Co-Expression Network and Integrative Analysis of Metabolome and Transcriptome Uncovers Biological Pathways for Fertility in Beef Heifers
Abstract
1. Introduction
2. Results
2.1. Transcriptome and Metabolome Profiles
2.2. Gene Network and Functional Over-Representation Analyses
2.3. Metabolite Network Analyses
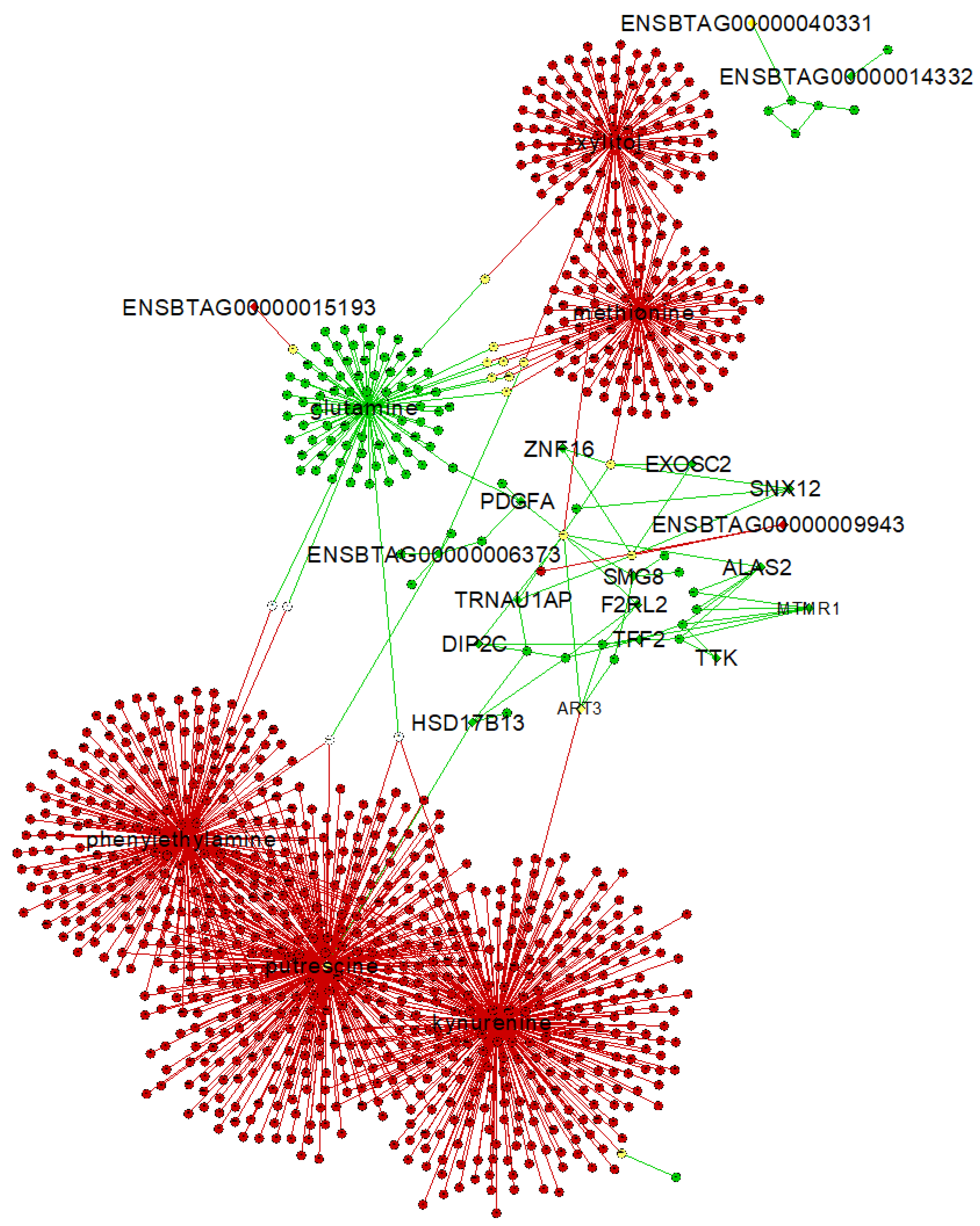
2.4. Gene–Metabolite Interaction Network
3. Discussion
4. Materials and Methods
4.1. Data Collection, Gene Expression, and Metabolite Profile
4.2. Gene and Metabolite Differential Expression Analyses
4.3. Gene and Metabolite Co-Expression Networks
4.4. Transcriptomic and Metabolomic Data Integration
- m: is the log-normalized metabolite abundance;
- b1: is the intercept;
- b2g: is the normalized and adjusted gene-expression level;
- b3p: is the phenotype (AI-P and NP);
- b4 (g:p): is the interaction between gene expression and phenotype; and
- e: is the residual effect associated with each observation.
