Metabolomics Markers of COVID-19 Are Dependent on Collection Wave
Abstract
:1. Introduction
2. Methods
2.1. Participant Sample Collection and Ethics
2.2. Metadata Collection
2.3. Targeted Metabolomics
2.4. Exploratory Data Analysis
2.5. Machine Learning Model Construction
3. Results
3.1. Metadata Summary
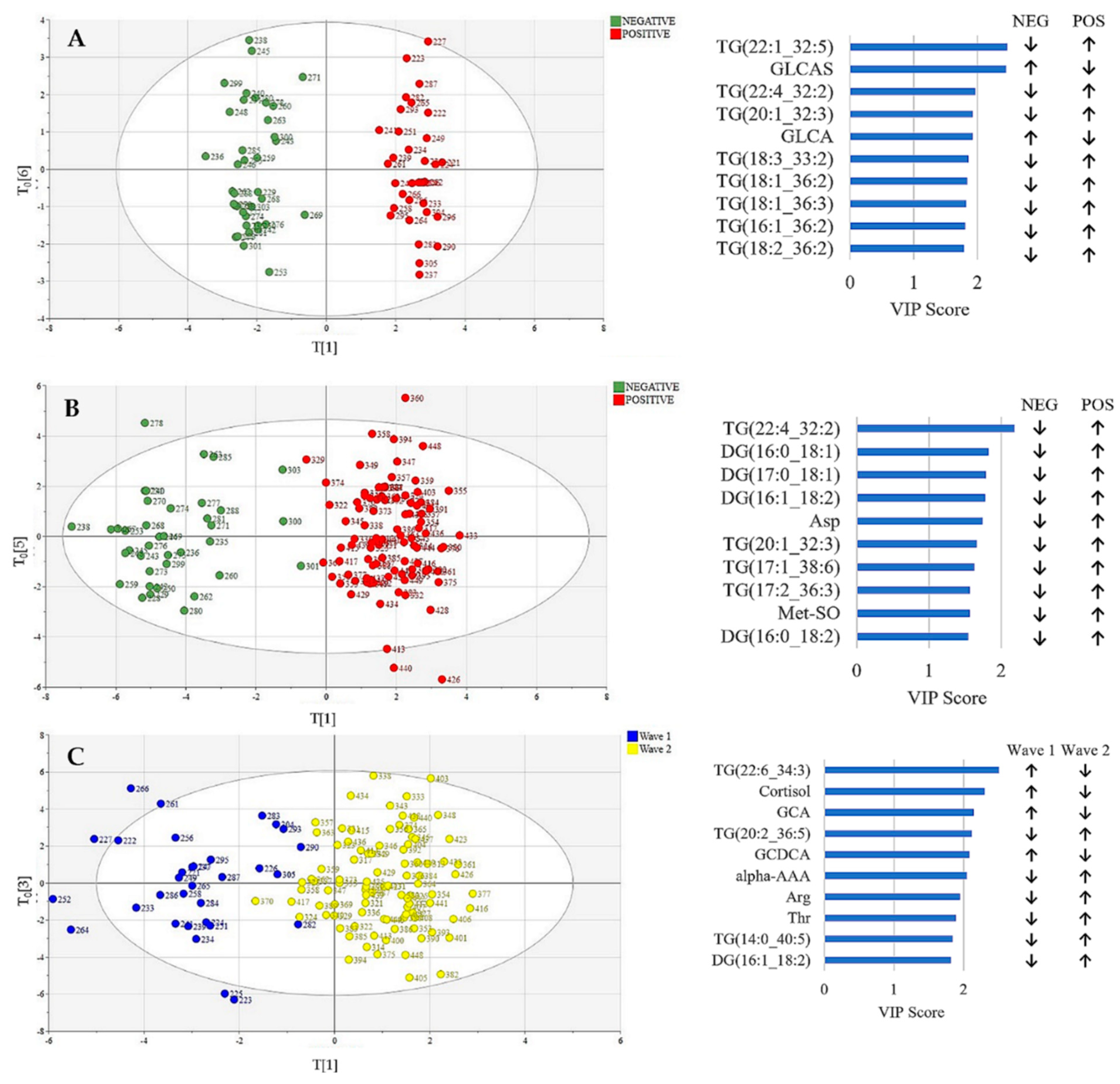
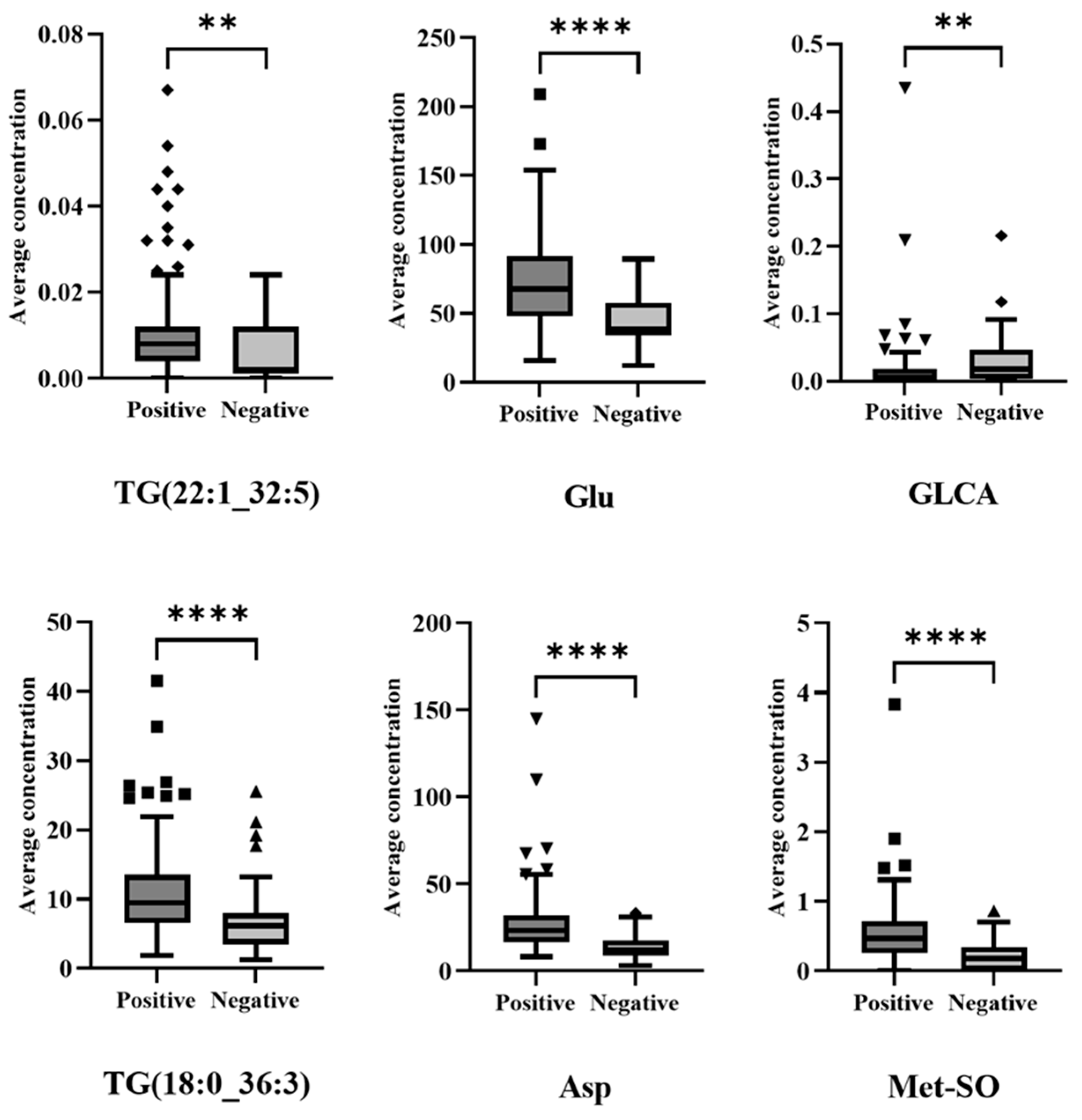
