Untargeted Metabolomics Reveals the Function of GPRC6A in Amino Acid and Lipid Metabolism in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Hematoxylin–Eosin (HE) Staining
2.3. Blood Collection and Serum Separation
2.4. Biochemical Parameter Determination
2.5. Metabolite Extraction
2.6. LC-MS/MS
2.7. Data Preprocessing and Annotation
2.8. KEGG Pathway Analysis
3. Results
3.1. Effect of Dietary Protein on GPRC6A+/+ and GPRC6A−/− Mice
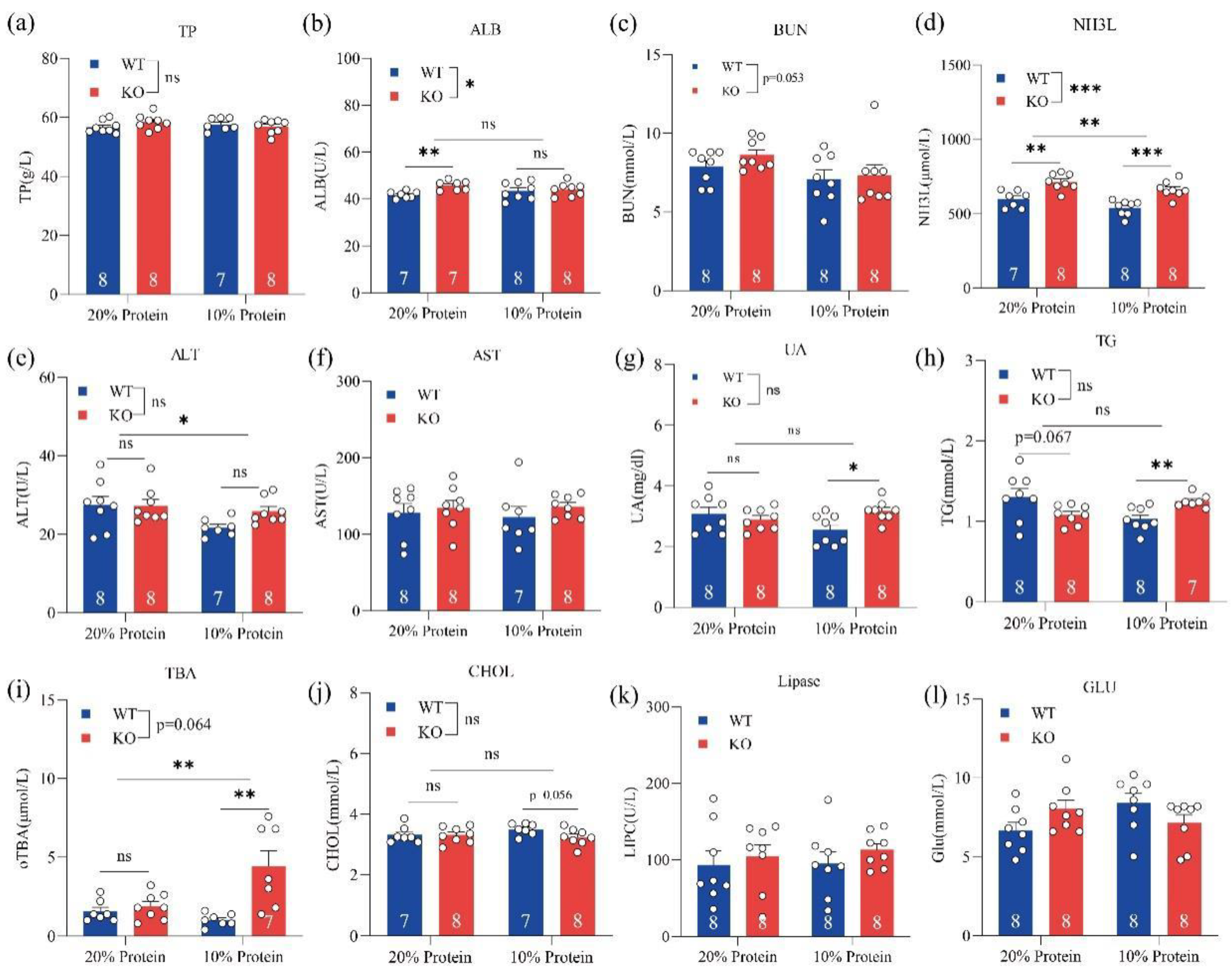
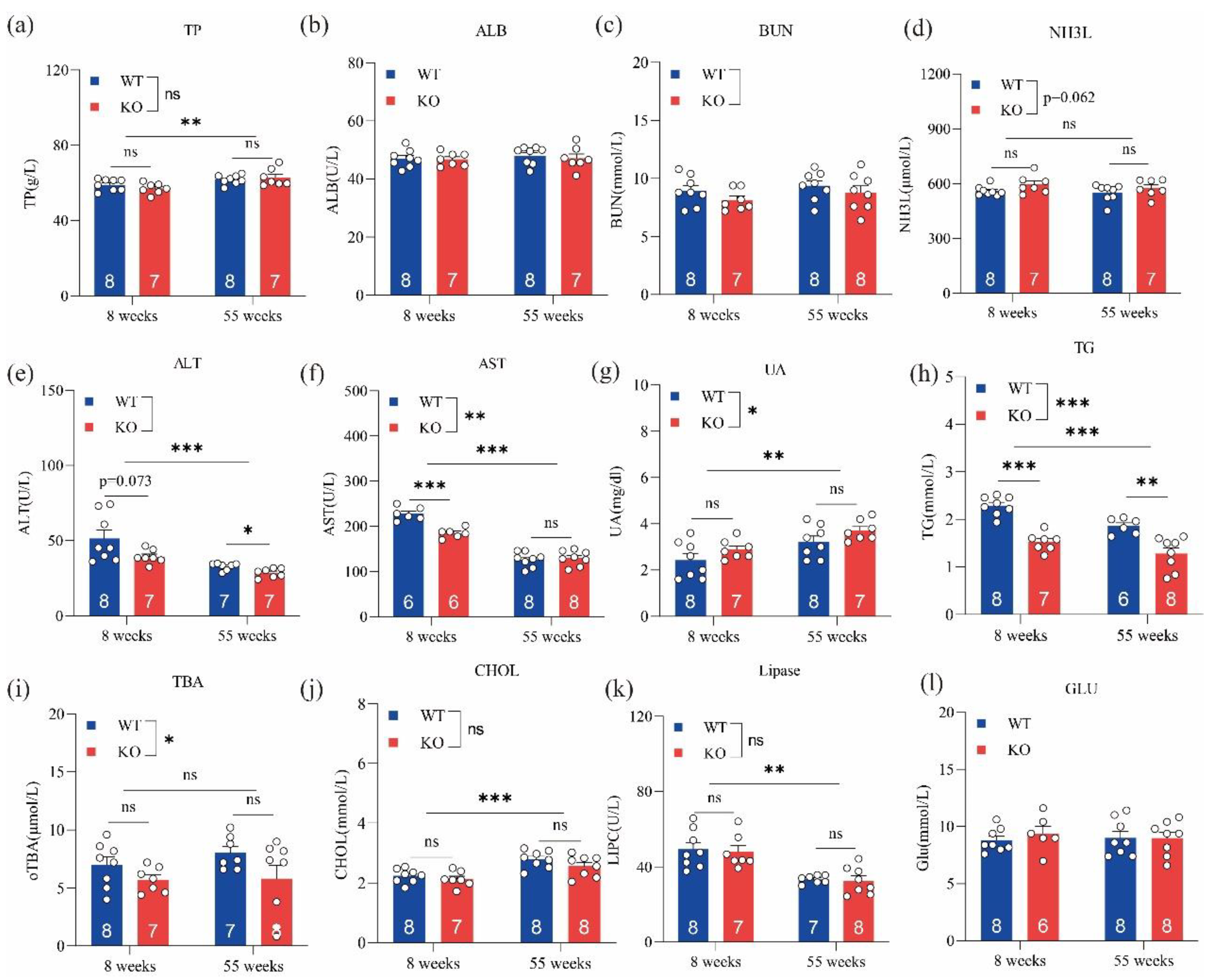
3.2. Loss of GPRC6A Altered Serum Biochemical Parameters
3.3. Loss of GPRC6A Altered Free Amino Acid Levels in Mice
3.4. Metabolomics Screening for Differentially Expressed Metabolites between GPRC6A +/+ and GPRC6A−/− Mice
3.5. Pathway Enrichment Analysis Based on the KEGG (Kyoto Encyclopedia of Genes and Genomes) Library
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Janssen, S.; Depoortere, I. Nutrient sensing in the gut: New roads to therapeutics? Trends Endocrinol. Metab. 2013, 24, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Reimann, F.; Tolhurst, G.; Gribble, F.M. G-protein-coupled receptors in intestinal chemosensation. Cell Metab. 2012, 15, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Conigrave, A.D.; Hampson, D.R. Broad-spectrum amino acid-sensing class C G-protein coupled receptors: Molecular mechanisms, physiological significance and options for drug development. Pharmacol. Ther. 2010, 127, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Wellendorph, P.; Bräuner-Osborne, H. Molecular cloning, expression, and sequence analysis of GPRC6A, a novel family C G-protein-coupled receptor. Gene 2004, 335, 37–46. [Google Scholar] [CrossRef]
- Kuang, N.; Yao, Y.; Lam, J.; Tsushima, R.G.; Hampson, D.R. Cloning and characterization of a Family C orphan G-protein coupled receptor. J. Neurochem. 2005, 93, 383–391. [Google Scholar] [CrossRef]
- Haid, D.; Widmayer, P.; Breer, H. Nutrient sensing receptors in gastric endocrine cells. J. Mol. Histol. 2011, 42, 355–364. [Google Scholar] [CrossRef]
- Pi, M.; Wu, Y.; I Lenchik, N.; Gerling, I.; Quarles, L.D. GPRC6A mediates the Effects of l-arginine on insulin secretion in mouse pancreatic islets. Endocrinology 2012, 153, 4608–4615. [Google Scholar] [CrossRef] [PubMed]
