Non-Esterified Fatty Acid Induces ER Stress-Mediated Apoptosis via ROS/MAPK Signaling Pathway in Bovine Mammary Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Flow Cytometer Detection of Apoptosis
2.3. Transmission Electron Microscopy (TEM)
2.4. Intracellular Reactive Oxygen Species Measurement
2.5. Real-Time Quantitative PCR Analysis
2.6. RNA-Seq and Transcriptome Analysis
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results
3.1. NEFA Induced ER Stress in BMECs
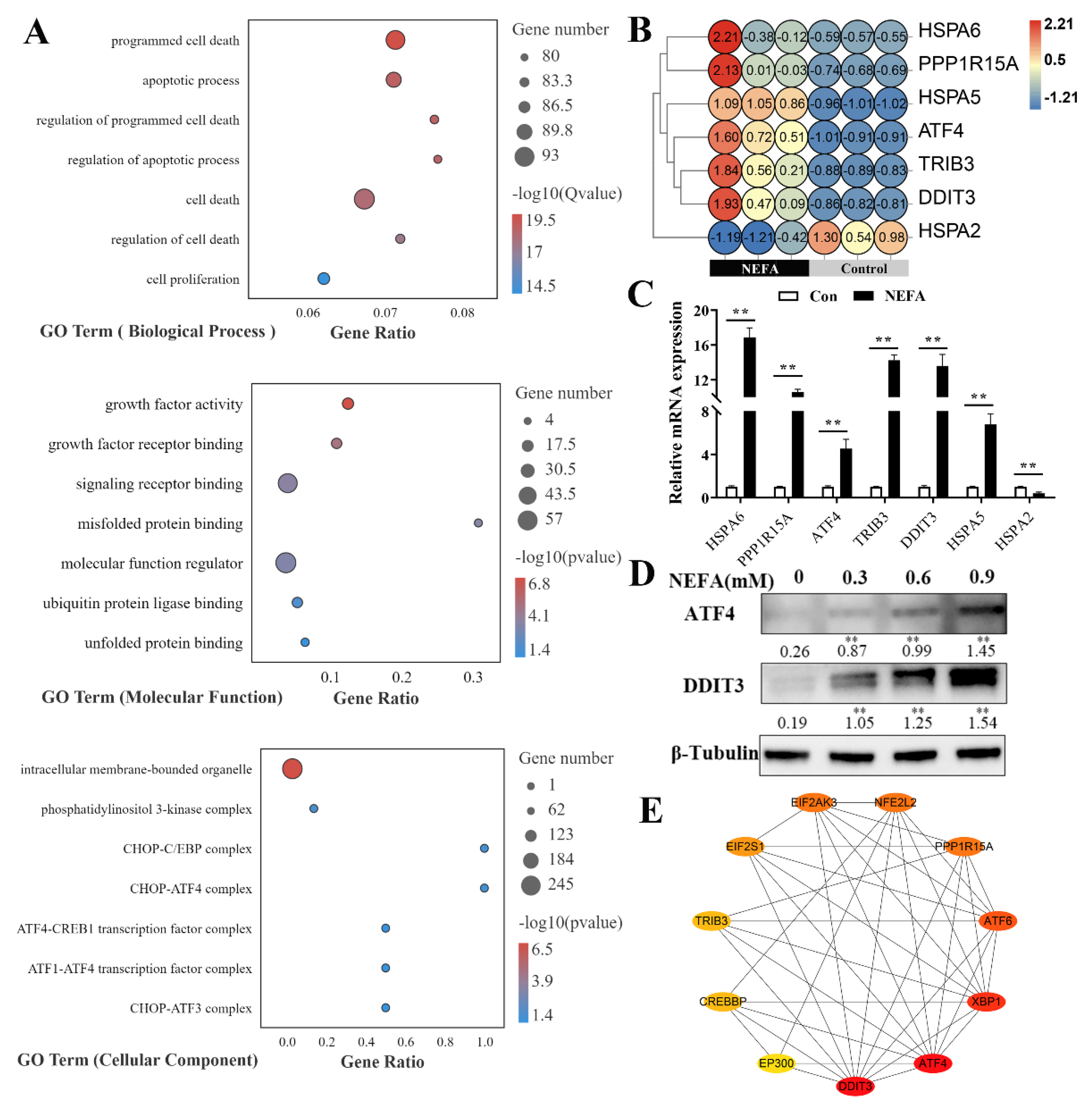
3.2. NEFA Activated the Three UPR Signaling Pathways in BMECs
3.3. NEFA Induced Apoptosis in BMECs
3.4. NEFA Induced Apoptosis Probably via ATF4-CHOP Axis
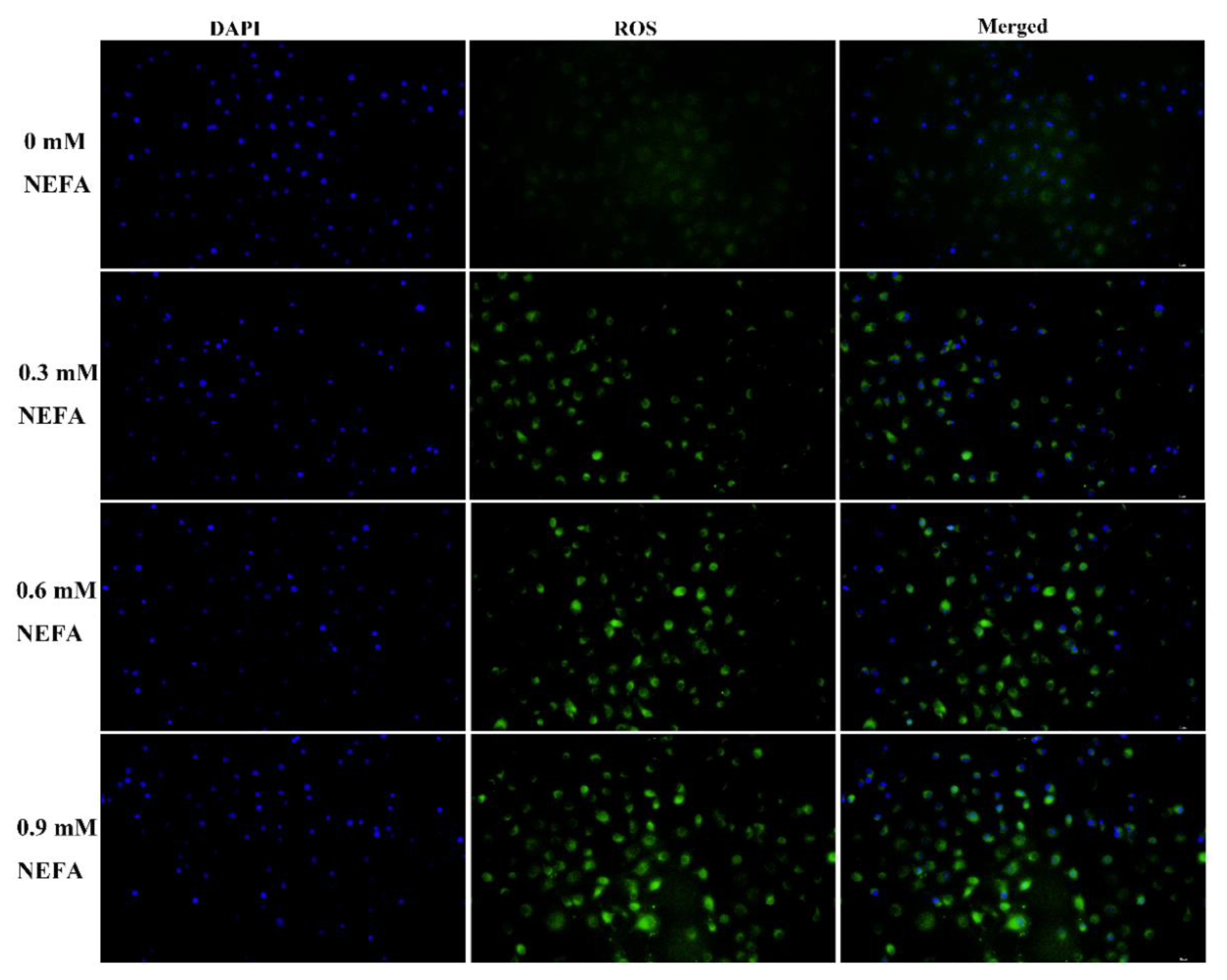
3.5. NEFA Caused Accumulation of ROS in BMECs
3.6. MAPK Signaling Pathway Was Involved in NEFA-Induced Apoptosis
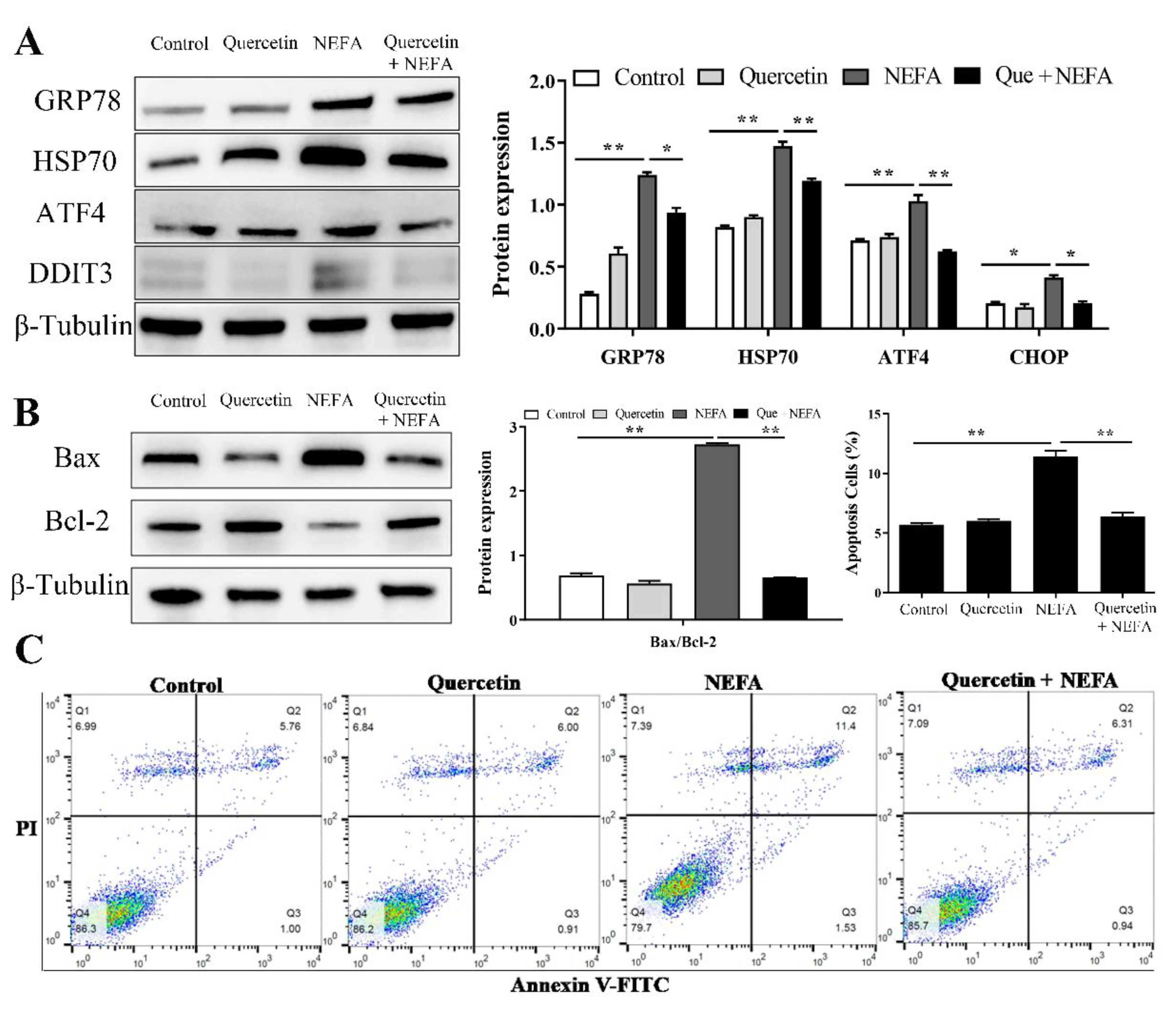
3.7. Quercetin Alleviated ER Stress-Mediated Apoptosis in NEFA Treated BMECs
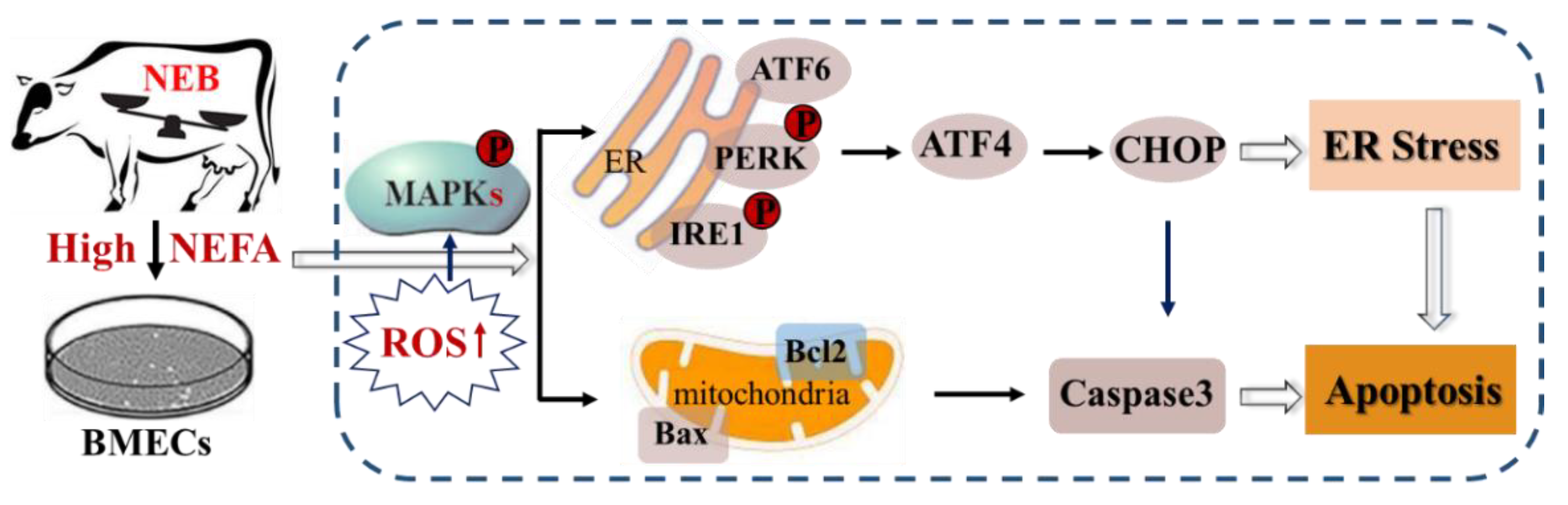
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wankhade, P.R.; Manimaran, A.; Kumaresan, A.; Jeyakumar, S.; Ramesha, K.P.; Sejian, V.; Rajendran, D.; Varghese, M.R. Metabolic and immunological changes in transition dairy cows: A review. Vet. World 2017, 10, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Irons, P.C.; Webb, E.C.; Chapwanya, A. Interactions between negative energy balance, metabolic diseases, uterine health and immune response in transition dairy cows. Anim. Reprod. Sci. 2014, 144, 60–71. [Google Scholar] [PubMed]
- Raboisson, D.; Mounié, M.; Maigné, E. Diseases, reproductive performance, and changes in milk production associated with subclinical ketosis in dairy cows: A meta-analysis and review. J. Dairy Sci. 2014, 97, 7547–7563. [Google Scholar] [PubMed]