4.5. Pathway Over-Representation Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moorey, S.E.; Biase, F.H. Beef Heifer Fertility: Importance of Management Practices and Technological Advancements. J. Anim. Sci. Biotechnol. 2020, 11, 97. [Google Scholar] [CrossRef]
- Hindman, M.S.; Engelken, T.J. Beef Heifer Development. In Bovine Reproduction; Wiley Online library: Hoboken, NJ, USA, 2021; pp. 359–365. [Google Scholar] [CrossRef]
- Wathes, D.C.; Brickell, J.S.; Bourne, N.E.; Swali, A.; Cheng, Z. Factors Influencing Heifer Survival and Fertility on Commercial Dairy Farms. Animal 2008, 2, 1135–1143. [Google Scholar] [CrossRef]
- Bach, À. Effects of Nutrition and Genetics on Fertility in Dairy Cows. Fertil. Dev. 2019, 31, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Herd, D.B.; Sprott, L.R. Body Condition, Nutrition and Reproduction of Beef Cows. AgriLife Ext. Tex. AM Syst. 1998, B-1526, 1–12. [Google Scholar]
- Pryce, J.E.; Coffey, M.P.; Simm, G. The Relationship Between Body Condition Score and Reproductive Performance. J. Dairy Sci. 2001, 84, 1508–1515. [Google Scholar] [CrossRef]
- Markusfeld, O.; Galon, N.; Ezra, E. Body Condition Score, Health, Yield and Fertility in Dairy Cows. Vet. Rec. 1997, 141, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Pence, M.; Ensley, D.; Berghaus, R.; Rossi, J.; Wilson, T.; Cannon, P.T. Improving Reproductive Efficiency Through the Use of Reproductive Tract Scoring in a Group of Beef Replacelllent Heifers. Bov. Pract. 2007, 41, 35–40. [Google Scholar]
- Anderson, K.J.D.; LeFever, D.G.; Brinks, J.S.; Odde, K.G. The Use of Reproductive Tract Scoring in Beef Heifers. Agric. Pract. 1991, 12, 19–26. [Google Scholar]
- Larson, R.L.; White, B.J.; Laflin, S. Beef Heifer Development. Vet. Clin. N. Am. Food Anim. Pract. 2016, 32, 285–302. [Google Scholar] [CrossRef]
- Neville, W.E.; Mullinix, B.G.; Smith, J.B.; McCormick, W.C. Growth Patterns for Pelvic Dimensions and Other Body Measurements of Beef Females. J. Anim. Sci. 1978, 47, 1080–1088. [Google Scholar] [CrossRef]
- Dickinson, S.E.; Elmore, M.F.; Kriese-Anderson, L.; Elmore, J.B.; Walker, B.N.; Dyce, P.W.; Rodning, S.P.; Biase, F.H. Evaluation of Age, Weaning Weight, Body Condition Score, and Reproductive Tract Score in Pre-Selected Beef Heifers Relative to Reproductive Potential. J. Anim. Sci. Biotechnol. 2019, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Wiggans, G.R.; VanRaden, P.M.; Cooper, T.A. The Genomic Evaluation System in the United States: Past, Present, Future. J. Dairy Sci. 2011, 94, 3202–3211. [Google Scholar] [CrossRef] [PubMed]
- Mamo, S.; Mehta, J.P.; McGettigan, P.; Fair, T.; Spencer, T.E.; Bazer, F.W.; Lonergan, P. RNA Sequencing Reveals Novel Gene Clusters in Bovine Conceptuses Associated with Maternal Recognition of Pregnancy and Implantation1. Biol. Reprod. 2011, 85, 1143–1151. [Google Scholar] [CrossRef]
- Graf, A.; Krebs, S.; Zakhartchenko, V.; Schwalb, B.; Blum, H.; Wolf, E. Fine Mapping of Genome Activation in Bovine Embryos by RNA Sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 4139–4144. [Google Scholar] [CrossRef] [PubMed]
- Forde, N.; Carter, F.; Fair, T.; Crowe, M.A.; Evans, A.C.O.; Spencer, T.E.; Bazer, F.W.; McBride, R.; Boland, M.P.; O’Gaora, P.; et al. Progesterone-Regulated Changes in Endometrial Gene Expression Contribute to Advanced Conceptus Development in Cattle. Biol. Reprod. 2009, 81, 784–794. [Google Scholar] [CrossRef]