- De Toni, L.; Guidolin, D.; De Filippis, V.; Peterle, D.; Rocca, M.S.; Di Nisio, A.; Ponce, M.D.R.; Foresta, C. SHBG141–161 domain-peptide stimulates GPRC6A-mediated response in leydig and beta-langerhans cell lines. Sci. Rep. 2019, 9, 19432. [Google Scholar] [CrossRef]
- Wauson, E.M.; Lorente-Rodríguez, A.; Cobb, M.H. Minireview: Nutrient sensing by g protein-coupled receptors. Mol. Endocrinol. 2013, 27, 1188–1197. [Google Scholar] [CrossRef]
- Clemmensen, C.; Smajilovic, S.; Wellendorph, P.; Bräuner-Osborne, H. The GPCR, class C, group 6, subtype A (GPRC6A) receptor: From cloning to physiological function. J. Cereb. Blood Flow Metab. 2014, 171, 1129–1141. [Google Scholar] [CrossRef]
- Wellendorph, P.; Hansen, K.B.; Balsgaard, A.; Greenwood, J.R.; Egebjerg, J.; Bräuner-Osborne, H. Deorphanization of GPRC6A: A promiscuous l-alpha-amino acid receptor with preference for basic amino acids. Mol. Pharmacol. 2004, 67, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, B.; Hansen, K.B.; Wellendorph, P.; Bräuner-Osborne, H. Pharmacological characterization of mouse GPRC6A, anL-alpha-amino-acid receptor modulated by divalent cations. Br. J. Pharmacol. 2007, 150, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Gloriam, D.E.; Wellendorph, P.; Johansen, L.D.; Thomsen, A.R.; Phonekeo, K.; Pedersen, D.S.; Bräuner-Osborne, H. Chemogenomic discovery of allosteric antagonists at the GPRC6A receptor. Chem. Biol. 2011, 18, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Rossol, M.; Pierer, M.; Raulien, N.; Quandt, D.; Meusch, U.; Rothe, K.; Schubert, K.; Schöneberg, T.; Schaefer, M.; Krügel, U.; et al. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nat. Commun. 2012, 3, 1329. [Google Scholar] [CrossRef] [PubMed]
- Pi, M.; Kapoor, K.; Wu, Y.; Ye, R.; Senogles, S.E.; Nishimoto, S.K.; Hwang, D.-J.; Miller, D.D.; Narayanan, R.; Smith, J.; et al. Structural and functional evidence for testosterone activation of GPRC6A in peripheral tissues. Mol. Endocrinol. 2015, 29, 1759–1773. [Google Scholar] [CrossRef] [PubMed]