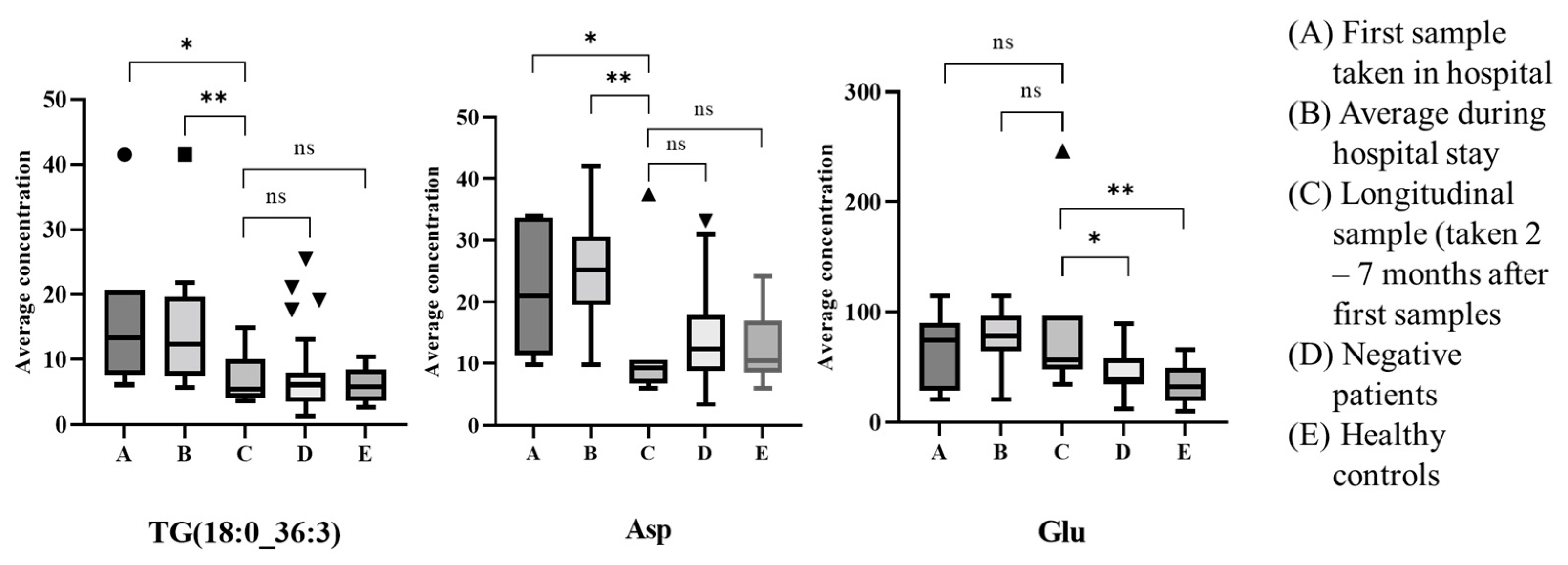
3.2. Metabolic Profiling: Exploratory Data Analysis
3.3. Metabolic Profiling: Machine Learning-Based Diagnosis Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phelan, A.L.; Katz, R.; Gostin, L.O. The Novel Coronavirus Originating in Wuhan, China: Challenges for Global Health Governance. JAMA 2020, 323, 709–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krammer, F. SARS-CoV-2 Vaccines in Development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Group, T.R.C. Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar]
- World Health Organization. COVID-19 Weekly Epidemiological Update. Available online: https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update (accessed on 28 July 2022).
