Abstract
The ability of microorganisms to detoxify xenobiotic compounds allows them to thrive in a toxic environment using carbon, phosphorus, sulfur, and nitrogen from the available sources. Biotransformation is the most effective and useful metabolic process to degrade xenobiotic compounds. Microorganisms have an exceptional ability due to particular genes, enzymes, and degradative mechanisms. Microorganisms such as bacteria and fungi have unique properties that enable them to partially or completely metabolize the xenobiotic substances in various ecosystems.There are many cutting-edge approaches available to understand the molecular mechanism of degradative processes and pathways to decontaminate or change the core structure of xenobiotics in nature. These methods examine microorganisms, their metabolic machinery, novel proteins, and catabolic genes. This article addresses recent advances and current trends to characterize the catabolic genes, enzymes and the techniques involved in combating the threat of xenobiotic compounds using an eco-friendly approach.
1. Introduction
In the industrial revolution and urbanization era, the global environment’s poisoning by a complex mixture of xenobiotics has become a major environmental threat worldwide [1,2]. Xenobiotic contaminants such as azodyes, phenolics, polycyclic aromatic hydrocarbons (PAHs), halogenated compounds, personal care products (PCPs), pharmaceuticals’ active compounds (PhACs), pesticides, nitroaromatic compounds, triazines, and chlorinated compounds adversely affect the environment by their long-term persistence and slow or no biodegradation in the ecosystems [3,4,5]. Once xenobiotics are discharged into the environment, they enter the food chain, causing harmful impacts at each trophic level and adversely affecting human and animal health. In 1960s, the discovery of DDT (dichloro-diphenyl-trichloroethane), and methyl mercury residues in fish and wildlife sparked public interest in the bioaccumulation of xenobiotic chemicals [6,7,8,9,10]. In addition, these pollutants have teratogenic, carcinogenic, mutagenic, and toxic effects on all organisms. Therefore, removing toxic undegradable xenobiotics from the environment is necessary [11,12]. These xenobiotic compounds have been degraded by physical and chemical methods such as coagulation, filtration, adsorption, chemical precipitation, electrolysis, and ozonation. However, it is not always cost-effective; lack of space, complicated procedures, stringent regulatory requirements imposed on decontamination by various countries, public dissatisfaction, waste disposal issues, and toxic by-products turn more hazardous than the parent compounds [2,13,14].
Over the past few decades, microbial-assisted degradation (bioremediation) of xenobiotic pollutants has evolved into the most effective, environment-friendly, cost-effective method for removing these noxious contaminants. Bioremediation is a method that involves the destruction, eradication, immobilization, or detoxification of a wide range of chemical waste and other harmful chemicals from the environment by an inclusive action of microorganisms. Bioremediation-related technologies include phytoremediation, rhizofilteration, bioaugmentation, biostimulation, landfarming, bioreactors, and composting. It is now gaining popularity; this method takes advantage of microorganisms’ metabolic capabilities to eliminate contaminants, making them the most appropriate and promising. Persistent organic pollutants (POPs) cleanup with microbial enzymes is eco-friendly, cost-effective, and inventive [15,16].
Various laws and rules have been formulated to address the problems of xenobiotics, and many patents have been adopted and are in use in the EU and around the world, with an increased focus on reducing xenobiotics from the environment in a way that is economically, environmentally, and socially acceptable and viable with reduced accumulation or generating other toxic components in nature [17]. Furthermore, patents are an accurate indicator of inventive activity and their implementation in the analysis of xenobiotics and other harmful products could help scientists, stakeholders (technologists, business leaders, attorneys), policymakers, and researchers to gain access to technology updates, develop new processes and products, design future research strategies, and make critical decisions for developing R&D investment plans for more significant economic and environmental growth [18]. This review aims to convey up-to-date knowledge on recently identified catabolic genes for xenobiotic pollutants using various omics technologies. In addition, this review gives a concise note on the role of microbial enzymes in the detoxification of xenobiotics and also highlights various patents filed for the transformation of xenobiotics from various environments.
2. Xenobiotic Pollution and Its Impact on the Environment
Xenobiotic pollution of the environment is a global concern caused by anthropogenic activities such as urbanization and population expansion. The enormous amounts of harmful compounds released into the environment result in widespread ecosystem contamination. Prominent substances such as polycyclic aromatic hydrocarbons (PAHs), heavy metal ions, pesticides, fertilizers, and oil derivatives are found in soil, sediment, and water [4].
During the Industrial Revolution, scientific and technological advances became a source of people’s over-exploitation of resources, which destroyed various ecosystems [19]. The irrational use of human, veterinary drugs and pharmaceutical waste is another well-known contributor to environmental contamination. Compared to other chemical compounds, medicines potentially impact aquatic flora and fauna. However, pharmaceuticals are believed to cause only a minimal risk of acute environmental toxicity. The scenario may differ for chronic effects; nevertheless, there is a substantial dearth of evidence about chronic effects and their toxicity. Furthermore, there is little or no evidence of multi-generational life cycle consequences, even though many aquatic creatures are exposed to toxicity throughout their life [20].
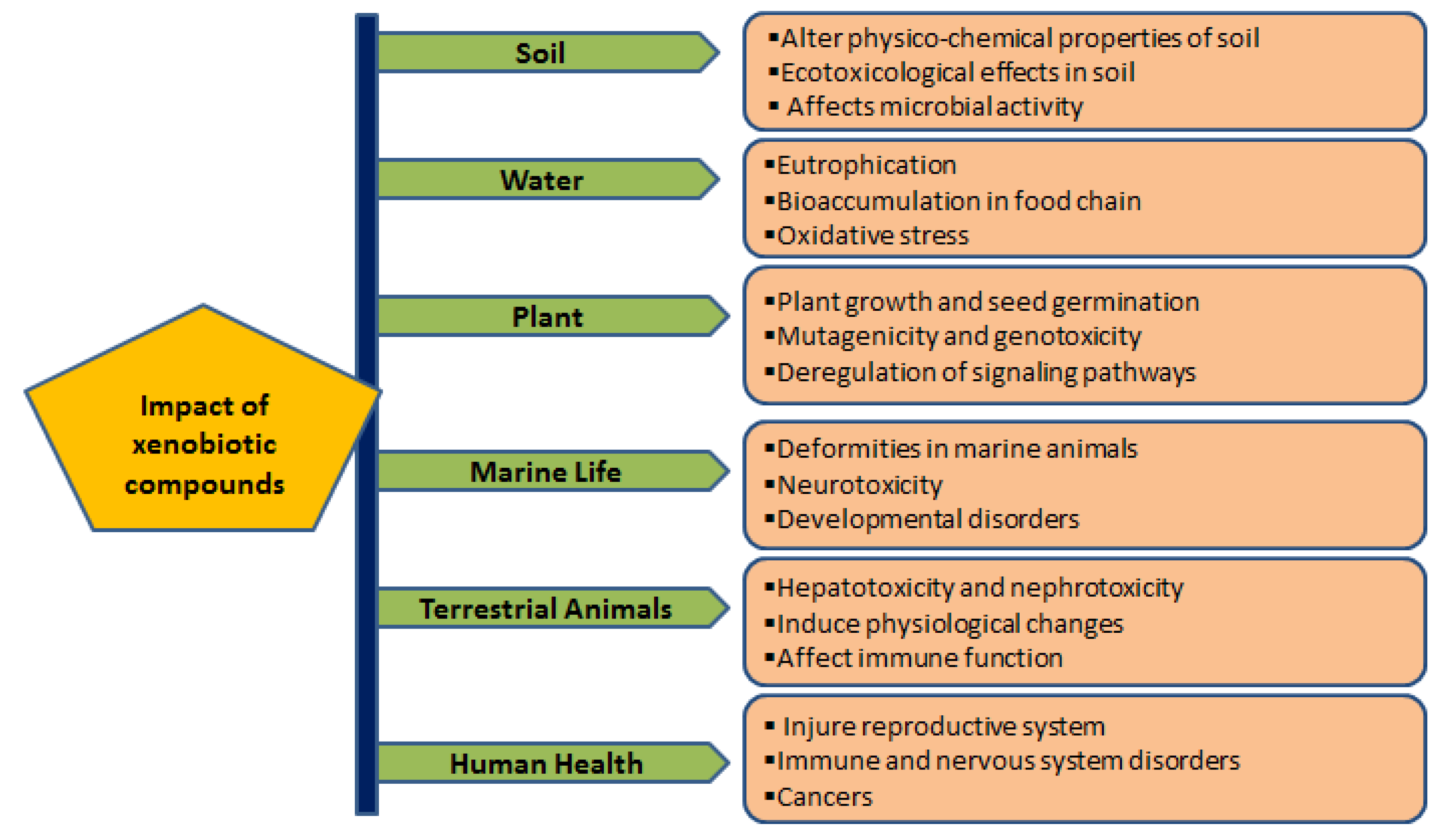
Major xenobiotic compounds have hazardous effects on the environment, plants, animals, and humans (Table 1; Figure 1).