- Pi, M.; Wu, Y.; Quarles, L.D. GPRC6A mediates responses to osteocalcin in beta-cells in vitro and pancreas in vivo. J. Bone Miner. Res. 2011, 26, 1680–1683. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Hanna, T.; Suda, N.; Karsenty, G.; Ducy, P. Osteocalcin Promotes beta-cell proliferation during development and adulthood through Gprc6a. Diabetes 2014, 63, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- De Toni, L.; Guidolin, D.; De Filippis, V.; Tescari, S.; Strapazzon, G.; Rocca, M.S.; Ferlin, A.; Plebani, M.; Foresta, C. Osteocalcin and Sex Hormone Binding Globulin Compete on a Specific Binding Site of GPRC6A. Endocrinology 2016, 157, 4473–4486. [Google Scholar] [CrossRef]
- Rueda, P.; Harley, E.; Lü, Y.; Stewart, G.D.; Fabb, S.; Diepenhorst, N.; Cremers, B.; Rouillon, M.-H.; Wehrle, I.; Géant, A.; et al. Murine GPRC6A mediates cellular responses to l-amino acids, but not osteocalcin variants. PLoS ONE 2016, 11, e0146846. [Google Scholar] [CrossRef]
- Pi, M.; Chen, L.; Huang, M.-Z.; Zhu, W.; Ringhofer, B.; Luo, J.; Christenson, L.; Li, B.; Zhang, J.; Jackson, P.D.; et al. GPRC6A null mice exhibit osteopenia, feminization and metabolic syndrome. PLoS ONE 2009, 3, e3858. [Google Scholar] [CrossRef]
- Wellendorph, P.; Johansen, L.D.; Jensen, A.A.; Casanova, E.; Gassmann, M.; Deprez, P.; Clément-Lacroix, P.; Bettler, B.; Bräuner-Osborne, H. No evidence for a bone phenotype in GPRC6A knockout mice under normal physiological conditions. J. Mol. Endocrinol. 2008, 42, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Smajilovic, S.; Clemmensen, C.; Johansen, L.D.; Wellendorph, P.; Holst, J.J.; Thams, P.G.; Ogo, E.; Bräuner-Osborne, H. The L-a-amino acid receptor GPRC6A is expressed in the islets of Langerhans but is not involved in L-arginine-induced insulin release. Amino Acids 2012, 44, 383. [Google Scholar] [CrossRef] [PubMed]
- Clemmensen, C.; Smajilovic, S.; Madsen, A.N.; Klein, A.B.; Holst, B.; Bräuner-Osborne, H. Increased susceptibility to diet-induced obesity in GPRC6A receptor knockout mice. J. Endocrinol. 2013, 217, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Clemmensen, C.; Pehmøller, C.; Klein, A.B.; Ratner, C.; Wojtaszewski, J.F.; Bräuner-Osborne, H. Enhanced voluntary wheel running in GPRC6A receptor knockout mice. Physiol. Behav. 2013, 118, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Pi, M.; Xu, F.; Ye, R.; Nishimoto, S.K.; Williams, R.W.; Lu, L.; Quarles, L.D. Role of GPRC6A in Regulating Hepatic Energy Metabolism in Mice. Sci. Rep. 2020, 10, 7216. [Google Scholar] [CrossRef]