- Chang, R.; Sun, X.; Jia, H.; Xu, Q.; Dong, Z.; Tang, Y.; Luo, S.; Jiang, Q.; Loor, J.J.; Xu, C. Inhibiting nuclear factor erythroid 2 related factor 2-mediated autophagy in bovine mammary epithelial cells induces oxidative stress in response to exogenous fatty acids. J. Anim. Sci. Biotechnol. 2022, 13, 48. [Google Scholar] [CrossRef] [PubMed]
- Adewuyi, A.A.; Gruys, E.; van Eerdenburg, F.J. Non esterified fatty acids (NEFA) in dairy cattle. A review. Vet. Q. 2005, 27, 117–126. [Google Scholar]
- Gessner, D.K.; Schlegel, G.; Ringseis, R.; Schwarz, F.J.; Eder, K. Up-regulation of endoplasmic reticulum stress induced genes of the unfolded protein response in the liver of periparturient dairy cows. BMC Vet. Res. 2014, 10, 46. [Google Scholar]
- Ringseis, R.; Gessner, D.K.; Eder, K. Molecular insights into the mechanisms of liver-associated diseases in early-lactating dairy cows: Hypothetical role of endoplasmic reticulum stress. J. Anim. Physiol. Anim. Nutr. 2015, 99, 626–645. [Google Scholar] [CrossRef]
- Song, Y.; Li, X.; Li, Y.; Li, N.; Shi, X.; Ding, H.; Zhang, Y.; Li, X.; Liu, G.; Wang, Z. Non-esterified fatty acids activate the ROS-p38-p53/Nrf2 signaling pathway to induce bovine hepatocyte apoptosis in vitro. Apoptosis 2014, 19, 984–997. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Li, J.; Wang, G.; Li, L. Non-Esterified Fatty Acid-Induced Reactive Oxygen Species Mediated Granulosa Cells Apoptosis Is Regulated by Nrf2/p53 Signaling Pathway. Antioxidants 2020, 9, 523. [Google Scholar] [CrossRef]
- Kitamura, M. The unfolded protein response triggered by environmental factors. Semin. Immunopathol. 2013, 35, 259–275. [Google Scholar]