- Almazeedi, S.; Al-Youha, S.; Jamal, M.H.; Al-Haddad, M.; Al-Muhaini, A.; Al-Ghimlas, F.; Al-Sabah, S. Characteristics, risk factors and outcomes among the first consecutive 1096 patients diagnosed with COVID-19 in Kuwait. EClinicalMedicine 2020, 24, 100448. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Blasco, H.; Bessy, C.; Plantier, L.; Lefevre, A.; Piver, E.; Bernard, L.; Marlet, J.; Stefic, K.; Benz-de Bretagne, I.; Cannet, P.; et al. The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 2020, 10, 16824. [Google Scholar] [CrossRef]
- Struwe, W.; Emmott, E.; Bailey, M.; Sharon, M.; Sinz, A.; Corrales, F.J.; Thalassinos, K.; Braybrook, J.; Mills, C.; Barran, P. The COVID-19 MS Coalition—accelerating diagnostics, prognostics, and treatment. Lancet 2020, 395, 1761–1762. [Google Scholar] [CrossRef]
- Skene, D.J.; Middleton, B.; Fraser, C.K.; Pennings, J.L.A.; Kuchel, T.R.; Rudiger, S.R.; Bawden, C.S.; Morton, A.J. Metabolic profiling of presymptomatic Huntington’s disease sheep reveals novel biomarkers. Sci. Rep. 2017, 7, 43030. [Google Scholar] [CrossRef] [Green Version]
- Menni, C.; McCallum, L.; Pietzner, M.; Zierer, J.; Aman, A.; Suhre, K.; Mohney, R.P.; Mangino, M.; Friedrich, N.; Spector, T.D.; et al. Metabolomic profiling identifies novel associations with Electrolyte and Acid-Base Homeostatic patterns. Sci. Rep. 2019, 9, 15088. [Google Scholar] [CrossRef] [Green Version]
- Spick, M.; Lewis, H.M.; Wilde, M.J.; Hopley, C.; Huggett, J.; Bailey, M.J. Systematic review with meta-analysis of diagnostic test accuracy for COVID-19 by mass spectrometry. Metabolism 2022, 126, 154922. [Google Scholar] [CrossRef]
- Collino, S.; Martin, F.P.J.; Rezzi, S. Clinical metabolomics paves the way towards future healthcare strategies. Br. J. Clin. Pharmacol. 2013, 75, 619–629. [Google Scholar] [CrossRef] [Green Version]
- Caterino, M.; Gelzo, M.; Sol, S.; Fedele, R.; Annunziata, A.; Calabrese, C.; Fiorentino, G.; D’Abbraccio, M.; Dell’Isola, C.; Fusco, F.M.; et al. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 2021, 11, 2941. [Google Scholar] [CrossRef] [PubMed]
- Kimhofer, T.; Lodge, S.; Whiley, L.; Gray, N.; Loo, R.L.; Lawler, N.G.; Nitschke, P.; Bong, S.; Morrison, D.L.; Begum, S.; et al. Integrative Modeling of Quantitative Plasma Lipoprotein, Metabolic, and Amino Acid Data Reveals a Multiorgan Pathological Signature of SARS-CoV-2 Infection. J. Proteome Res. 2020, 19, 4442–4454. [Google Scholar] [CrossRef] [PubMed]
- López-Hernández, Y.; Monárrez-Espino, J.; Oostdam, A.S.H.V.; Delgado, J.E.C.; Zhang, L.; Zheng, J.; Valdez, J.J.O.; Mandal, R.; González, F.d.O.; Moreno, J.C.B.; et al. Targeted metabolomics identifies high performing diagnostic and prognostic biomarkers for COVID-19. Sci. Rep. 2021, 11, 14732. [Google Scholar] [CrossRef]