- Pi, M.; Nishimoto, S.K.; Quarles, L.D. GPRC6A: Jack of all metabolism (or master of none). Mol. Metab. 2017, 6, 185–193. [Google Scholar] [CrossRef]
- Haid, D.C.; Jordan-Biegger, C.; Widmayer, P.; Breer, H. Receptors responsive to protein breakdown products in G-cells and D-cells of mouse, swine and human. Front. Physiol. 2012, 3, 65. [Google Scholar] [CrossRef]
- Teng, B.; Huang, C.; Cheng, C.-L.; Udduttula, A.; Yu, X.-F.; Liu, C.; Li, J.; Yao, Z.-Y.; Long, J.; Miao, L.-F.; et al. Newly identified peptide hormone inhibits intestinal fat absorption and improves NAFLD through its receptor GPRC6A. J. Hepatol. 2020, 73, 383–393. [Google Scholar] [CrossRef]
- Zhang, M.; Nie, X.; Yuan, Y.; Wang, Y.; Ma, X.; Yin, J.; Bao, Y. Osteocalcin Alleviates Nonalcoholic Fatty Liver Disease in Mice through GPRC6A. Int. J. Endocrinol. 2021, 9178616. [Google Scholar] [CrossRef]
- Ye, R.; Pi, M.; Cox, J.V.; Nishimoto, S.K.; Quarles, L.D. CRISPR/Cas9 targeting of GPRC6A suppresses prostate cancer tumorigenesis in a human xenograft model. J. Exp. Clin. Cancer Res. 2017, 36, 90. [Google Scholar] [CrossRef]
- Oury, F.; Sumara, G.; Sumara, O.; Ferron, M.; Chang, H.; Smith, C.E.; Hermo, L.; Suarez, S.; Roth, B.L.; Ducy, P.; et al. Endocrine regulation of male fertility by the skeleton. Cell 2011, 144, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Mukai, S.; Mizokami, A.; Otani, T.; Sano, T.; Matsuda, M.; Chishaki, S.; Gao, J.; Kawakubo-Yasukochi, T.; Tang, R.; Kanematsu, T.; et al. Adipocyte-specific GPRC6A ablation promotes diet-induced obesity by inhibiting lipolysis. J. Biol. Chem. 2021, 296, 100274. [Google Scholar] [CrossRef] [PubMed]
- Pettini, F.; Visibelli, A.; Cicaloni, V.; Iovinelli, D.; Spiga, O. Multi-Omics Model Applied to Cancer Genetics. Int. J. Mol. Sci. 2021, 22, 5751. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Pi, M.; Zhang, L.; Lei, S.-F.; Huang, M.-Z.; Zhu, W.; Zhang, J.; Shen, H.; Deng, H.-W.; Quarles, L.D. Impaired osteoblast function inGPRC6ANull mice. J. Bone Miner. Res. 2009, 25, 1092–1102. [Google Scholar] [CrossRef]
- Kinsey-Jones, J.S.; Alamshah, A.; McGavigan, A.K.; Spreckley, E.; Banks, K.; Monteoliva, N.C.; Norton, M.; Bewick, G.A.; Murphy, K.G. GPRC6a is not required for the effects of a high-protein diet on body weight in mice. Obesity 2015, 23, 1194–1200. [Google Scholar] [CrossRef]