- Badr, C.E.; Hewett, J.W.; Breakefield, X.O.; Tannous, B.A. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE 2007, 2, e571. [Google Scholar] [CrossRef] [PubMed]
- Patil, C.; Walter, P. Intracellular signaling from the endoplasmic reticulum to the nucleus: The unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001, 13, 349–355. [Google Scholar] [CrossRef]
- Ron, D. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 2002, 110, 1383–1388. [Google Scholar] [CrossRef]
- Gessner, D.K.; Koch, C.; Romberg, F.J.; Winkler, A.; Dusel, G.; Herzog, E.; Most, E.; Eder, K. The effect of grape seed and grape marc meal extract on milk performance and the expression of genes of endoplasmic reticulum stress and inflammation in the liver of dairy cows in early lactation. J. Dairy Sci. 2015, 98, 8856–8868. [Google Scholar] [CrossRef] [PubMed]
- Yonekura, S.; Tsuchiya, M.; Tokutake, Y.; Mizusawa, M.; Nakano, M.; Miyaji, M.; Ishizaki, H.; Haga, S. The unfolded protein response is involved in both differentiation and apoptosis of bovine mammary epithelial cells. J. Dairy Sci. 2018, 101, 3568–3578. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Koizumi, Y.; Hayashi, S.; Hanaoka, M.; Tokutake, Y.; Yonekura, S. The role of unfolded protein response in differentiation of mammary epithelial cells. Biochem. Biophys. Res. Commun. 2017, 484, 903–908. [Google Scholar] [CrossRef]
- Boussabbeh, M.; Prola, A.; Ben Salem, I.; Guilbert, A.; Bacha, H.; Lemaire, C.; Abis-Essefi, S. Crocin and quercetin prevent PAT-induced apoptosis in mammalian cells: Involvement of ROS-mediated ER stress pathway. Environ. Toxicol. 2016, 31, 1851–1858. [Google Scholar] [CrossRef]
- Feng, K.; Chen, Z.; Pengcheng, L.; Zhang, S.; Wang, X. Quercetin attenuates oxidative stress-induced apoptosis via SIRT1/AMPK-mediated inhibition of ER stress in rat chondrocytes and prevents the progression of osteoarthritis in a rat model. J. Cell. Physiol. 2019, 234, 18192–18205. [Google Scholar] [CrossRef]