- Bauersachs, S.; Ulbrich, S.E.; Gross, K.; Schmidt, S.E.M.; Meyer, H.H.D.; Wenigerkind, H.; Vermehren, M.; Sinowatz, F.; Blum, H.; Wolf, E. Embryo-Induced Transcriptome Changes in Bovine Endometrium Reveal Species-Specific and Common Molecular Markers of Uterine Receptivity. Reproduction 2006, 132, 319–331. [Google Scholar] [CrossRef]
- Mazzoni, G.; Pedersen, H.S.; Rabaglino, M.B.; Hyttel, P.; Callesen, H.; Kadarmideen, H.N. Characterization of the Endometrial Transcriptome in Early Diestrus Influencing Pregnancy Status in Dairy Cattle after Transfer of in Vitro-Produced Embryos. Physiol. Genom. 2020, 52, 269–279. [Google Scholar] [CrossRef]
- Mesquita, F.S.; Ramos, R.S.; Pugliesi, G.; Andrade, S.C.S.; Van Hoeck, V.; Langbeen, A.; Oliveira, M.L.; Gonella-Diaza, A.M.; Gasparin, G.; Fukumasu, H.; et al. Endometrial Transcriptional Profiling of a Bovine Fertility Model by Next-Generation Sequencing. Genom. Data 2016, 7, 26–28. [Google Scholar] [CrossRef] [PubMed]
- Chankeaw, W.; Lignier, S.; Richard, C.; Ntallaris, T.; Raliou, M.; Guo, Y.; Plassard, D.; Bevilacqua, C.; Sandra, O.; Andersson, G.; et al. Analysis of the Transcriptome of Bovine Endometrial Cells Isolated by Laser Micro-Dissection (1): Specific Signatures of Stromal, Glandular and Luminal Epithelial Cells. BMC Genom. 2021, 22, 451. [Google Scholar] [CrossRef]
- Binelli, M.; Scolari, S.C.; Pugliesi, G.; Van Hoeck, V.; Gonella-Diaza, A.M.; Andrade, S.C.S.; Gasparin, G.R.; Coutinho, L.L. The Transcriptome Signature of the Receptive Bovine Uterus Determined at Early Gestation. PLoS ONE 2015, 10, e0122874. [Google Scholar] [CrossRef]
- Moorey, S.E.; Walker, B.N.; Elmore, M.F.; Elmore, J.B.; Rodning, S.P.; Biase, F.H. Rewiring of Gene Expression in Circulating White Blood Cells Is Associated with Pregnancy Outcome in Heifers (Bos Taurus). Sci. Rep. 2020, 10, 16786. [Google Scholar] [CrossRef]
- Dickinson, S.E.; Griffin, B.A.; Elmore, M.F.; Kriese-Anderson, L.; Elmore, J.B.; Dyce, P.W.; Rodning, S.P.; Biase, F.H. Transcriptome Profiles in Peripheral White Blood Cells at the Time of Artificial Insemination Discriminate Beef Heifers with Different Fertility Potential. BMC Genom. 2018, 19, 129. [Google Scholar] [CrossRef] [PubMed]
- Geary, T.W.; Burns, G.W.; Moraes, J.G.N.; Moss, J.I.; Denicol, A.C.; Dobbs, K.B.; Ortega, M.S.; Hansen, P.J.; Wehrman, M.E.; Neibergs, H.; et al. Identification of Beef Heifers with Superior Uterine Capacity for Pregnancy. Biol. Reprod. 2016, 95, 47. [Google Scholar] [CrossRef]
- Phillips, K.M.; Read, C.C.; Kriese-Anderson, L.A.; Rodning, S.P.; Brandebourg, T.D.; Biase, F.H.; Marks, M.L.; Elmore, J.B.; Stanford, M.K.; Dyce, P.W. Plasma Metabolomic Profiles Differ at the Time of Artificial Insemination Based on Pregnancy Outcome, in Bos Taurus Beef Heifers. Sci. Rep. 2018, 8, 13196. [Google Scholar] [CrossRef]