- Schroeder, M.; Schaumburg, B.; Müller, Z.; Parplys, A.; Jarczak, D.; Nierhaus, A.; Kloetgen, A.; Schneider, B.; Peschka, M.; Stoll, F.; et al. Sex hormone and metabolic dysregulations are associated with critical illness in male Covid-19 patients. Emerg. Microbes Inections 2021, 10, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Rees, C.A.; Rostad, C.A.; Mantus, G.; Anderson, E.J.; Chahroudi, A.; Jaggi, P.; Wrammert, J.; Ochoa, J.B.; Ochoa, A.; Basu, R.K.; et al. Altered amino acid profile in patients with SARS-CoV-2 infection. Proc. Natl. Acad. Sci. USA 2021, 118, 4–6. [Google Scholar] [CrossRef]
- Matsuyama, T.; Yoshinaga, S.K.; Shibue, K.; Mak, T.W. Comorbidity-associated glutamine deficiency is a predisposition to severe COVID-19. Cell Death Differ. 2021, 28, 3199–3213. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, S.; Singh, M.; Kirtipal, N.; Kang, S.G. SARS-CoV-2 and Glutamine: SARS-CoV-2 Triggered Pathogenesis via Metabolic Reprograming of Glutamine in Host Cells. Front. Mol. Biosci. 2021, 7, 627842. [Google Scholar] [CrossRef]
- Castañé, H.; Iftimie, S.; Baiges-gaya, G.; Rodríguez-tom, E.; Jiménez-Franco, A.; Ana, F.; Garrido, P.; Castro, A.; Jordi, C.; Joven, J.; et al. Machine learning and semi-targeted lipidomics identify distinct serum lipid signatures in hospitalized COVID-19-positive and COVID-19-negative patients Helena Casta n. Metabolism 2022, 131, 155197. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Valdes, A.M.; Polidori, L.; Antonelli, M.; Penamakuri, S.; Nogal, A.; Panayiotis, L.; Anna, M.; Jane, C.F.; Christina, H.; et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: A prospective observational study from the ZOE COVID Study. Lancet 2022, 399, 1618–1624. [Google Scholar] [CrossRef]
- Whitaker, M.; Elliott, J.; Bodinier, B.; Barclay, W. Variant-specific symptoms of COVID-19 among 1,542,510 people in England. medRxiv 2022. [Google Scholar] [CrossRef]
- Iftimie, S.; López-Azcona, A.F.; Vallverdú, I.; Hernández-Flix, S.; de Febrer, G.; Parra, S.; Hernández-Aguilera, A.; Riu, F.; Joven, J.; Andreychuk, N.; et al. First and second waves of coronavirus disease-19: A comparative study in hospitalized patients in Reus, Spain. PLoS ONE 2021, 16, e0248029. [Google Scholar] [CrossRef] [PubMed]
- Cacciapaglia, G.; Cot, C.; Sannino, F. Second wave COVID-19 pandemics in Europe: A temporal playbook. Sci. Rep. 2020, 10, 15514. [Google Scholar] [CrossRef]
- Pitt, A.; Trivedi, D.; Rankin, N.; Burgess, K.; Bailey, M.J.; Emmott, E. COVID-19-MSC Metabolomics and Lipidomics Protocol. 2020. Available online: https://covid19-msc.org/metabolomics-and-lipidomics-protocol/ (accessed on 28 July 2022).