- Loor, J.J.; Everts, R.E.; Bionaz, M.; Dann, H.M.; Morin, D.E.; Oliveira, R.; Rodriguez-Zas, S.L.; Drackley, J.K.; Lewin, H.A. Nutrition-induced ketosis alters metabolic and signaling gene networks in liver of periparturient dairy cows. Physiol. Genom. 2007, 32, 105–116. [Google Scholar] [CrossRef]
- Zhang, B.; Li, M.; Yang, W.; Loor, J.J.; Wang, S.; Zhao, Y.; Guo, H.; Ma, X.; Xia, C.; Xu, C. Orai calcium release-activated calcium modulator 1 (ORAI1) plays a role in endoplasmic reticulum stress in bovine mammary epithelial cells challenged with physiological levels of ketone bodies. J. Dairy Sci. 2020, 103, 4691–4701. [Google Scholar]
- Shi, Z.; Song, Y.; Gao, X.; Loor, J.J.; Aboragah, A.; Yu, H.; Fang, Z.; Zhu, Y.; Du, X.; Li, X.; et al. Disruption of endoplasmic reticulum homeostasis exacerbates liver injury in clinically ketotic cows. J. Dairy Sci. 2021, 104, 9130–9141. [Google Scholar] [PubMed]
- Invernizzi, G.; Naeem, A.; Loor, J.J. Short communication: Endoplasmic reticulum stress gene network expression in bovine mammary tissue during the lactation cycle. J. Dairy Sci. 2012, 95, 2562–2566. [Google Scholar]
- Huang, Y.; Zhao, C.; Kong, Y.; Tan, P.; Liu, S.; Liu, Y.; Zeng, F.; Yuan, Y.; Zhao, B.; Wang, J. Elucidation of the mechanism of NEFA-induced PERK-eIF2α signaling pathway regulation of lipid metabolism in bovine hepatocytes. J. Steroid Biochem. Mol. Biol. 2021, 211, 105893. [Google Scholar]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [PubMed]
- Sano, R.; Reed, J.C. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta 2013, 1833, 3460–3470. [Google Scholar] [PubMed]