- Kusama, K.; Bai, R.; Matsuno, Y.; Ideta, A.; Sakurai, T.; Nagaoka, K.; Hori, M.; Imakawa, K. Characterization of Serum Metabolome and Proteome Profiles Identifies SNX5 Specific for Pregnancy Failure in Holstein Heifers. Life 2022, 12, 309. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Salvetti, P.; Gatien, J.; Carrocera, S.; Martín-González, D.; Muñoz, M. Blood Plasma Metabolomics Predicts Pregnancy in Holstein Cattle Transferred with Fresh and Vitrified/Warmed Embryos Produced in Vitro. J. Proteome Res. 2020, 19, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, T.M.; Gonçalves, R.F.; Melo, C.F.O.R.; de Oliveira, D.N.; Lima, E.d.O.; Visintin, J.A.; de Achilles, M.A.; Catharino, R.R. A Metabolomic Overview of Follicular Fluid in Cows. Front. Vet. Sci. 2018, 5, 10. [Google Scholar] [CrossRef]
- Aranciaga, N.; Morton, J.D.; Berg, D.K.; Gathercole, J.L. Proteomics and Metabolomics in Cow Fertility: A Systematic Review. Reproduction 2020, 160, 639–658. [Google Scholar] [CrossRef]
- Gómez, E.; Muñoz, M.; Gatien, J.; Carrocera, S.; Martín-González, D.; Salvetti, P. Metabolomic Identification of Pregnancy-Specific Biomarkers in Blood Plasma of BOS TAURUS Beef Cattle after Transfer of in Vitro Produced Embryos. J. Proteom. 2020, 225, 103883. [Google Scholar] [CrossRef]
- Reverter, A.; Chan, E.K.F. Combining Partial Correlation and an Information Theory Approach to the Reversed Engineering of Gene Co-Expression Networks. Bioinformatics 2008, 24, 2491–2497. [Google Scholar] [CrossRef]
- Choueiry, F.; Singh, S.; Sircar, A.; Laliotis, G.; Sun, X.; Chavdoula, E.; Zhang, S.; Helmig-Mason, J.; Hart, A.; Epperla, N.; et al. Integration of Metabolomics and Gene Expression Profiling Elucidates IL4I1 as Modulator of Ibrutinib Resistance in ABC-Diffuse Large B Cell Lymphoma. Cancers 2021, 13, 2146. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.; Pan, W.; Gawad, C.; Fan, H.C.; Kerchner, G.A.; Wyss-Coray, T.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Quake, S.R. Noninvasive in Vivo Monitoring of Tissue-Specific Global Gene Expression in Humans. Proc. Natl. Acad. Sci. USA 2014, 111, 7361–7366. [Google Scholar] [CrossRef]
- Basu, M.; Wang, K.; Ruppin, E.; Hannenhalli, S. Predicting Tissue-Specific Gene Expression from Whole Blood Transcriptome. Sci. Adv. 2021, 7, eabd6991. [Google Scholar] [CrossRef]
- Carmelo, V.A.O.; Banerjee, P.; da Silva Diniz, W.J.; Kadarmideen, H.N. Metabolomic Networks and Pathways Associated with Feed Efficiency and Related-Traits in Duroc and Landrace Pigs. Sci. Rep. 2020, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Michou, V.I.; Kanavaros, P.; Athanassiou, V.; Chronis, G.B.; Stabamas, S.; Tsilivakos, V. Fraction of the Peripheral Blood Concentration of CD56+/CD16−/CD3− Cells in Total Natural Killer Cells as an Indication of Fertility and Infertility. Fertil. Steril. 2003, 80, 691–697. [Google Scholar] [CrossRef]
- Thum, M.Y.; Bhaskaran, S.; Abdalla, H.I.; Ford, B.; Sumar, N.; Shehata, H.; Bansal, A.S. An Increase in the Absolute Count of CD56dimCD16+CD69+ NK Cells in the Peripheral Blood Is Associated with a Poorer IVF Treatment and Pregnancy Outcome. Hum. Reprod. 2004, 19, 2395–2400. [Google Scholar] [CrossRef] [PubMed]