Table 1.
Major xenobiotic compounds and their effects.
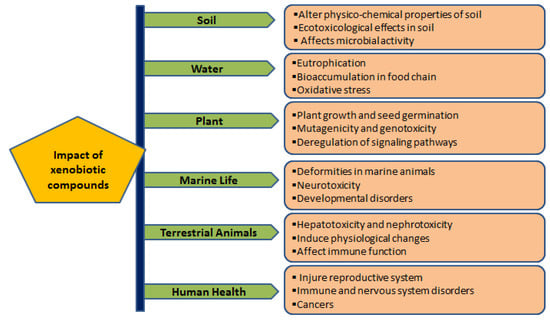
Figure 1.
Hazardous effects are caused by direct or indirect exposure of xenobiotic compounds on the environment, plants, animals and human health. Xenobiotics impose ecotoxicological effects on soil organisms, reduce microbial activity, and change the soil’s physico-chemical properties. Releasing xenobiotic compounds to aquatic systems (fresh and marine water) causes eutrophication and severe threats to faunal diversity, including deformities and developmental disorders. In addition, continuous exposure to xenobiotics adversely affects the immune, reproductive and nervous systems and sometimes causes various cancers.
2.1. Impact of Xenobiotics on Soil
Xenobiotics such as dioxins, 1,1,1-trichloro-2,2-bis (4-chlorophenyl) ethane (DDT), polychlorinated biphenyls (PCBs), chlordane, polycyclic aromatic hydrocarbons (PAHs), and nitroaromatics are the primary threat to the soil ecosystems of developed nations. However, there are reports that a few other pollutants such as benzene, nitrobenzene, toluene, xylene, aniline, ethylbenzene, trinitrotoluene/dibenzofurans, and chlorinated solvents could be xenobiotic, especially in the soil ecosystem [32]. Cosmetics and personal care products also contribute as xenobiotic pollutants, especially parabens in soil and air [33] and azodyes in soil, due to one or more aromatic rings and azo bonds [34].
Anthropogenic activities that stimulate these chemical compounds in soil include industrial activities, fuel combustion, military movement, use of pesticides, fertilizers, and soil modifications in high-production agricultural practices that cause detrimental effects [35,36]. Chemical characteristics of xenobiotics and site conditions influence their bioavailability, and distribution in soil, with soil organic matter(SOM) playing an important role [37]. Pesticides (herbicides, insecticides, fungicides, algaecides, bactericides, etc.) are chemicals used for crop protection and management and are the most widely used toxins in the environment over the last century. Millions of tonnes of pesticides are produced and spread each year around the world [38]. Environmental factors such as temperature, soil pH, and moisture significantly impact the behavior of persistent organic pollutants (POPs) in the soil. One possible strategy is binding xenobiotic compounds to soil organic matter (SOM). Many xenobiotics and their degraded products resemble humic precursors and are frequently used in humification. It has been suggested that this naturally existing process is used to neutralize environmental contaminants found in soil. Inorganic minerals interact well with xenobiotics and play a crucial role in xenobiotic transformation [39].
2.2. Impact of Xenobiotics on Water
The diffusive and point contributions of anthropogenic activities such as urban industrial production, transportation, building construction, and housing pollute surface and groundwater in urban areas. The presence of chemical substances and indicators of human activity in urban water systems has been the subject of numerous kinds of research [40]. In sewage treatment plants, some common xenobiotics sensors must be treated with municipal wastewater before being discharged into aquatic systems. Several trace metals, xenobiotic substances, and synthetic organic chemicals, such as PAHs, phthalates, and pesticides, are also noticed in different water bodies [41]. Xenobiotic substances can enter water bodies through different sources. These include (a) airborne particulate deposition; (b) surface water running from roads and land surfaces; (c) continuous inputs from commercial and sewage effluents, as well as fossil fuel products; (d) solid waste burning [42]. Xenobiotics substances also reach the water table through the leaching process, which affects the biological integrity of aquatic ecosystems [20]. The presence of xenobiotic pollutants induces oxidative stress among aquatic organisms. A recent study by Ibor et al. [43] observed a significant increase in oxidative stress response in the fish fauna of an artificial Eleyele lake, Nigeria.
A study reported that xenobiotic compounds alter the homeostasis in fishes and cause oxidative stress by producing large numbers of reactive oxygen species and suppressing the antioxidant system [44].
2.3. Impact of Xenobiotics on Plants
Xenobiotics affect the plant’s physiological and morphological characteristics in many ways; for example, particulate matter from the automobile sector changes the photosynthetic pigments, protein, cysteine contents, leaf area, and the foliar surface of plants [45]. The extensive range of xenobiotics with diverse structures and designs causes changes in gene expression, regulation, and signal transduction in the higher plants. Xenobiotics, such as phytohormone analogs, have intrinsic interactions with plant hormone receptors and signaling pathways [46]. Metals that are needed for plant growth, such as Cu, Zn, Fe and Mo, have deleterious effects at high concentrations, but metals that are not essential for plant growth, such as Pb, Cd, Hg and As, have adverse effects even at low concentrations in plant growth [47]. Xenobiotics induce DNA damage in the case of plants due to the production of reactive oxygen species and oxidative stress. The signaling pathways get deregulated due to xenobiotic toxicity in plants by influencing various signaling receptors such as G-Protein coupled receptor and receptor tyrosine kinase [48].
2.4. Impact of Xenobiotics on Marine Life
Xenobiotics negatively impact several metabolic processes of marine animals, particularly in developing fish embryos, causing morphological and functional abnormalities, retarded growth leading to death. Altered body shape, body abnormalities, hatching delays, and death have also been recorded in fishes [49]. Dyes and paints are also considered xenobiotics because they restrict sunlight penetration and inhibit gas exchange even if they are present in the traces [50]. Pesticides and herbicides are significant sources of xenobiotic pollution in marine life. Chemicals, including organophosphorus, nitrophenols, morpholine, synthetic pyrethroids, and carbamates, are often used in agricultural and daily life; later on, these substances enter various water bodies, including the sea and ocean. Insecticide such as β-Cypermethrin is a severe threat to the life of marine life and invertebrates [51].
2.5. Impact of Xenobiotics on Terrestrial Animals
Xenobiotic exposure is also possible due to application or inoculation of pharmacologic drugs or other chemicals as part of a typical conditioning or experimental operation. The consumption pattern and disposition of xenobiotics determine their toxicity. In addition, the mechanical and chemical properties also play a vital role in determining the toxicity of these xenobiotics’ compounds [52]. The xenobiotics and their metabolites may induce physiological changes in animals by altering immunological functions, cardiovascular indices, or organ systems. For example, ivermectin, a popular anthelmintic and acaricide, is harmful to some dog breeds and mouse strains due to a lack of p-glycoprotein [53]. Compared to controls, pazufloxacin and meloxicam cause oxidative damage in rabbits, including decreased glutathione content and considerable lipid peroxidation [54].
2.6. Impact of Xenobiotics on Human Health
Xenobiotics pollute the environment, so their assimilation by living species has increased dramatically in recent decades. Introducing these substances into ecosystems may increase allergic reactions, organism mortality, genetic alterations, immune system lowering, metabolic disorders, and disruptions in natural ecosystem processes [55]. Humans are exposed to a wide range of xenobiotics, such as medications and non-essential exogenous substances, throughout their lives by ingesting, breathing, dermal contact, or any other intravenous route of exposure that may represent a risk to human health [56]. Xenobiotics may alter the human gut microbiome leading to dysbiosis, which is indirectly linked to various undesirable health outcomes. The continuing biotransformation process consistently seeks to balance the metabolic activation of xenobiotics to the detoxification of their mutagenic metabolites, as it evolved to neutralize and remove body-invading agents. When this balance is disrupted, chronic diseases and DNA damage in the human body can occur. The toxicity of xenobiotics varies significantly between individuals. These oscillations are caused by the organism’s enhanced sensitivity and intraspecific variability. A large spectrum of substances is utterly foreign to the human body. These chemicals have harmful and irritating effects on various human organs and systems directly and indirectly [57].