- Iurlaro, R.; Muñoz-Pinedo, C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016, 283, 2640–2652. [Google Scholar] [PubMed] [Green Version]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.H.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar]
- Yu, X.X.; Zhu, M.Y.; Wang, J.R.; Li, H.; Hu, P.; Qing, Y.J.; Wang, X.Y.; Wang, H.Z.; Wang, Z.Y.; Xu, J.Y.; et al. LW-213 induces cell apoptosis in human cutaneous T-cell lymphomas by activating PERK-eIF2α-ATF4-CHOP axis. Acta Pharmacol. Sin. 2021, 42, 290–300. [Google Scholar]
- Li, X.; Zheng, J.; Chen, S.; Meng, F.D.; Ning, J.; Sun, S.L. Oleandrin, a cardiac glycoside, induces immunogenic cell death via the PERK/elF2α/ATF4/CHOP pathway in breast cancer. Cell Death Dis. 2021, 12, 314. [Google Scholar]
- Wang, X.; Zhuang, Y.; Fang, Y.; Cao, H.; Zhang, C.; Xing, C.; Guo, X.; Li, G.; Liu, P.; Hu, G.; et al. Endoplasmic reticulum stress aggravates copper-induced apoptosis via the PERK/ATF4/CHOP signaling pathway in duck renal tubular epithelial cells. Environ. Pollut. 2021, 272, 115981. [Google Scholar]
- Tabas, I.; Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011, 13, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Li, R.; Dai, P.; Li, Z.; Li, Y.; Li, C. Deoxynivalenol induced apoptosis and inflammation of IPEC-J2 cells by promoting ROS production. Environ. Pollut. 2019, 251, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wu, X.; Cui, Y.; Liu, P.; Xiao, B.; Zhang, X.; Zhang, J.; Sun, Z.; Song, M.; Shao, B.; et al. Mitophagy and apoptosis mediated by ROS participate in AlCl (3)-induced MC3T3-E1 cell dysfunction. Food Chem. Toxicol. 2021, 155, 112388. [Google Scholar] [CrossRef]