- Topaloglu, A.K.; Reimann, F.; Guclu, M.; Yalin, A.S.; Kotan, L.D.; Porter, K.M.; Serin, A.; Mungan, N.O.; Cook, J.R.; Ozbek, M.N.; et al. TAC3 and TACR3 Mutations in Familial Hypogonadotropic Hypogonadism Reveal a Key Role for Neurokinin B in the Central Control of Reproduction. Nat. Genet. 2009, 41, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Tusset, C.; Noel, S.D.; Trarbach, E.B.; Silveira, L.F.G.; Jorge, A.A.L.; Brito, V.N.; Cukier, P.; Seminara, S.B.; Mendonça, B.B.d.; Kaiser, U.B.; et al. Mutational Analysis of TAC3 and TACR3 Genes in Patients with Idiopathic Central Pubertal Disorders. Arq. Bras. Endocrinol. Metabol. 2012, 56, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Blasco, V.; Pinto, F.M.; Fernández-Atucha, A.; Prados, N.; Tena-Sempere, M.; Fernández-Sánchez, M.; Candenas, L. Altered Expression of the Kisspeptin/KISS1R and Neurokinin B/NK3R Systems in Mural Granulosa and Cumulus Cells of Patients with Polycystic Ovarian Syndrome. J. Assist. Reprod. Genet. 2019, 36, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.; Navarro, V.M. Novel Biology of Tachykinins in Gonadotropin-Releasing Hormone Secretion. Semin. Reprod. Med. 2019, 37, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Kahnamouyi, S.; Nouri, M.; Farzadi, L.; Darabi, M.; Hosseini, V.; Mehdizadeh, A. The Role of Mitogen-Activated Protein Kinase–Extracellular Receptor Kinase Pathway in Female Fertility Outcomes: A Focus on Pituitary Gonadotropins Regulation. Ther. Adv. Endocrinol. Metab. 2018, 9, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Liebanas, E.; Nahar, K.; Bertuzzi, G.; Keller, A.; Betsholtz, C.; Mäe, M.A. Adult-Induced Genetic Ablation Distinguishes PDGFB Roles in Blood-Brain Barrier Maintenance and Development. J. Cereb. Blood Flow Metab. 2022, 42, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.E.; Detzel, C.; Skinner, M.K. Platelet-Derived Growth Factor Modulates the Primordial to Primary Follicle Transition. Reproduction 2006, 131, 1007–1015. [Google Scholar] [CrossRef]
- Schmahl, J.; Rizzolo, K.; Soriano, P. The PDGF Signaling Pathway Controls Multiple Steroid-Producing Lineages. Genes Dev. 2008, 22, 3255–3267. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, P.; Gerhardt, H.; Liebner, S.; Abramsson, A.; Enge, M.; Hellström, M.; Bäckström, G.; Fredriksson, S.; Landegren, U.; Nyström, H.C.; et al. Endothelial PDGF-B Retention Is Required for Proper Investment of Pericytes in the Microvessel Wall. Genes Dev. 2003, 17, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Hou, X.; Xia, J.; Qian, X.; Miele, L.; Sarkar, F.H.; Wang, Z. Emerging Roles of PDGF-D in EMT Progression during Tumorigenesis. Cancer Treat. Rev. 2013, 39, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex Networks Orchestrate Epithelial–Mesenchymal Transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Gui, T.; Sun, Y.; Shimokado, A.; Muragaki, Y. The Roles of Mitogen-Activated Protein Kinase Pathways in TGF- β -Induced Epithelial-Mesenchymal Transition. J. Signal Transduct. 2012, 2012, 1–10. [Google Scholar] [CrossRef]
- Korkina, L.G. Phenylpropanoids as Naturally Occurring Antioxidants: From Plant Defense to Human Health. Cell. Mol. Biol. 2007, 53, 15–25. [Google Scholar] [PubMed]
- Ly, C.; Yockell-Lelièvre, J.; Ferraro, Z.M.; Arnason, J.T.; Ferrier, J.; Gruslin, A. The Effects of Dietary Polyphenols on Reproductive Health and Early Development. Hum. Reprod. Update 2015, 21, 228–248. [Google Scholar] [CrossRef] [PubMed]