3. Omics Approaches to Combat Xenobiotic Pollution
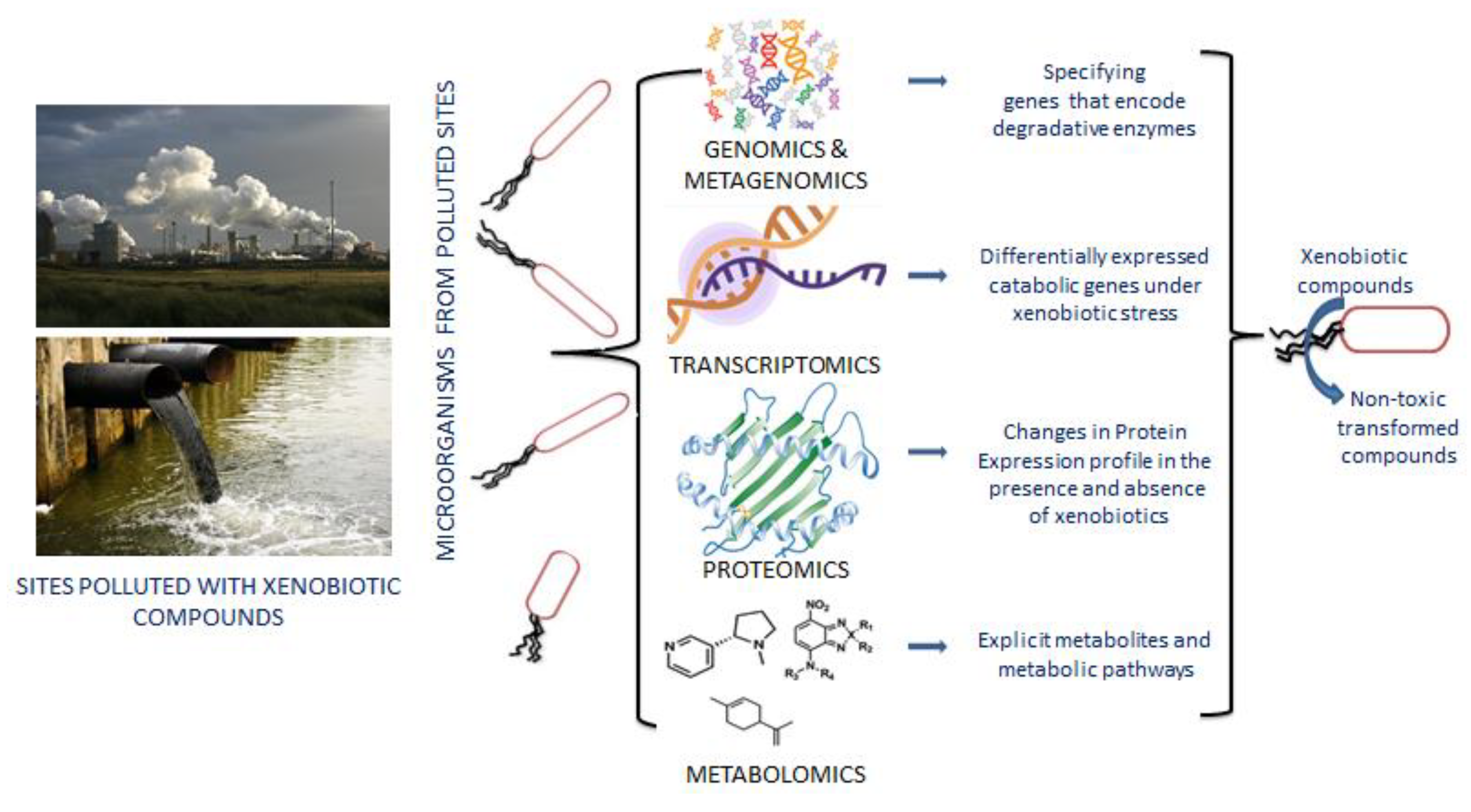
Human activities regularly emit xenobiotics into the environment, causing pollution and harming human and natural ecosystems. However, certain xenobiotic-degrading bacteria and fungi have been identified. Most of the xenobiotic-degrading bacterial strains rely only on xenobiotics for their carbon source and energy, making them great models for studying bacterial adaptability and evolution in the environment (Figure 2) [58]. Initially, bacterial strains with metabolic properties were isolated and cultured to degrade pollutants. However, very few microbes are cultivable with xenobiotic degradative potential; few of them have been isolated and characterized in the recent past with incomparable biodegradation ability such as Alcaligenes [59], Pseudomonas [60], Enterobacter, Achromobacter, Hyphomicrobiaceae, Microbacterium [61], Micrococcus and Rhodococcus [62], Aeromonas [63], Sphingobium [64], Aspergillus and Purpureocillium [65], Penicillium and Trichoderma [66], Rhodotorula and Candida [67] etc. Hence, new culture-independent approaches such as metagenomics are gaining momentum to identify non-cultivable microbes with xenobiotic degradation potential [68,69]. Few relevant xenobiotic degrading microorganisms were identified with culture-independent approaches, such as Sphingopyxis, Afipia, Oligotropha, Rhodopseudomonas, Mesorhizobium, and Stenotrophomonas [70]. The dominance of Thalassolituus and Oleispira have also been identified as vital oil-degrading bacteria through metagenomics and the metatranscriptomic approach [71].
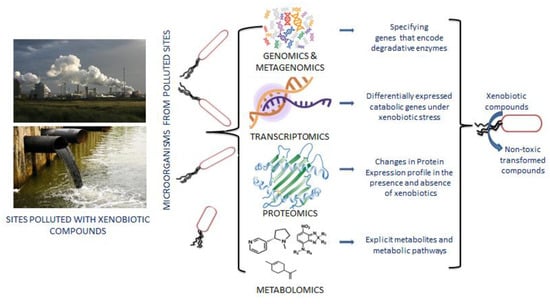
Figure 2.
Distinct features of multi-omics technologies in the transformation of xenobiotic compounds. Genomics and metagenomics identify detoxifying enzymes from the whole genome or metagenome sequencing data. RNA seq or transcriptomics data indicate up- and down-regulated genes in response to xenobiotic exposure. Proteomics techniques help to compare the changes in protein profile in the presence and absence of toxic compounds.
3.1. Genomics and Metagenomics
Genome sequencing of uncultured microorganisms helps to find new genes associated with the microbe and gives details of the degradation potential of these microbial communities. Genomics determines the genetic information and metagenomics determines the genetic sequences of a community of an organism in total. Internal transcribed spacer (ITS) regions distinguish fungal DNA from other organisms in the ribosomal genes. Plants or bacteria do not share these regions. Thus, ITS amplicon sequencing helps identify fungal species able to degrade xenobiotic compounds [72]. Functional metagenomics studies demonstrated that Burkholderia, Bradyrhizobium, Koribacter and Acidomicrobium were the most abundant genera in soil contaminated with pesticides [73]. This study also reported the abundance of phosphodiesterase encoding genes that plays a vital role in organophosphorus degradation. Whole-genome sequencing studies of atrazine-degrading Pseudomonas sp. Strain ADPe, Variovorax sp. Strain 38R, Arthrobacter sp. Strain TES, Chelatobacter sp. Strain SR38 [74] using Illumina HiSeq 3000 platform unravel the genetic changes in the strains during environmental challenges.
The Gordonia sp. 1D genome analysis revealed the existence of two alkane hydroxylase gene clusters, dibenzothiophene cleavage genes, and intermediates in the metabolism of salicylate and gentisate-naphthalene. In hot climates, the highly effective thermotolerant strain Gordonia sp. 1D can be employed to remediate oil-contaminated soils [75]. Complete genome sequence data for several significant microbial strains, including Shewanella oneidensis MR-1, Pseudomonas aeruginosa KT2440, Deinococcus indicus R1, and Dehalococcoides mccartyi WBC-2, have already been provided, which is crucial for efficient bioremediation (http://www.tigr.org, accessed on 20 February 2022).
The metagenomic approach is called ecogenomics, community, or environmental genomics [68]. Metagenomic approaches can link microbial identity, functional diversity, and the role of essential genes, for which metagenomic libraries are constructed. Although sequence-driven and function-driven approaches are used for diversity screening, novel gene identification and functions are being studied in a new approach called function-driven metagenomics. Low recovery of active clones is the main limitation of this approach [76,77,78].
3.2. Transcriptomics and Metatranscriptomics
A subset of genes transcribed to RNA is referred as transcriptome and links the genome, the proteome, and the cellular phenotypes. The mRNA expression level, which is upregulated or downregulated in an organism, can be determined using RNA sequencing and DNA microarrays [79,80]. The mRNA expression level changes with the environmental conditions which the organisms inhabit; the high cost, tremendous efforts, and a smaller number of genes to be analyzed limits the use of DNA microarray [79]. Also, when interpreting the microarray data statistically, there are chances of false results [81]. RNA sequencing has the edge on DNA microarrays due to a more comprehensive quantitative range of expression [82]. Hence, many studies are now relying upon this particular approach. The transcriptomic study of a DDT-resistant Trichoderma hamatum FBL 587 showed upregulation of around 1706 genes involved in DDT degradation and upregulation of many DDT-metabolizing enzymes such as FAD-dependent monooxygenases, epoxide hydrolases, glycosyl- and glutathione-transferases [83]. Lima-Morales et al. [84] investigated the catabolic gene diversity of BTEX-contaminated soil under continuous long-term pollutant stress to identify the occurrence of important genes for catabolic pathways. The RNA-seq and coexpression network analysis approach was used to reveal the metabolism of hexabromocyclododecane degradation in Rhodopseudomonas palustris [85]. Lima-Morales et al. further confirmed the over-expression of hexabromocyclododecane degradation enzymes such as glutathione-S-transferase, haloacid dehalogenases, cytochrome p450, dioxygenases and transcriptional regulator LysR by qRT-PCR. The mechanism of breakdown of organophosphorous pesticide phoxim by Bacillus amyloliquefaciens YP6 and its biodegradation pathway was proposed based on the transcriptomic data [86]. They observed the upregulation of oxidase, hydrolase and NADPH- cytochrome P450 reductase genes for hydrolysis, oxidation and dealkylation of phoxim. Metatranscriptomic analysis of a two-cell Canadian biobed system identified diverse xenobiotic-degrading bacterial phyla such as Sphingopyxis, Mesorhizobium, Oligotropha, Stenotrophomonas, Afipia and Pseudomonas having an important role in the degradation of xenobiotics [70].