- Kratzel, A.; Todt, D.; V’kovski, P.; Steiner, S.; Mitra, L.G.; Thao, T.T.N.; Ebert, N.; Holwerda, M.; Steinmann, J.; Niemeyer, D.; et al. Efficient inactivation of SARS-CoV-2 by WHO-recommended hand rub formulations and alcohols. Emerg. Infect. Dis. 2020, 26, 1592. [Google Scholar] [CrossRef]
- Eriksson, L.; Jaworska, J.; Worth, A.P.; Cronin, M.T.D.; McDowell, R.M.; Gramatica, P. Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ. Health Perspect. 2003, 111, 1361–1375. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Xiao, H.; Zhang, L.; Feng, D.; Zhang, F.; Jiang, M.; Sui, Q.; Jia, L. Fast determination of oxide content in cement raw meal using NIR spectroscopy with the SPXY algorithm. Anal. Methods. 2019, 11, 3936–3942. [Google Scholar] [CrossRef]
- Wang, K.; Chen, T.; Lau, R. Bagging for robust non-linear multivariate calibration of spectroscopy. Chemom. Intell. Lab. Syst. 2011, 105, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Galvao, R.; Araujo, M.; Jose, G.; Pontes, M.; Silva, E.; Saldanha, T. A method for calibration and validation subset partitioning. Talanta 2005, 67, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Mathieu, B.; Peter, P.; Ron, W.; Vincent, D.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Rigatti, S.J. Random Forest. J. Insur. Med. 2017, 47, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Georén, S.K.; Tcacencu, I.; Wikström, A.C.; Stierna, P. Early timing of low-dose dexamethasone decreases inflammation in a murine model of eosinophilic airway disease. Acta Oto-Laryngol. 2009, 129, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Matthay, M.A.; Thompson, B.T. Dexamethasone in hospitalised patients with COVID-19: Addressing uncertainties. Lancet Respir. Med. 2020, 8, 1170–1172. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Keiji, T.; Ana Maria, C.; Mark, J.C. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, L.; Cao, R.; Ma, J.; Lv, L.; Li, W.; Zhu, Y.; Wu, Z.; Wang, M.; Zhou, Y.; Zhong, W. Pathological features of COVID-19-associated liver injury—a preliminary proteomics report based on clinical samples. Signal Transduct. Target. Ther. 2021, 6, 2020–2022. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Wang, H.; Cui, G.; Lu, H.; Wang, L.; Luo, H.; Chen, X.; Ren, H.; Sun, R.; Liu, W.; et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 2021, 70, 1253–1265. [Google Scholar] [CrossRef]
- Sokol, H.; Contreras, V.; Maisonnasse, P.; Desmons, A.; Delache, B.; Sencio, V.; Machelart, A.; Brisebarre, A.; Humbert, L.; Deryuter, L.; et al. SARS-CoV-2 infection in nonhuman primates alters the composition and functional activity of the gut microbiota. Gut Microbes 2021, 13, 1893113. [Google Scholar] [CrossRef]
- Åsvold, B.O.; Grill, V.; Thorstensen, K.; Bjørgaas, M.R. Association between posttest dexamethasone and cortisol concentrations in the 1 mg overnight dexamethasone suppression test. Endocr. Connect. 2012, 1, 62–67. [Google Scholar] [CrossRef]
- Maltais-Payette, I.; Boulet, M.M.; Prehn, C.; Adamski, J.; Tchernof, A. Circulating glutamate concentration as a biomarker of visceral obesity and associated metabolic alterations. Nutr. Metab. 2018, 15, 78. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, X.; Yin, P.; Wu, R.; Zhou, L.; Xu, G.; Niu, J. Serum metabolome and targeted bile acid profiling reveals potential novel biomarkers for drug-induced liver injury. Medicine 2019, 98, 16717. [Google Scholar] [CrossRef] [PubMed]