- Kaminskyy, V.O.; Zhivotovsky, B. Free radicals in cross talk between autophagy and apoptosis. Antioxid. Redox Signal. 2014, 21, 86–102. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, M.; Xu, S.; Khan, M.A.; Shi, Y.; Qu, W.; Gao, J.; Liu, G.; Kastelic, J.P.; Han, B. Mycoplasma bovis-generated reactive oxygen species and induced apoptosis in bovine mammary epithelial cell cultures. J. Dairy Sci. 2020, 103, 10429–10445. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; An, G.; Park, H.; Lim, W.; Song, G. Diflubenzuron leads to apoptotic cell death through ROS generation and mitochondrial dysfunction in bovine mammary epithelial cells. Pestic. Biochem. Physiol. 2021, 177, 104893. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, C.; Xu, X.; Zhao, X.; Han, Z.; Liu, D.; Bo, R.; Li, J.; Liu, Z. Ferulic acid inhibits LPS-induced apoptosis in bovine mammary epithelial cells by regulating the NF-κB and Nrf2 signalling pathways to restore mitochondrial dynamics and ROS generation. Vet. Res. 2021, 52, 104. [Google Scholar] [CrossRef]
- Xian, M.; Cao, H.; Cao, J.; Shao, X.; Zhu, D.; Zhang, N.; Huang, P.; Li, W.; Yang, B.; Ying, M.; et al. Bortezomib sensitizes human osteosarcoma cells to adriamycin-induced apoptosis through ROS-dependent activation of p-eIF2α/ATF4/CHOP axis. Int. J. Cancer 2017, 141, 1029–1041. [Google Scholar] [CrossRef]
- Cui, Y.; Zhou, X.; Chen, L.; Tang, Z.; Mo, F.; Li, X.C.; Mao, H.; Wei, X.; Wang, C.; Wang, H. Crosstalk between Endoplasmic Reticulum Stress and Oxidative Stress in Heat Exposure-Induced Apoptosis Is Dependent on the ATF4-CHOP-CHAC1 Signal Pathway in IPEC-J2 Cells. J. Agric. Food Chem. 2021, 69, 15495–15511. [Google Scholar] [CrossRef]
- Zangouei, A.S.; Barjasteh, A.H.; Rahimi, H.R.; Mojarrad, M.; Moghbeli, M. Role of tyrosine kinases in bladder cancer progression: An overview. Cell Commun. Signal. 2020, 18, 127. [Google Scholar] [PubMed]