3.3. Proteomics and Metaproteomics
Proteomics is the study of all the proteins expressed in an organism, and metaproteomics/community proteomics is the large-scale study of identifying and quantifying proteins from microbial communities [87]. Protein synthesis, protein-protein interaction, mRNA turnover, and gene expression-related studies can be performed using Proteomics.
A comparative proteomic analysis study of the strain Burkholderia zhejiangensis CEIB, S4–3 in the absence and presence of methyl parathion, revealed the changes in protein expression profile through 2D-PAGE [88]. The MALDI-TOF approach was used to identify 72 differentially expressed proteins; 35 and 37 in the absence and presence of methyl parathion, respectively. They also concluded that these proteins are involved in catabolism of aromatic compounds and detoxification of xenobiotics. The metaproteomic approach used by An et al. [89] indicated the upregulation of 430 proteins which are mainly involved in the detoxification of Direct Black G azo dye, such as peroxidase, aldehyde dehydrogenase and oxidoreductase activity proteins.
3.4. Metabolomics
This approach involves the analyses of primary and secondary proteinaceous metabolites produced by microbial cells under defined physiological conditions. Metabolites produced by microbes play an essential role in intra-species and inter-species interactions. Various methods can study metabolomics, such as metabolic flux analysis, metabolite profiling, metabolic fingerprinting, and target analysis, to identify and quantify a wide array of cellular metabolites [90].
Metabolomics, or global profiling of metabolite content, is a potent tool used to investigate toxicant effects on organisms. The metabolic approach involves analyzing primary and secondary proteinaceous metabolites inside the cells, tissues, or bio-fluids. Metabolomics is the study of metabolites in biological matrices under specified conditions. Metabolomics has recently been utilized in environmental studies to investigate metabolic alterations in humans and other creatures exposed to various contaminants. Thus, metabolomics has become an essential technique in research to investigate xenobiotics’ molecular effects [91].
In the metabolism of any xenobiotic compound, a series of metabolic pathways utilizing a variety of enzymes is needed [92]. Recent genome analyses of bacterial strains that digest xenobiotics have suggested that they arose recently by gathering genes for xenobiotic degradation, with mobile genetic components playing a pivotal role in gene recruitment [93]. However, the origins of such bacterial strains’ genes and evolutionary processes are mainly unclear. The xenobiotic degrading enzymes are valuable for studying protein evolution since they have a wide range of activities and their characteristics vary substantially with a limited number of mutations [94].
The metabolomics approach was used to study the degradation mechanism of carbaryl and other N-methyl carbamates pesticides in Burkholderia sp. strain C3 and the findings of this study demonstrated Burkholderia sp. C3’s metabolic adaptation to carbaryl in comparison to glucose and nutrient broth. The metabolic changes were most prominently linked to the biosynthesis and metabolism of amino acids, sugars, PAH lipids and cofactors [95]. In addition, a comparative metabolic approach was used to examine the microbial breakdown of cyfluthrin by Photobacterium ganghwense [96]. Soil metabolomics is an efficient method for elucidating the intricate molecular networks and metabolic pathways utilized by the soil microbial community. This method can also be used to identify soil pollution biomarkers [97].
The metabolomic characterization of two potent marine bacterial isolates, Mycobacterium sp. DBP42 and Halomonas sp. ATBC 28, is capable of degrading phthalate and plasticizers such ATBC, DBP and DEHP. They concluded that DBP is degraded by sequential elimination of the ester side chains and produces monobutyl phthalate first then phthalate and two butanol molecules by employing a metabolomics approach [98]. Drechslera sp. strain 678, is capable of degrading a common additive used in gasoline, known as methyl tertiary-butyl ether (MtBE), the organic extracts obtained from the culture filtrate of strain 678 were examined. The presence of two major bioactive metabolites, monocerin and an alkyl substituted epoxycyclohexanone derivative with good antifungal activity and bioremediation, was revealed by metabolomic analysis [99].
Metabolomics and bioinformatics technologies and databases have improved the knowledge of microbial communities, their catabolic pathways, and the genes encoding catabolic enzymes. Thus, it is an effective method for identifying novel metabolic pathways and describing metabolic networks. It has been used to evaluate variation in metabolic and catabolic gene expressions, analyze the physiology of microbial communities in varied environments, and uncover the bacterial species for xenobiotic pollutant destruction. The advance in various omics technologies such as whole genome sequencing, shotgun metagenome sequencing, transcriptomics analysis and metabolomics identified many xenobiotic-degrading microorganisms and their catabolic genes (Table 2). A recent study on the transcriptomics of Fusarium verticillioides identified genes (FDB1 and FDB2) and four associated putative gene clusters involved in the degradation of lactam and lactone xenobiotics. The study also reported the induction of a gene cluster involved in the biosynthesis of vitamin B6 upon exposure to 2-benzoxazolinone and it helps the fungus to combat the ROS generated during the metabolization of xenobiotic compounds [100]. The omics approaches clarify our understanding that many putative gene clusters are induced not only to catabolize the xenobiotics directly but also that their expressions are related to many intermediates generated during the degradation pathways.

Table 2.
List of catabolic genes identified recently for xenobiotic pollutants through various omics approaches.
3.4.1. Analytical Approaches for Metabolite Screening and Their Use in the Detection and Degradation of Xenobiotics
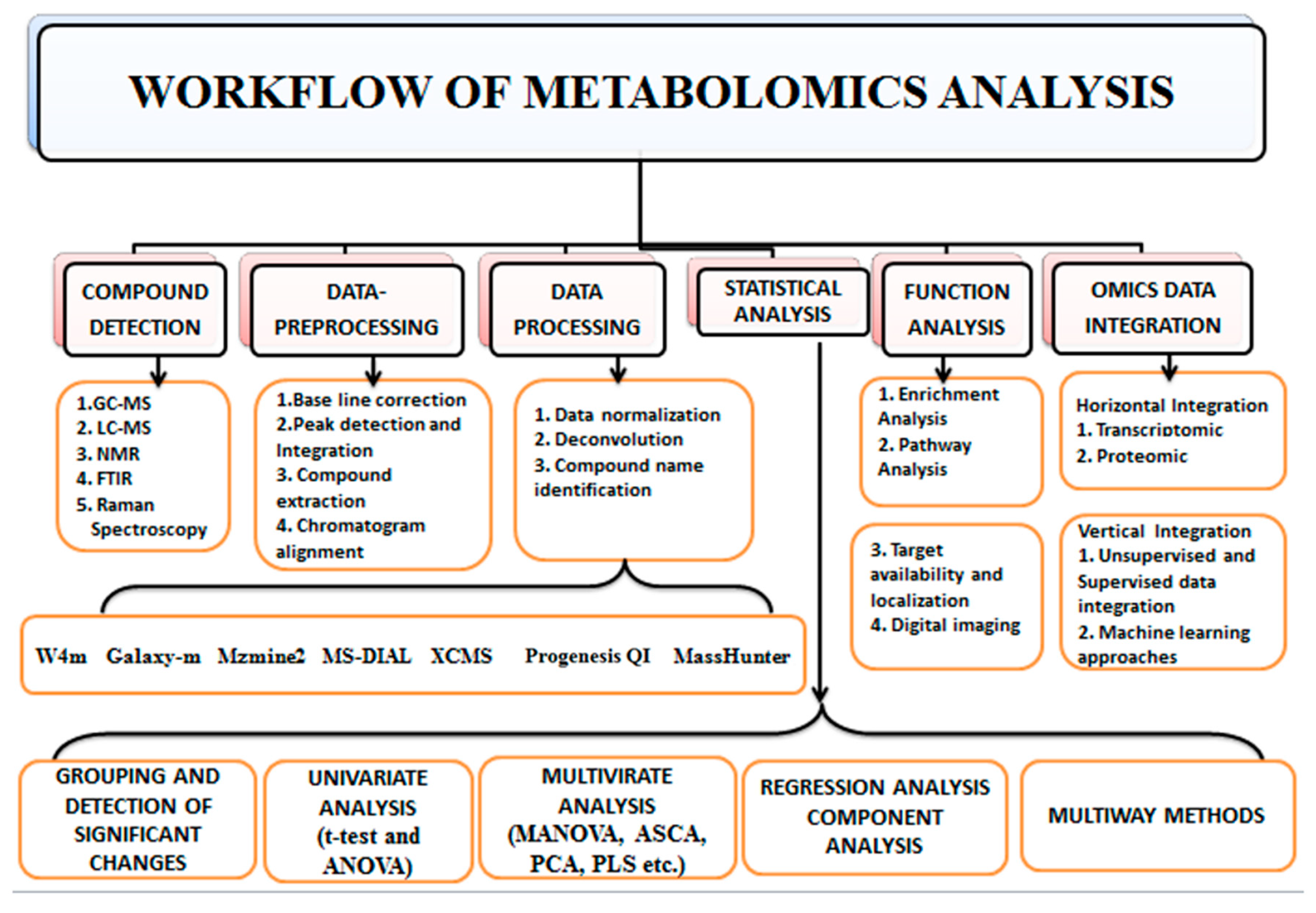
The characteristics of metabolomics data require the implementation of several tools of bioinformatics by a particular workflow. Various approaches are utilized to separate and characterize distinct metabolite classes (Figure 3). The major analytical techniques of metabolomic investigations are high-throughput techniques such as GC (Gas chromatography), HPLC (High-performance liquid chromatography), UPLC (Ultra-performance liquid chromatography), and CE (Capillary electrophoresis) with MS (mass spectroscopy) and NMR spectroscopy which enable the isolation, detection, characterization, and quantification of such metabolites and associated metabolic pathways [51,106]. Plumb et al. [107] first combined the multivariate data analysis and LC-MS to detect xenobiotics metabolites; numerous xenobiotic investigations have used UPLCMS-based metabolomics for further studies. Among different analytical techniques, LC-MS (Liquid chromatography-mass spectroscopy) and NMR have been employed extensively in metabolomic studies [108,109,110]. Many analytical procedures are generally required to achieve comprehensive data due to the metabolites’ diverse chemical characteristics. A single extract of metabolites from biological materials can contain thousands of metabolites. In untargeted metabolomics, it is typically required to segregate metabolites using an analytical column based on their chemical characteristics [106].