- Behl, T.; Rana, T.; Alotaibi, G.H.; Shamsuzzaman, M.; Naqvi, M.; Sehgal, A.; Singh, S.; Sharma, N.; Almoshari, Y.; Abdellatif, A.A.H.; et al. Polyphenols Inhibiting MAPK Signalling Pathway Mediated Oxidative Stress and Inflammation in Depression. Biomed. Pharmacother. 2022, 146, 112545. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, H.N.; Sales, K.J.; Catalano, R.D.; Norman, J.E. Inflammatory Pathways in Female Reproductive Health and Disease. Reproduction 2009, 138, 903–919. [Google Scholar] [CrossRef] [PubMed]
- Waters, W.W.; Chen, P.L.; McArthur, N.H.; Moreno, P.A.; Harms, P.G. Calcium/Calmodulin-Dependent Protein Kinase II Involvement in Release of Gonadotropin-Releasing Hormone. Neuroendocrinology 1998, 67, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, H.C.; Evans, J.; Johnson, N.; Infusini, G.; Webb, A.; Rombauts, L.J.R.; Vollenhoven, B.J.; Salamonsen, L.A.; Edgell, T.A. Idiopathic Infertility in Women Is Associated with Distinct Changes in Proliferative Phase Uterine Fluid Proteins. Biol. Reprod. 2018, 98, 752–764. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Mu, C.; Wang, Z.; Wang, Y.; Li, Y.; Mai, Y.; Li, S.; Xu, H.; Gu, B.; Luo, L.; et al. Diagnostic and Prognostic Values of Serum EpCAM, TGM2, and HE4 Levels in Endometrial Cancer. Front. Oncol. 2020, 10, 1697. [Google Scholar] [CrossRef] [PubMed]
- Beazley, K.E.; Nurminskaya, M. Effects of Dietary Quercetin on Female Fertility in Mice: Implication of Transglutaminase 2. Reprod. Fertil. Dev. 2016, 28, 974. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Camlin, N.J.; Holt, J.E.; Teixeira, J.M.; McLaughlin, E.A.; Tanwar, P.S. Germ Cell Specific Overactivation of WNT/Βcatenin Signalling Has No Effect on Folliculogenesis but Causes Fertility Defects Due to Abnormal Foetal Development. Sci. Rep. 2016, 6, 27273. [Google Scholar] [CrossRef]
- Li, H.; Shi, L.; Liang, S.; Fang, C.; Xu, Q.; Lu, G.; Wang, Q.; Cheng, J.; Shen, J.; Shen, M. Moxibustion Alleviates Decreased Ovarian Reserve in Rats by Restoring the PI3K/AKT Signaling Pathway. J. Integr. Med. 2022, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Chang, Y.; Wei, L.; Chen, J.; Li, J.; Goldsmith, S.; Silber, S.; Liang, X. Apoptosis of Mural Granulosa Cells Is Increased in Women with Diminished Ovarian Reserve. J. Assist. Reprod. Genet. 2019, 36, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L.; Bridgham, J.T.; Swenson, J.A. Activation of the Akt/Protein Kinase B Signaling Pathway Is Associated with Granulosa Cell Survival1. Biol. Reprod. 2001, 64, 1566–1574. [Google Scholar] [CrossRef]
- Brown, C.; LaRocca, J.; Pietruska, J.; Ota, M.; Anderson, L.; Duncan Smith, S.; Weston, P.; Rasoulpour, T.; Hixon, M.L. Subfertility Caused by Altered Follicular Development and Oocyte Growth in Female Mice Lacking PKBalpha/Akt11. Biol. Reprod. 2010, 82, 246–256. [Google Scholar] [CrossRef]
- Chu, D.; Zhang, Z.; Zhou, Y.; Li, Y.; Zhu, S.; Zhang, J.; Zhao, Q.; Ji, G.; Wang, W.; Zheng, J. NDRG4, a Novel Candidate Tumor Suppressor, Is a Predictor of Overall Survival of Colorectal Cancer Patients. Oncotarget 2015, 6, 7584–7596. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Gu, Y.; Zhang, X.; Wang, J.-M.; He, Y.-P.; Shi, Y.; Sun, Z.-G.; Shi, H.-J.; Wang, J. Uterine Expression of NDRG4 Is Induced by Estrogen and Up-Regulated during Embryo Implantation Process in Mice. PLoS ONE 2016, 11, e0155491. [Google Scholar] [CrossRef] [PubMed]
- Ipsa, E.; Cruzat, V.F.; Kagize, J.N.; Yovich, J.L.; Keane, K.N. Growth Hormone and Insulin-Like Growth Factor Action in Reproductive Tissues. Front. Endocrinol. 2019, 10, 777. [Google Scholar] [CrossRef] [PubMed]