- Gardi, C.; Bauerova, K.; Stringa, B.; Kuncirova, V.; Slovak, L.; Ponist, S.; Drafi, F.; Bezakova, L.; Tedesco, I.; Acquaviva, A.; et al. Quercetin reduced inflammation and increased antioxidant defense in rat adjuvant arthritis. Arch. Biochem. Biophys. 2015, 583, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yuan, Q.; Xu, G.; Chen, H.; Lei, H.; Su, J. Effects of Quercetin on Proliferation and H₂O₂-Induced Apoptosis of Intestinal Porcine Enterocyte Cells. Molecules 2018, 23, 2012. [Google Scholar] [CrossRef]
- Natsume, Y.; Ito, S.; Satsu, H.; Shimizu, M. Protective effect of quercetin on ER stress caused by calcium dynamics dysregulation in intestinal epithelial cells. Toxicology 2009, 258, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Eisvand, F.; Tajbakhsh, A.; Seidel, V.; Zirak, M.R.; Tabeshpour, J.; Shakeri, A. Quercetin and its role in modulating endoplasmic reticulum stress: A review. Phytother. Res. 2022, 36, 73–84. [Google Scholar] [PubMed]




| a Gene | Accession Number | Primer Sequences (5′-3′) | (%)PCR Efficiency | R2 | Fragment Size (bp) |
|---|---|---|---|---|---|
| GRP78 | NM_001075148.1 | F: TCCAGGCTGGTGTGCTCTCT R: TCGTCAGGGGTCGTTCACCT | 94 | 0.9977 | 222 |
| DDIT3 | NM_001078163.1 | F: TCGGGGCACCTGTGTTTCAC R: CTCTGGTGGTCCTGGCTCCT | 96 | 0.9981 | 112 |
| PPP1R15A | NM_001046178.2 | F: CTGATGGGCCTTCTCAGCCG R: TGCTTGGCTTCCAGGTTGGG | 95 | 0.9958 | 248 |
| HSPA2 | NM_174344.1 | F: GGCCATGAACCCCACCAACA R: ACTCCACCTGCACTTTGGGC | 93 | 0.9966 | 143 |
| TRIB3 | NM_001076103.1 | F: CCCCTGGCTGTTCCAGCAAA R: GGGCCCACTTCGAGCTTGTT | 102 | 0.9992 | 105 |
| ATF4 | NM_001034342.2 | F: TTGGGGGCTGAAGAGAGCCT R: CAGCCATTCGGAGGAGCCTG | 95 | 0.9975 | 114 |