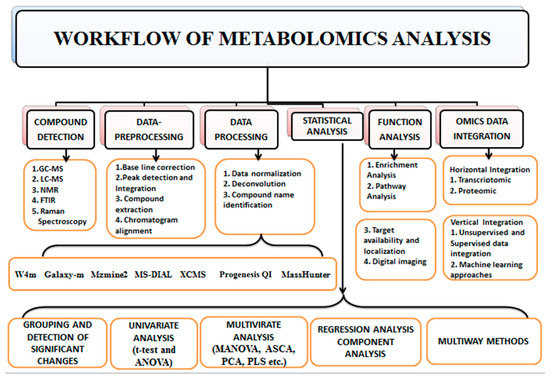
Figure 3.
Workflow of Metabolomics. The first step of the metabolomics workflow is compound detection; by employing mass spectrometry, NMR, FTIR, etc. The second step is data pre-processing, which aims to improve the signal-to-noise ratio and quality of spectra by noise reduction, baseline correction, peak detection and integration. The third step is data processing through data normalization to reduce technical bias through various software such as MZmine, XCMS, Progenesis QI, etc. The fourth step is a statistical analysis to detect the expressed metabolite, followed by the fifth step, which is function analysis that interconnects metabolites to biological pathways. The final step is integrating metabolomics data to omics data (omics data integration) to understand the mechanism of action.
Many researchers have found these techniques very helpful in identifying substances and metabolites useful in the detection and degradation of xenobiotics, ref. [111] identified three oxidative products and two cellular metabolites by Gas Chromatography-Mass Spectrometry capable of debromination and mineralizing 2, 4, 6-tribromophenol (TBP). Chen and Kim [108] used LC-MS, for metabolomic investigations of XIT (xenobiotic-induced toxicities). Rodríguez-Robledo et al. [112] determined endocrine disruptors atrazine and propazine metabolites in seminal human plasma by LC-ESI-MS/MS. Lee et al. [113] analyzed the proteome of the PAH-degrading bacterium Sphingobium chungbukense. This strain displayed exceptional aromatic compound destruction capabilities and it was also observed that 2-DE and MALDI-TOF-MS effectively analyze xenobiotic chemicals such as phenanthrene, naphthalene, and biphenyls (PNB), and their related proteins. The 5-carboxylated diclofenac could be a crucial intermediary for the complete biodegradation of diclofenac (xenobiotic) via 2,6- dichloroanailine and 3-(carboxymethyl)-4-hydroxybenzoic acid by a microbial consortium. The carboxylated diclofenac intermediate could be extracted and identified by LC-MS/MS-TOF [114]. Bhattacharyya et al. [115] implemented modified QuEChERS-GC-MS-LC-MS/MS technique for screening several classes of multiple pesticides in betelvine and estimating public risk.
3.4.2. Miscellaneous Methods Used in Detection of Xenobiotics
Appropriate extraction and analytical methods for the separation and determination of xenobiotic and derivative mixtures are critical, and they must be fast, accurate, and affordable [17]. In the recent past, there has been noticeable progress in the development of sample preparation techniques such as quick, easy, cheap, effective, rugged, and safe (QuEChERS), dispersive liquid-liquid microextraction (DLLME), focused ultrasonic solid-liquid extraction (FUSLE), solid phase extraction (SPE), solid phase microextraction (SPME), stir bar sorptive extraction (SBSE), hollow-fiber liquid phase microextraction (HFLPME) and many others [116].
QuEChERS analyzes multi-residue pesticides, antibiotics, hormones, mycotoxins, polycyclic aromatic hydrocarbons, and persistent organic pollutants such as dioxins and polychlorinated biphenyls in food and environmental matrices. QuEChERS is paired with GC–MS or LC–MS for high selectivity, sensitivity, and specificity [117]. Solid phase extraction (SPE) encompasses preparation strategies for organic pollutants from environmental matrices. Pharmaceuticals, pesticides, carbamate, bisphenols, and phthalate acid esters are analyzed using this technique [118]. In contrast, solid-phase microextraction (SPME) allows simultaneous sampling and sample preparation and is used to analyze pesticides, polycyclic aromatic hydrocarbons, phenols, amines, and polychlorinated bisphenols in food and environmental samples [119]. The stir bar sorptive extraction (SBSE) is used to determine pesticides, pharmaceuticals, polycyclic aromatic hydrocarbons, phenols, alkylphenols, chlorophenols, bisphenol A, and mycotoxins present in the environment and food [120].
HFLPME with a porous hollow-fiber membrane is used to analyse lead, arsenic, medicines, and other organic substances in environmental, clinical, and biological samples, petroleum products, pharmaceuticals, and food. It works with chromatography, electrophoresis, molecular and atomic spectrometry, and electrochemistry instruments [121]. DLLME is applied for organic compounds such as phthalate esters or parabens and metal ions such as cadmium, selenium, and lead. Pesticide analysis is used to look for chlorophenols and endocrine-disrupting phenols and medicines [122]. FUSLE can identify inorganic, organometallic, and organic substances in environmental samples, such as polycyclic aromatic hydrocarbons, PCBs, phthalate esters, and nonylphenols. It can also detect endocrine disruptors (bisphenol A and alkylphenols) in sewage sludge [123].