- Charalambous, M.; Cowley, M.; Geoghegan, F.; Smith, F.M.; Radford, E.J.; Marlow, B.P.; Graham, C.F.; Hurst, L.D.; Ward, A. Maternally-Inherited Grb10 Reduces Placental Size and Efficiency. Dev. Biol. 2010, 337, 1–8. [Google Scholar] [CrossRef]
- Ooi, J.; Yajnik, V.; Immanuel, D.; Gordon, M.; Moskow, J.J.; Buchberg, A.M.; Margolis, B. The Cloning of Grb10 Reveals a New Family of SH2 Domain Proteins. Oncogene 1995, 10, 1621–1630. [Google Scholar]
- Shiura, H.; Miyoshi, N.; Konishi, A.; Wakisaka-Saito, N.; Suzuki, R.; Muguruma, K.; Kohda, T.; Wakana, S.; Yokoyama, M.; Ishino, F.; et al. Meg1/Grb10 Overexpression Causes Postnatal Growth Retardation and Insulin Resistance via Negative Modulation of the IGF1R and IR Cascades. Biochem. Biophys. Res. Commun. 2005, 329, 909–916. [Google Scholar] [CrossRef]
- Rosa, P.R.A.d.; Bohrer, R.C.; Ludke, C.A.; Cesaro, M.P.D.; Pereira, G.R.; Mondadori, R.G.; Antoniazzi, A.Q.; Gonçalves, P.B.D. Grb10 Characterization in Bovine Cumulus Oocyte Complexes from Different Follicle Sizes. Ciência Rural. 2015, 45, 898–904. [Google Scholar] [CrossRef][Green Version]
- Wu, Y.; Zhang, Z.; Zhang, J.B.; Deng, Q. Association of Growth Factor Receptor-Bound Protein 10 Gene Polymorphism with Superovulation Traits in Changbaishan Black Cattle. Genet. Mol. Res. 2016, 15, 1–9. [Google Scholar] [CrossRef]
- He, J.; Li, Y.; Wang, Y.; Zhang, H.; Ge, S.; Fan, X. Targeted Silencing of the ADP-Ribosyltransferase 3 Gene Inhibits the Migration Ability of Melanoma Cells. Oncol. Lett. 2018, 15, 7053–7059. [Google Scholar] [CrossRef]
- Lavergne, C.L.J.; Tao, Y.; Ren, Y.; Lafleur, N.; Liu, X.J. Systemic L-Ornithine Supplementation Specifically Increases Ovarian Putrescine Levels during Ovulation in Mice. Biol. Reprod. 2022, 106, 792–801. [Google Scholar] [CrossRef]
- Liu, D.; Mo, G.; Tao, Y.; Wang, H.; Liu, X.J. Putrescine Supplementation during in Vitro Maturation of Aged Mouse Oocytes Improves the Quality of Blastocysts. Reprod. Fertil. Dev. 2017, 29, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A.-B. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int. J. Tryptophan Res. 2017, 10, 117864691769193. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.M.; Dawes, M.A.; Mathias, C.W.; Acheson, A.; Hill-Kapturczak, N.; Dougherty, D.M. L-Tryptophan: Basic Metabolic Functions, Behavioral Research and Therapeutic Indications. Int. J. Tryptophan Res. 2009, 2, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A.-B. Tryptophan Metabolism, Disposition and Utilization in Pregnancy. Biosci. Rep. 2015, 35, e00261. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Liu, H.; Bai, M.; Gao, J.; Wu, X.; Yin, Y. Redox Properties of Tryptophan Metabolism and the Concept of Tryptophan Use in Pregnancy. Int. J. Mol. Sci. 2017, 18, 1595. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Cesar, A.S.M.; Regitano, L.C.A.; Koltes, J.E.; Fritz-Waters, E.R.; Lanna, D.P.D.; Gasparin, G.; Mourão, G.B.; Oliveira, P.S.N.; Reecy, J.M.; Coutinho, L.L. Putative Regulatory Factors Associated with Intramuscular Fat Content. PLoS ONE 2015, 10, e0128350. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2018. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 30 June 2022).
