, 655, 615, 463, 301, 275, 169 | Excoecariphenol C Isomer | N/A |
| 23 | 8.73 | 787.0977 (2.2) | C34H28O22 | 635, 617, 593, 465, 449, 169 | Tetragalloyl-glucose | a CCMSLIB00004719474 |
| 24 | 8.76 | 593.1528 (3.7) | C27H30O15 | 285, 284, 255, 227, 151 | Kaempferol 3-O-rhamnosyl-glucoside | a CCMSLIB00005743498 |
| 25 | 8.87 | 579.1376 (4.0) | C26H28O15 | 285, 284, 255, 227, 151 | Kaempferol 3-O-xylosyl-glucoside | a CCMSLIB00004706607 |
| 26 | 8.87 | 463.0898 (4.5) | C21H20O12 | 301, 300, 271, 255, 243, 179, 151 | Quercetin 3-O-glucoside Isomer II | a CCMSLIB00004684243 |
| 27 | 8.93 | 491.0852 (5.3) | C22H20O13 | 313, 298, 285, 270 | Di-O-Methyl ellagic acid O-glucoside | a CCMSLIB00004715986 |
| 28 | 9.41 | 579.1350 (0.0) | C26H28O15 | 301, 300, 271, 255, 243, 179, 151 | Quercetin 3-O-rhamnosyl-xyloside | a CCMSLIB00004678837 |
| 29 | 9.61 | 433.0765 (1.4) | C20H18O11 | 300, 301, 271, 255, 243, 179, 151 | Quercetin 3-O-xyloside | a CCMSLIB00004718550 |
| 30 | 9.70 | 447.0935 (1.8) | C21H20O11 | 285, 284, 255, 227, 151 | Kaempferol 3-O-glucoside Isomer I | a CCMSLIB00004683728 |
| 31 | 9.95 | 603.0945 (6.8) | C27H24O16 | 451, 433, 301, 275, 169 | Galloyl-HHDP-dideoxyglucose | N/A |
| 32 | 10.15 | 603.1013 (4.5) | C27H24O16 | 451, 433, 211, 169 | Trigalloyl-dideoxyglucose | N/A |
| 33 | 10.24 | 447.0914 (2.9) | C21H20O11 | 285, 284, 255, 227, 151 | Kaempferol 3-O-glucoside Isomer II | a CCMSLIB00004683728 |
| 34 | 10.61 | 563.1431 (5.3) | C26H27O14 | 285, 284, 255, 227, 151 | Kaempferol 3-O-rhamnosyl-xyloside | [28] |
| 35 | 10.69 | 417.0836 (3.4) | C20H18O10 | 285, 284, 255, 227, 151 | Kaempferol 3-O-xyloside | a CCMSLIB00005739911 |
| 36 | 10.78 | 461.0736 (3.5) | C21H18O12 | 315, 300 | Methylellagic acid O-rhamnoside | [26] |
| 37 | 11.01 | 951.0743 (0.3) | C41H28O27 | 907, 781, 737, 649, 615, 605, 497, 479, 435, 335, 301, 291, 275, 273, 247, 169 | Phyllanthusiin A Isomer | [25] |
| 38 | 12.10 | 937.0962 (1.6) | C41H30O26 | 785, 767, 635, 615, 465, 301, 275, 249, 169 | Trigalloyl-HHDP-glucose | [29] |
| 39 | 12.29 | 923.0801 (1.1) | C40H28O26 | 879, 825, 655, 621, 615, 577, 523, 451, 407, 301, 275, 249, 169 | Phyllanthusiin U Isomer | N/A |
| 40 | 14.00 | 301.0334 (4.7) | C15H10O7 | 273, 257, 229,179, 151, 121, 107 | Quercetin | a CCMSLIB00004691125 |
| 41 | 14.91 | 477.1018 (3.1) | C22H22O12 | 314, 315 | Methylquercetin 3-O-glucoside | a CCMSLIB00004678842 |
| 42 | 16.91 | 285.0399 (0.0) | C15H10O6 | 267, 255, 243, 239, 229, 227, 185, 163, 151 | Kaempferol | a CCMSLIB00004691748 |
| 43 | 18.14 | 763.1154 (0.9) | C36H28O19 | 615, 593, 463, 445, 301, 275, 249, 169 | Galloyl-Cinnamoyl-HHDP-glucose | N/A |
| 44 | 19.04 | 343.0450 (1.2) | C17H12O8 | 328, 313, 298, 285, 270, 257, 242 | Tri-O-methylellagic acid | [27] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago, J.C.C.; Albuquerque, C.A.B.; Muribeca, A.d.J.B.; Sá, P.R.C.; Pamplona, S.d.G.S.R.; Silva, C.Y.Y.e.; Ribera, P.C.; Fontes-Júnior, E.d.A.; da Silva, M.N. Margaritaria nobilis L.F. (Phyllanthaceae): Ethnopharmacology and Application of Computational Tools in the Annotation of Bioactive Molecules. Metabolites 2022, 12, 681. https://doi.org/10.3390/metabo12080681
Santiago JCC, Albuquerque CAB, Muribeca AdJB, Sá PRC, Pamplona SdGSR, Silva CYYe, Ribera PC, Fontes-Júnior EdA, da Silva MN. Margaritaria nobilis L.F. (Phyllanthaceae): Ethnopharmacology and Application of Computational Tools in the Annotation of Bioactive Molecules. Metabolites. 2022; 12(8):681. https://doi.org/10.3390/metabo12080681
Chicago/Turabian StyleSantiago, Johan Carlos C., Carlos Alberto B. Albuquerque, Abraão de Jesus B. Muribeca, Paulo Roberto C. Sá, Sônia das Graças Santa R. Pamplona, Consuelo Yumiko Y. e Silva, Paula Cardoso Ribera, Enéas de Andrade Fontes-Júnior, and Milton Nascimento da Silva. 2022. "Margaritaria nobilis L.F. (Phyllanthaceae): Ethnopharmacology and Application of Computational Tools in the Annotation of Bioactive Molecules" Metabolites 12, no. 8: 681. https://doi.org/10.3390/metabo12080681
APA StyleSantiago, J. C. C., Albuquerque, C. A. B., Muribeca, A. d. J. B., Sá, P. R. C., Pamplona, S. d. G. S. R., Silva, C. Y. Y. e., Ribera, P. C., Fontes-Júnior, E. d. A., & da Silva, M. N. (2022). Margaritaria nobilis L.F. (Phyllanthaceae): Ethnopharmacology and Application of Computational Tools in the Annotation of Bioactive Molecules. Metabolites, 12(8), 681. https://doi.org/10.3390/metabo12080681