4. Role of Microorganisms in Xenobiotic Degradation
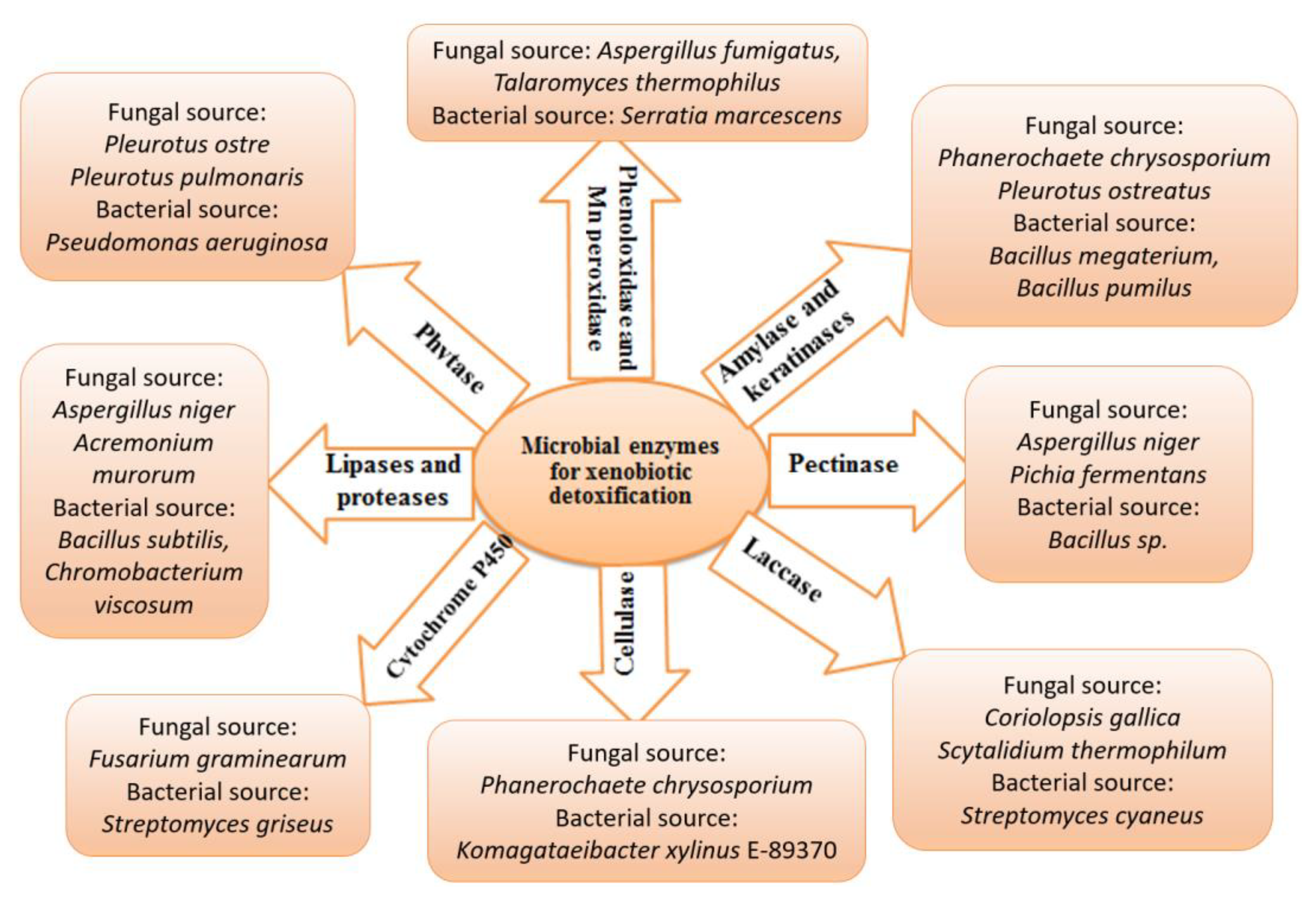
Chemical contamination can be cleaned up using biological organisms in a process known as bioremediation. The biotransformation of xenobiotics in soils, sediments, and water bodies relies heavily on microorganisms. Bioremediation uses the biological systems of living creatures (bacteria, fungi, and plants) and enzymes [124,125]. Microorganisms have an incredible ability to catabolize with the help of various genes, enzymes, and degradation pathways involved in biodegradation. Numerous microbes such as Alcaligenes, Cellulosimicrobium, Microbacterium, Micrococcus, Methanospirillum, Aeromonas, Sphingobium, Flavobacterium, Rhodococcus, Aspergillus, Penicillium, Trichoderma, Streptomyces, Rhodotorula, Candida and Aureobasidium have been isolated, characterized and have exhibited an excellent ability to biodegrade a variety of xenobiotic pollutants found in soil/water settings [79]. However, few representative microbial enzymes are involved in detoxifying xenobiotics, including cytochrome P450s, laccases, cellulase, phytase, proteases, and lipases shown in Figure 4. These enzymes can degrade aromatic hydrocarbons, dyes and halogenated compounds through various mechanisms.
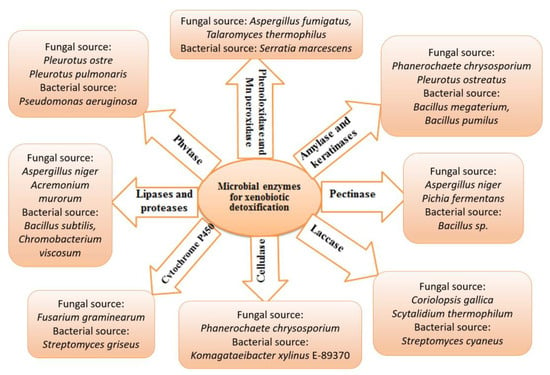
Figure 4.
Microbial enzymes in xenobiotic management. This figure summarizes a few representative enzymes and their corresponding microbial sources involved in xenobiotic detoxification.
4.1. Xenobiotic Degrading Enzymes Associated with Bacteria
Bacteria are known for their extraordinary capacity to multiply rapidly in large numbers and withstand harsh environmental conditions [126]. Recent genomic investigations of strains of bacteria that digest xenobiotics suggest that they evolved by accumulating genes for xenobiotic destruction. Bacterial species such as Pseudomonas, Escherichia, Sphingobium, Pandoraea, Rhodococcus, Gordonia, Bacillus, Moraxella, Micrococcus (aerobic bacteria), Pelatomaculum, Desulfotomaculum, Syntrophobacter, Syntrophus, Desulphovibrio, Methanospirillum, Methanosaeta (anaerobic bacteria), etc., have been isolated from soil and characterized for their biodegradation potential of xenobiotic compounds (DDT, lindane, PCBs, TNT and crystal violet) [127]. The human intestinal microbiota has a direct xenobiotic-metabolizing potential, but it can also affect the expression of host metabolizing genes and the activity of host enzymes [79]. Based on the examination of 16S rRNA and gyrB gene sequences, strain 1D of thermotolerant bacteria isolated from oil-contaminated soil at a refinery was identified as Gordonia sp. [72].
Aromatic compounds (xenobiotics) act as an electron-donating substrate in the lack of oxygen (anaerobic condition), and microbes grow by oxidizing these substances in the existence of an electron acceptor. Enzymatic biodegradation begins with selecting an enzyme for a bioremediation application; it must be capable of degrading the target pollutants into less-toxic products [127]. Many bacteria species can potentially change the hazardous xenobiotic substances into less or nontoxic substances with the help of specific enzymes present inside them.
The present review aims to report recent investigations on microbial degradation of aliphatic and aromatic hydrocarbons. The biodegradation of different types of hydrocarbons requires distinct enzymes’ due structural variation of these xenobiotic compounds at a molecular level [128]. The degradation of aliphatic hydrocarbons occurs either through monooxygenases which add single oxygen to the terminal methyl functional group or dioxygenase, which adds two oxygen atoms resulting in the peroxide formation converted to a fatty acid. The fatty acid molecule oxidizes to form TCA cycle intermediates that further metabolize to CO2 and H2O. The aromatic hydrocarbons are slowly degradable due to low solubility, production of toxic metabolites and metabolite repression [129]. At first, these compounds are converted to cis-dihydrodiols and cleaved by dioxygenase enzymes either through ortho- or meta-cleavage pathways. Then, the fission of aromatic rings occurs between the hydroxyl groups in ortho-cleavage and adjacent to hydroxyl groups in meta-cleavage pathways, finally leading to intermediates of central pathways. A few recently isolated bacteria and their associated enzymes responsible for aliphatic and aromatic hydrocarbons along with their mechanism of action are listed in Table 3.

Table 3.
Bacterial Enzymes involved in the transformation of various aliphatic and aromatic hydrocarbons.
4.2. Xenobiotic Degrading Enzymes Associated with Fungi
In addition to bacteria, fungi have a role in organic pollutant remediation. They have unique characteristics that make them ideal microorganisms for bioremediation procedures. They can reduce pollutant concentrations by physically adsorbing various contaminants via a thick cell wall composed of polymers such as chitin and cellulose. The fungal decomposition of xenobiotic compounds has highlighted the importance of the intracellular enzymatic system’s involvement in xenobiotic transformation (Table 4) [79]. These fungi benefit various activities, including biofuel degrading, environmental management, and industries such as food, paper, beverages, textiles, etc.

Table 4.
Fungi and their working enzymes involved in Xenobiotic transformation.
5. Practical Use of Microorganisms in Bioremediation of Xenobiotics
The patents are highly relevant to xenobiotic degradation; many such patents were retrieved from different databases on the basis of priority of filing and properties relevant in use to handle xenobiotics. Therefore, the search includes publicly available databases, i.e., Espacenet, DPMA, USPTO, JPO, EPO, PatFT, WIPO which cover databases produced by the Canadian Intellectual Property Office, German patents, German Patent and Trademark Office, European Patents and Chinese Patents etc (Table 5).
Regarding the environmental threats of xenobiotic compounds, there are many proven methods and products in the form of patents and process patents [99,146,147,148,149]. However, with the fast-growing technologies and human needs, many products are being designed globally, and many are not entirely degradable; therefore, scientists are working on those with long shelf-life and poor degradative nature.

Table 5.
Various patents and their properties used in the field of Xenobiotics.
Table 5.