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, Scaling, and Transformations: Improving the Biological Information Content of Metabolomics Data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- Jauhiainen, A.; Madhu, B.; Narita, M.; Narita, M.; Griffiths, J.; Tavaré, S. Normalization of Metabolomics Data with Applications to Correlation Maps. Bioinformatics 2014, 30, 2155–2161. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Goenawan, I.H.; Bryan, K.; Lynn, D.J. DyNet: Visualization and Analysis of Dynamic Molecular Interaction Networks. Bioinformatics 2016, 32, 2713–2715. [Google Scholar] [CrossRef] [PubMed]
- Fuller, T.F.; Ghazalpour, A.; Aten, J.E.; Drake, T.A.; Lusis, A.J.; Horvath, S. Weighted Gene Coexpression Network Analysis Strategies Applied to Mouse Weight. Mamm. Genome 2007, 18, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Afonso, J.; Fortes, M.R.S.; Reverter, A.; Diniz, W.J.d.S.; Cesar, A.S.M.; Lima, A.O.d.; Petrini, J.; de Souza, M.M.; Coutinho, L.L.; Mourão, G.B.; et al. Genetic Regulators of Mineral Amount in Nelore Cattle Muscle Predicted by a New Co-Expression and Regulatory Impact Factor Approach. Sci. Rep. 2020, 10, 8436. [Google Scholar] [CrossRef]
- Siddiqui, J.K.; Baskin, E.; Liu, M.; Cantemir-Stone, C.Z.; Zhang, B.; Bonneville, R.; McElroy, J.P.; Coombes, K.R.; Mathé, E.A. IntLIM: Integration Using Linear Models of Metabolomics and Gene Expression Data. BMC Bioinform. 2018, 19, 81. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, P.; Rodning, S.P.; Diniz, W.J.S.; Dyce, P.W. Co-Expression Network and Integrative Analysis of Metabolome and Transcriptome Uncovers Biological Pathways for Fertility in Beef Heifers. Metabolites 2022, 12, 708. https://doi.org/10.3390/metabo12080708
Banerjee P, Rodning SP, Diniz WJS, Dyce PW. Co-Expression Network and Integrative Analysis of Metabolome and Transcriptome Uncovers Biological Pathways for Fertility in Beef Heifers. Metabolites. 2022; 12(8):708. https://doi.org/10.3390/metabo12080708
Chicago/Turabian StyleBanerjee, Priyanka, Soren P. Rodning, Wellison J. S. Diniz, and Paul W. Dyce. 2022. "Co-Expression Network and Integrative Analysis of Metabolome and Transcriptome Uncovers Biological Pathways for Fertility in Beef Heifers" Metabolites 12, no. 8: 708. https://doi.org/10.3390/metabo12080708
APA StyleBanerjee, P., Rodning, S. P., Diniz, W. J. S., & Dyce, P. W. (2022). Co-Expression Network and Integrative Analysis of Metabolome and Transcriptome Uncovers Biological Pathways for Fertility in Beef Heifers. Metabolites, 12(8), 708. https://doi.org/10.3390/metabo12080708