- Kuerschner, L.; Moessinger, C.; Thiele, C. Imaging of lipid biosynthesis: How a neutral lipid enters lipid droplets. Traffic 2008, 9, 338–352. [Google Scholar] [CrossRef]
- Staels, B.; Handelsman, Y.; Fonseca, V. Bile Acid Sequestrants for Lipid and Glucose Control. Curr. Diabetes Rep. 2010, 10, 70–77. [Google Scholar] [CrossRef]
- Griffin, J.W.D.; Bradshaw, P.C. Effects of a high protein diet and liver disease in an in silico model of human ammonia metabolism. Theor. Biol. Med. Model. 2019, 16, 11. [Google Scholar] [CrossRef]
- Ezaki, J.; Matsumoto, N.; Takeda-Ezaki, M.; Komatsu, M.; Takahashi, K.; Hiraoka, Y.; Taka, H.; Fujimura, T.; Takehana, K.; Yoshida, M.; et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 2011, 7, 727–736. [Google Scholar] [CrossRef]
- Jin, M.; Klionsky, D.J. The amino acid transporter SLC38A9 regulates MTORC1 and autophagy. Autophagy 2015, 11, 1709–1710. [Google Scholar] [CrossRef] [PubMed]
- Wyant, G.A.; Abu-Remaileh, M.; Wolfson, R.L.; Chen, W.W.; Freinkman, E.; Danai, L.V.; Heiden, M.G.V.; Sabatini, D.M. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 2017, 171, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Guo, X.; Yu, S.; Sun, G.; Yang, H.; Li, Z.; Sun, Y. Association between Serum Uric Acid and Elevated Alanine Aminotransferase in the General Population. Int. J. Environ. Res. Public Health 2016, 13, 841. [Google Scholar] [CrossRef] [PubMed]
- Atiq, M.; Holt, A.F.; Safdar, K.; Weber, F.; Ravinuthala, R.; Jonas, M.E.; Neff, G.W. Adult Onset Urea Cycle Disorder in a patient with presumed hepatic encephalopathy. J. Clin. Gastroenterol. 2008, 42, 213–214. [Google Scholar] [CrossRef]


| Ingredient (% by Weight) | 10% Protein (LP) | 20% Protein (NP) |
|---|---|---|
| Corn Sugar | 46.06 | 39.75 |
| Casein Lactic | 10.00 | 20.00 |
| Granular Sugar | 11.74 | 10.00 |
| Dextrin | 15.30 | 13.20 |
| Solka Floc-40 | 5.00 | 5.00 |
| AIN-93 Mineral Mix | 3.50 | 3.50 |
| AIN-93 Vitamin Mix | 1.00 | 1.00 |
| l-Cystine | 0.15 | 0.30 |
| Choline Bitartrate | 0.25 | 0.25 |
| Soy Oil | 7.00 | 7.00 |
| Sum | 100 | 100 |
| AAs (μM) | WT–NP | KO–NP | WT–LP | KO–LP | p-Value | |
|---|---|---|---|---|---|---|
| Genotype | Protein | |||||