Various patents and their properties used in the field of Xenobiotics.
| Patent | Patent No. | Country | Application | Novelties/Inventions | References |
|---|---|---|---|---|---|
| Microbial degradation of waste/sludge | 0 274 856 A1 | England; European Patent | Biotransformation and/or mineralisation of each determined constituent of the waste | This study revealed the use of the defined assorted culture of bacteria isolated through enrichment on major individual constituents of an effluent, followed by mixing the isolates to detoxify the complex non-degradable effluent. | [150] |
| Microbial removal of xenobiotic dyes | DD290004A5 | Germany; German Patent | Microbial degradation of xenobiotic dyes from triphenylmethane compounds | This invention is unique in terms of its way of selecting and using oleophilic microorganisms that ensure the degradation of xenobiotic dyes, in particular, those of triphenylmethane compounds | [151] |
| Microbial detoxification of xenobiotics using yeast | US4968620A | Peoria, United States; United States Patent | Detoxification of a variety of xenobiotics, including insecticides, herbicides, mycotoxins, and plant toxins (allelochemicals) | This invention provides insight into symbiotic yeast i.e., cigarette beetle (Lasioderma serricorne) NRRLY-18546 that detoxify pesticides, herbicides, mycotoxins, and plant poisons (allelochemicals) | [152] |
| Two-phase partitioning bioreactor for the degradation of a xenobiotech (organic and aqueous) | CA2216327A1 | Canada; Canadian Intellectual Property Office | Causing the microorganism to metabolize the xenobiotic in the aqueous phase | The novelty of the invention is the two-phase concentration of xenobiotic compounds using bioreactors | [153] |
| Bioremediation of Xenobiotics Including Methyl Tert-Butylether | US 6,194,197 B1 | United States; United States Patent | Degradation of Methyl Tert-Butylether (MTBE) | The novelty of this patent suggests that the co-metabolism of MTBE by graphium and other microbial species having a non-specific P-450 cytochrome oxidase could be used for the remediation of MTBE contamination | [154] |
| Treatment of contaminated groundwater using immobilized cells | WO 01/32566 Al | United States; Australian Patent | Creating a “bio-trench” or “bio-curtain” to clean contaminated groundwater | A method of removing contaminated groundwater is provided which places a biological permeable barrier in the path of the groundwater flow to contact the contaminated groundwater with encapsulated microorganisms which act to decontaminate the contaminated groundwater | [155] |
| Environmental remediation of organic compounds | EP 0 822 253 B1 | Tokyo-Japan; European Patent | Biodegrading of chlorinated organic compounds such as trichloroethylene (TCE) and dichloroethylene (DCE) | Processes for making harmful chemical substances harmless or less harmful by effecting a chemical change in the substances by biological methods, i.e., processes of utilizing enzymes or microorganisms as whole | [156] |
| Microbial decomposition of xenobiotics | DE10125365A1 | Germany; German Patent | Degradation of the herbicide Isoproturon | Effective method for decomposing xenobiotics (X) using a physiologically compatible combination of at least one fungus (A) with mono-/di-oxygenase activity and at least one fungus (B) with glutathione-S-transferase (GST) activity. An independent claim is also included for a combination of decomposing (X) containing (A) and (B). | [157] |
| Anaerobic microbial degradation of phthalic acid esters | WO2006136173A2 | Denmark; World Intellectual Property Organization International Bureau | Degradation of phthalic acid esters | A process for anaerobic microbial degradation of phthalic acid esters, comprising the step of adding to a bioreactor at least one bacterial strain, which as a pure isolate capable of anaerobic degradation of phthalic acid esters. | [158] |
| bioremediation of chlorinated organic compound using recombinant bacteria | US 7,989,194B2 | Chile; United States Patent | Degradation or mineralization of pollutants such as polychlorobiphenyls (PCBs), | Wautersia eutropha strain JMS34, a recombinant bacterium that can completely degrade or mineralize pollutants such as polychlorobiphenyls (PCBs), bioremediation of PCB-contaminated environments that contain a bacterial inoculum of this recombinant strain. | [148] |
| Method for simultaneous biological removal of nitrogen compounds and xenobiotics of wastewaters | WO2013166611 | Prilly, Switzerland; European Patent | Removal of nitrogen compounds and xenobiotics of wastewaters using aerobic granular biomass | According to the present invention, it can provide a kind of when in order to handle the method that contains ammonia-state nitrogen waste water and carry out promotion when biological nitrogen is removed nitration reaction. | [159] |
| Purification of soil contamination using bacterial strain | EP 2 788 512 B1 | Warszawa-Poland; European Patent | Removal of contaminants from soil, as well as a method of soil treatment | The present solution is a natural method of removing hazardous pollutants from the environment without introducing synthetic products. | [149] |
| Soil and Plant remediation using Atrazine degrading bacteria | CN104762227A | China; Chinese Patent | atrazine degradation- | The bacterium Arthrobacter ureafaciens liulou 1 (CGMCC 9667) possesses a unique combination of high atrazine-degrading activity and can colonize plant roots after seed inoculation and traits of plant growth-promoting bacterium. | [160] |
| Xenobiotic metabolism and associated enzyme | US 2019/0100792 A1 | United States; United States Patent | Probes for specifically identifying target active enzymes involved in xenobiotic metabolism | The activity-based probes labeled only their target active enzymes involved in xenobiotic metabolism and therefore provide a measurement of true protein functional activity rather than transcript or protein abundance. | [150] |
| Bioremediation of xenobiotics in the honey bee hive | US2021378263A1 | United States; United States Patent | GE bacteria can hydrolyze ester bonds or remove a carboxyl group | Described herein are engineered cells, enzymes, methods of use, and bee bread incorporating engineered cells and enzymes as described herein to address honey bee hive contamination | [161] |
| In-vitro model of the human gut microbiome to understand the Impact of xenobiotics | US20200370005 | United States Patent | Modifications of xenobiotics by intrinsic gut microbiota | The model facilitates metabolic modeling and enables a better understanding of the structure and function of the human gut microbiome and modifications of xenobiotics by intrinsic gut microbiota, such as biotransformation and bioaccumulation. | [162] |
6. Conclusions and Future Perspective
Omics approaches are an effective way to understand environmental toxicology and its remediation by employing a hybrid or integrated approach to decipher various effects of xenobiotics and other pollutants on flora, fauna including various ecosystems. The advantages include a better understanding of catabolic genes, degradative enzymes and involved metabolic pathways. In xenobiotic-contaminated soil/water ecosystems, microbial communities have the potential to play an influential role in mediating the successful biodegradation processes. Various molecular techniques provide potential measures to tackle the in-depth assessment of microbial communities at all levels, from the gene to molecule and organism to ecosystem. Many microbes with strong catabolic capability have been identified and described. The omics technique has uncovered many enzymes, especially those produced by unculturable microbes. These innovative steps have discovered various biocatalysts that are organically fitted to industrial restrictions. In this review, several patents have been discussed that employed either single isolates or mixed microbial strains to biotransform xenobiotics from contaminated environments. Resistant microbial technologies must be considered from a practical perspective; however, there is still some controversy on their field applications.
However, more research is required to accomplish exceptional advancements in bioremediation by developing novel genetically modified strains with potent catabolizing genes to have xenobiotics-free ecosystems. Furthermore, the combined approach of green nanotechnology and microbe-mediated bioremediation must be given close attention to combat xenobiotic pollution. Sustainable policies should be developed frequently using contemporary technologies; they need support from government, policymakers, and stakeholders.