- Bizkarguenaga, M.; Bruzzone, C.; Gil-Redondo, R.; SanJuan, I.; Martin-Ruiz, I.; Barriales, D.; Palacios, A.; Pasco, S.T.; González-Valle, B.; Laín, A.; et al. Uneven metabolic and lipidomic profiles in recovered COVID-19 patients as investigated by plasma NMR metabolomics. NMR Biomed. 2022, 35, e4637. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhou, L.; Sun, X.; Yan, Z.; Hu, C.; Wu, J.; Xu, L.; Li, X.; Liu, H.; Yin, P.; et al. Altered Lipid Metabolism in Recovered SARS Patients Twelve Years after Infection. Sci. Rep. 2017, 7, 9110. [Google Scholar]


| All Patients | Positive Patients | |||||
|---|---|---|---|---|---|---|
| Negative | Positive | p-Value | Wave 1 | Wave 2 | p-Value | |
| N | 41 | 123 | 32 | 91 | ||
| Age (mean, standard deviation; years) | 62.4 ± 19.9 | 61.6 ± 16.9 | 0.696 | 61.7 ± 19.7 | 61.8 ± 15.9 | 0.890 |
| Male/Female (n) | 16/22 | 81/45 | 0.023 | 17/15 | 64/30 | 0.140 |
| Time between positive RT-PCR test and sampling (mean, standard deviation; years) | N/A | 9 ± 13 | - | 11 ± 14 | 9 ± 13 | 0.253 |
| Treated for Hypertension (n) | 16 | 44 | 0.570 | 10 | 34 | 0.670 |
| Treated for High Cholesterol (n) | 7 | 12 | 0.133 | 5 | 7 | 0.296 |
| Treated for Type 2 Diabetes Mellitus (n) | 11 | 36 | 1.000 | 10 | 26 | 0.820 |
| Treated for Ischemic Heart Disease (n) | 7 | 19 | 0.618 | 5 | 14 | 1.000 |
| Current Smoker (n) | 1 | 4 | 1.000 | 2 | 2 | 0.267 |
| Ex-Smoker (n) | 12 | 38 | 0.844 | 4 | 34 | 0.014 |
| Medical Acute Dependency admission (n) | 7 | 66 | 0.000 | 12 | 54 | 0.023 |
| Intensive Care Unit admission (n) | 1 | 16 | 0.125 | 4 | 12 | 1.000 |
| Did Not Survive Admission (n) | 1 | 7 | 0.683 | 2 | 5 | 1.000 |
| Lymphocytes (mean, standard deviation; cells/μL) | 0.98 ± 0.49 | 0.88 ± 0.75 | 0.065 | 0.61 ± 0.34 | 0.96 ± 0.83 | 0.034 |
| C-Reactive Protein (mean, standard deviation; mg/L) | 111.30 ± 99.91 | 101.12 ± 102.01 | 0.46 | 164.7 ± 125.1 | 79.5 ± 83.1 | 0.000 |
| Eosinophils (mean, standard deviation; 100/μL) | 0.34 ± 0.39 | 0.12 ± 0.23 | 0.000 | 0.24 ± 0.36 | 0.07 ± 0.16 | 0.000 |
| Bilateral Chest X-Ray changes (n) | 5 | 81 | 0.000 | 18 | 63 | 0.292 |
| Continuous Positive Airway Pressure (n) | 5 | 44 | 0.014 | 9 | 35 | 0.397 |
| O2 required (n) | 12 | 76 | 0.003 | 17 | 59 | 0.404 |
| Dexamethasone treatment | 1 | 65 | 0.000 | 0 | 65 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewis, H.-M.; Liu, Y.; Frampas, C.F.; Longman, K.; Spick, M.; Stewart, A.; Sinclair, E.; Kasar, N.; Greener, D.; Whetton, A.D.; et al. Metabolomics Markers of COVID-19 Are Dependent on Collection Wave. Metabolites 2022, 12, 713. https://doi.org/10.3390/metabo12080713
Lewis H-M, Liu Y, Frampas CF, Longman K, Spick M, Stewart A, Sinclair E, Kasar N, Greener D, Whetton AD, et al. Metabolomics Markers of COVID-19 Are Dependent on Collection Wave. Metabolites. 2022; 12(8):713. https://doi.org/10.3390/metabo12080713
Chicago/Turabian StyleLewis, Holly-May, Yufan Liu, Cecile F. Frampas, Katie Longman, Matt Spick, Alexander Stewart, Emma Sinclair, Nora Kasar, Danni Greener, Anthony D. Whetton, and et al. 2022. "Metabolomics Markers of COVID-19 Are Dependent on Collection Wave" Metabolites 12, no. 8: 713. https://doi.org/10.3390/metabo12080713
APA StyleLewis, H.-M., Liu, Y., Frampas, C. F., Longman, K., Spick, M., Stewart, A., Sinclair, E., Kasar, N., Greener, D., Whetton, A. D., Barran, P. E., Chen, T., Dunn-Walters, D., Skene, D. J., & Bailey, M. J. (2022). Metabolomics Markers of COVID-19 Are Dependent on Collection Wave. Metabolites, 12(8), 713. https://doi.org/10.3390/metabo12080713