| His | 60.5 ± 1.8 | 59.3 ± 3 | 65 ± 2.6 | 62.2 ± 2.7 | 0.432 | 0.152 |
| Arg | 114.6 ± 2.6 | 140.2 ± 3.9 ** | 146.9 ± 7.8 | 145.9 ± 7.5 | 0.059 | 0.005 |
| Asn | 51.1 ± 2 | 62 ± 5.2 | 55.1 ± 2.6 | 60.5 ± 1.4 | 0.013 | 0.693 |
| Gln | 480.8 ± 10.9 | 447.3 ± 19 | 556.8 ± 7.8 | 463 ± 8.7 *** | <0.001 | 0.001 |
| Ser | 102.3 ± 4.4 | 129.3 ± 5.1 | 136.9 ± 4.6 | 129.7 ± 3.1 | 0.166 | 0.019 |
| Gly | 249.2 ± 11.1 | 267.3 ± 8.5 | 281.8 ± 13 | 263.1 ± 8.1 | 0.979 | 0.192 |
| Asp | 12.2 ± 1 | 14 ± 0.9 | 11.7 ± 0.8 | 11.5 ± 0.7 | 0.366 | 0.087 |
| Glu | 50.2 ± 1.8 | 66.6 ± 3.5 ** | 52.8 ± 2.7 | 56.1 ± 1.5 | <0.001 | 0.124 |
| Thr | 149.4 ± 6.5 | 175.1 ± 9.3 * | 186.2 ± 7.6 | 194.6 ± 6.9 | 0.03 | 0.001 |
| Ala | 285.5 ± 5.5 | 344 ± 30.6 | 377.3 ± 21.7 | 321.7 ± 8.2 * | 0.942 | 0.102 |
| Pro | 76.1 ± 4 | 96.7 ± 10.7 | 88.4 ± 3.2 | 93.7 ± 3.5 | 0.025 | 0.407 |
| Lys | 266.5 ± 12.5 | 301.3 ± 15.6 | 348.4 ± 16.1 | 325 ± 8.4 | 0.673 | <0.001 |
| Cys | 2.1 ± 0.3 | 1.7 ± 0.1 | 1.7 ± 0.1 | 1.9 ± 0.2 | 0.79 | 0.61 |
| Tyr | 51.1 ± 2 | 79 ± 4.5 *** | 50.5 ± 2.2 | 64.2 ± 2 *** | <0.001 | 0.009 |
| Met | 50.6 ± 1.9 | 63 ± 3.4 ** | 58 ± 1.5 | 57.3 ± 1.2 | 0.006 | 0.673 |
| Val | 212.1 ± 6.8 | 198.2 ± 9.6 | 188.2 ± 9.5 | 209.5 ± 7.4 | 0.665 | 0.462 |
| Ile | 96.3 ± 2.9 | 86.3 ± 5 | 83.9 ± 3.6 | 94.6 ± 2.6 | 0.927 | 0.593 |
| Leu | 170.6 ± 6.5 | 148.9 ± 7.5 | 150.1 ± 7.7 | 159.7 ± 8.6 | 0.435 | 0.531 |
| Phe | 78.2 ± 1.7 | 79 ± 3.3 | 72.7 ± 1.8 | 78.9 ± 2.9 | 0.173 | 0.263 |
| Trp | 70 ± 3 | 90.2 ± 3.5 *** | 90.5 ± 4.6 | 89.3 ± 5.3 | 0.033 | 0.027 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Su, J.; Gao, H.; Li, J.; Feng, Z.; Yin, Y. Untargeted Metabolomics Reveals the Function of GPRC6A in Amino Acid and Lipid Metabolism in Mice. Metabolites 2022, 12, 776. https://doi.org/10.3390/metabo12090776
He Y, Su J, Gao H, Li J, Feng Z, Yin Y. Untargeted Metabolomics Reveals the Function of GPRC6A in Amino Acid and Lipid Metabolism in Mice. Metabolites. 2022; 12(9):776. https://doi.org/10.3390/metabo12090776
Chicago/Turabian StyleHe, Yumin, Jingyun Su, Hongrui Gao, Jianzhong Li, Zemeng Feng, and Yulong Yin. 2022. "Untargeted Metabolomics Reveals the Function of GPRC6A in Amino Acid and Lipid Metabolism in Mice" Metabolites 12, no. 9: 776. https://doi.org/10.3390/metabo12090776