| HSPA6 | XM_002685850.5 | F: AGGCCCAGAGAGACAGGGTG R: CCACAACTGCTGCCCACAGA | 94 | 0.9968 | 280 |
| PERK | NM_001098086.1 | F: ACATGCTGTCCCCATCCCCT R: ATTGCTGGGCAAAGGGCTGT | 101 | 0.9956 | 182 |
| EIF2α | NM_175813.2 | F: CATGCGCAGTGTGGTCAAGC R: CACAACAAGGTCCCACGCCA | 94 | 0.9962 | 110 |
| IRE-1α | XM_024980954.1 | F: ACTGAGAGGGACCGGCAGTT R: TGGTCTGTTGCAGCAGGGTG | 95 | 0.9979 | 127 |
| XBP1 | NM_001034727.3 | F: GACCAAGGGGAATGGAGCGG R: GAAGGGGAGGCCGGTAAGGA | 95 | 0.9984 | 290 |
| ATF6α | XM_024989877.1 | F: TGTGAGGGGAAGTAGGGGGC R: GGAGTGGTCCCCTGGGAAGT | 93 | 0.9963 | 175 |
| GAPDH | NM_001034034.2 | F: CATGACCACTTTGGCATCGT R: CCATCCACAGTCTTCTGGGT | 96 | 0.9985 | 133 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Huang, J.; Huan, C.; Li, L.; Li, C. Non-Esterified Fatty Acid Induces ER Stress-Mediated Apoptosis via ROS/MAPK Signaling Pathway in Bovine Mammary Epithelial Cells. Metabolites 2022, 12, 803. https://doi.org/10.3390/metabo12090803
Yan Y, Huang J, Huan C, Li L, Li C. Non-Esterified Fatty Acid Induces ER Stress-Mediated Apoptosis via ROS/MAPK Signaling Pathway in Bovine Mammary Epithelial Cells. Metabolites. 2022; 12(9):803. https://doi.org/10.3390/metabo12090803
Chicago/Turabian StyleYan, Yexiao, Junpeng Huang, Changchao Huan, Lian Li, and Chengmin Li. 2022. "Non-Esterified Fatty Acid Induces ER Stress-Mediated Apoptosis via ROS/MAPK Signaling Pathway in Bovine Mammary Epithelial Cells" Metabolites 12, no. 9: 803. https://doi.org/10.3390/metabo12090803
APA StyleYan, Y., Huang, J., Huan, C., Li, L., & Li, C. (2022). Non-Esterified Fatty Acid Induces ER Stress-Mediated Apoptosis via ROS/MAPK Signaling Pathway in Bovine Mammary Epithelial Cells. Metabolites, 12(9), 803. https://doi.org/10.3390/metabo12090803