Author Contributions
Conceptualization, R.M., N.P., A.K., M.A.A., A.K.P. and S.K.; methodology, R.M., N.P., A.K., M.A.A., S.K., G.R., S.S.B., J.U. and M.N.A.; software, R.M., N.P., A.K., M.A.A., S.K., G.R., S.S.B., J.U. and M.N.A.; resources, R.M., N.P. and M.N.A.; data curation, R.M., N.P., A.K., M.A.A., S.K., G.R., S.S.B., J.U., A.K.P. and M.N.A.; writing—original draft preparation, R.M., N.P., A.K., M.A.A., S.K., G.R., S.S.B. and M.N.A.; writing—review and editing, R.M., N.P., A.K., M.A.A., S.K., G.R., S.S.B., J.U. and M.N.A.; supervision, R.M., N.P., and M.N.A.; project administration, R.M., N.P. and M.N.A.; funding acquisition, R.M., N.P., J.U. and M.N.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors are grateful to the Department of Biotechnology (DBT), Ministry of Science and Technology, New Delhi, India (Vide order number BT/PR24972/2017) for financial assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Malla, M.A.; Dubey, A.; Yadav, S.; Kumar, A.; Hashem, A.; Abd-Allah, E.F. Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front. Microbiol. 2018, 9, 1132. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.; Srinivasan, L.R. Enzymes and operons mediating xenobiotic degradation in bacteria. Crit. Rev. Microbiol. 2001, 27, 133–166. [Google Scholar] [CrossRef] [PubMed]
- Godheja, J.; Shekhar, S.K.; Siddiqui, S.A.; Modi, D.R. Xenobiotic compounds present in soil and water: A review on remediation strategies. J. Environ. Anal. Toxicol. 2016, 6, 2161. [Google Scholar] [CrossRef]
- Embrandiri, A.; Kiyasudeen, S.K.; Rupani, P.F.; Ibrahim, M.H. Environmental xenobiotics and its effect on natural ecosystem. In Plant Response to Xenobiotic; Singh, A., Prasad, S.M., Singh, R.P., Eds.; Springer: Singapore, 2016; pp. 1–18. [Google Scholar]
- Dhakal, K.; Gadupudi, G.S.; Lehmler, H.J.; Ludewig, G.; Duffel, M.W.; Robertson, L.W. Sources and toxicities of phenolic polychlorinated biphenyls (OH-PCBs). Environ. Sci. Pollut. Res. Int. 2018, 25, 16277–16290. [Google Scholar] [CrossRef]
- Baun, A.; Sørensen, S.N.; Rasmussen, R.F.; Hartmann, N.B.; Koch, C.B. Toxicity and bioaccumulation of xenobiotic organic compounds in the presence of aqueous suspensions of aggregates of nano-C60. Aquat. Toxicol. 2012, 86, 379–387. [Google Scholar] [CrossRef]
- Ashauer, R.; Hintermeister, A.; O’Connor, I.; Elumelu, M.; Hollender, J.; Escher, B.I. Significance of xenobiotic Metabolism for bioaccumulation kinetics of organic chemicals in Gammarus pulex. Environ. Sci. Technol. 2012, 46, 3498–3508. [Google Scholar] [CrossRef]
- Maurya, P.K. Bioaccumulation of xenobiotics compound of pesticides in riverine system and its control technique: A critical review. J. Ind. Pollut. Control. 2016, 32, 580–594. [Google Scholar]
- Derby, A.P.; Fuller, N.W.; Huff Hartz, K.E.; Segarra, A.; Connon, R.E.; Brander, S.M.; Lydy, M.J. Trophic transfer, bioaccumulation and transcriptomic effects of permethrin in inland silversides, Menidiaberyllina, under future climate scenarios. Environ. Pollut. 2021, 275, 116545. [Google Scholar] [CrossRef]
- Zhou, Y.; Yao, L.; Pan, L.; Wang, H. Bioaccumulation and function analysis of glutathione S-transferase isoforms in Manila clam (Ruditapes philippinarum) exposed to different kinds of PAHs. J. Environ. Sci. 2022, 112, 129–139. [Google Scholar] [CrossRef]
- Elekwachi, O. Global use of bioremediation technologies for decontamination of ecosystems. J. Bioremed. Biodegrad. 2014, 5, 1–30. [Google Scholar] [CrossRef]
- Liu, L.; Bilal, M.; Duan, X.H.; Iqbal, H.M.N.; Iqbal, N. Mitigation of environmental pollution by genetically engineered bacteria-current challenges and future perspectives. Sci. Total Environ. 2019, 667, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, S.; Poudel, D.K.; Marahatha, R.; Dawadi, S.; Khadayat, K.; Phuyal, S.; Shrestha, S.; Gaire, S.; Basnet, K.; Khadka, U.; et al. Microbial Enzymes Used in Bioremediation. J. Chem. 2021, 8849512, 1–17. [Google Scholar] [CrossRef]
- Charles, S.; Ratier, A.; Baudrot, V.; Multari, G.; Siberchicot, A.; Wu, D.; Lopes, C. Taking full advantage of modelling to better assess environmental risk due to xenobiotics—the all-in-one facility MOSAIC. Environ. Sci. Pollut. Res. Int. 2022, 29, 29244–29257. [Google Scholar] [CrossRef]
- Perelo, L.W. In situ and Bioremediation of organic pollutants in aquatic sediments. J. Hazard. Mater. 2010, 177, 81–89. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques- classification based on site of application: Principle, advantages, limitations, and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef] [PubMed]
- Štefanac, T.; Grgas, D.; LandekaDragičević, T. Xenobiotics-Division and Methods of Detection: A Review. J. Xenobiot. 2021, 11, 130–141. [Google Scholar] [CrossRef]
- Saraswat, S. Patent Analysis on Bioremediation of Environmental Pollutants. J. Bioremed. Biodeg 2014, 5, 251. [Google Scholar] [CrossRef]
- Sikandar, A.; Shehzadi, K.; Arshad, Q.; Munir, K. Phytoremediation: An analytical technique for the assessment of biodegradation of organic xenobiotic pollutants: A review. Int. J. Sci. Res. 2013, 4, 2250–2253. [Google Scholar]
- Fent, K.; Weston, A.A.; Caminada, D. Ecotoxicology of human pharmaceuticals. Aquat. Toxicol. 2006, 76, 122–159. [Google Scholar] [CrossRef]
- Faroon, O.; Ruiz, P. Polychlorinated biphenyls: New evidence from the last decade. Toxicol. Ind. Health 2016, 32, 1825–1847. [Google Scholar] [CrossRef]
- Yadav, A. Potential Impact of Polychlorinated Biphenyls on Plant and Environmental Health. Int. J. Plant. Soil. Sci. 2020, 32, 51–57. [Google Scholar]
- Kroeze, C.; Reijnders, L. Halocarbons and global warming. Sci. Total Environ. 1992, 111, 1–24. [Google Scholar] [CrossRef]
- Piers, M.D.F.; Joshi, M. The role of halocarbons in the climate change of the troposphere and stratosphere. Clim. Chang. 2005, 71, 249–266. [Google Scholar]
- Griffin, G.J.L. Synthetic polymers and the living environment. Pure Appl. Chem. 1980, 52, 399–407. [Google Scholar] [CrossRef]
- Moore, C.J. Synthetic polymers in the marine environment: A rapidly increasing, long-term threat. Environ. Res. 2008, 108, 131–139. [Google Scholar] [CrossRef]
- Patneedi, C.B.; Prasadu, K.D. Impact of Pharmaceutical Wastes on Human Life and Environment. Rasyan J. Chem. 2015, 8, 67–70. [Google Scholar]
- Honda, M.; Suzuki, N. Toxicities of polycyclic aromatic hydrocarbons for aquatic animals. Int. J. Environ. Res. Public Health 2020, 17, 1363. [Google Scholar] [CrossRef]
- Curtis, S.W.; Terrell, M.L.; Jacobson, M.H.; Cobb, D.O.; Jiang, V.S.; Neblett, M.F.; Gerkowicz, S.A.; Spencer, J.B.; Marder, M.E.; Barr, D.B.; et al. Thyroid hormone levels associate with exposure to polychlorinated biphenyls and polybrominated biphenyls in adults exposed as children. J. Environ. Health 2019, 18, 1–12. [Google Scholar] [CrossRef]
- Wang, X.; Sial, M.U.; Bashir, M.A.; Bilal, M.; Raza, Q.U.A.; Ali Raza, H.M.; Rehim, A.; Geng, Y. Pesticides Xenobiotics in Soil Ecosystem and Their Remediation Approaches. Sustainability 2022, 14, 3353. [Google Scholar] [CrossRef]
- Sall, M.L.; Diaw, A.K.D.; Gningue-Sall, D.; Efremova Aaron, S.; Aaron, J.J. Toxic heavy metals: Impact on the environment and human health, and treatment with conducting organic polymers, a review. Environ. Sci. Pollut. Res. 2022, 27, 29927–29942. [Google Scholar] [CrossRef]
- Salem, H.M.; Abdel-Salam, A.; Abdel-Salam, M.A.; Seleiman, M.F. Soil Xenobiotics and their Phyto-chemical remediation. In Xenobiotics in the Soil Environment; Hashmi, M., Kumar, V., Varma, A., Eds.; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Juliano, C.; Magrini, G.A. Cosmetic ingredients as emerging pollutants of environmental and health concern. A Mini-Review. Cosmetics 2017, 4, 11. [Google Scholar] [CrossRef]
- Priyanka, J.V.; Rajalakshmi, S.; Kumar, P.S.; Krishnaswamy, V.G.; Al Farraj, D.A.; Elshikh, M.S.; Gawwad, M.R.A. Bioremediation of soil contaminated with toxic mixed reactive azo dyes by co-cultured cells of Enterobacter cloacae and Bacillus subtilis. Environ. Res. 2022, 204, 112136. [Google Scholar] [CrossRef] [PubMed]
- Alkorta, I.; Garbisu, C. Phytoremediation of organic contaminants in soils. Bioresour. Technol. 2001, 79, 273–276. [Google Scholar] [CrossRef]
- Samanta, S.K.; Singh, O.V.; Jain, R.K. Polycyclic aromatic hydrocarbons: Environmental pollution and Bioremediation. Trends Biotechnol. 2002, 20, 243–248. [Google Scholar] [CrossRef]
- Borpatragohain, B.; Sahoo, S.; Rai, A. An overview on the impact, interaction and fate of xenobiotic-soil organic matter complexes. Int. J. Commun. Syst. 2019, 7, 4935–4941. [Google Scholar]
- Schwitzguébel, J.P.; Meyer, J.; Kidd, P. Pesticide’s removal using plants: Phytodegradation versus Phyto stimulation. In Phytoremediation and Rhizoremediation Theoretical Background; Mackova, M., Dowling, D.N., Macek, T., Eds.; Springer: Dordrecht, The Netherlands, 2006; pp. 179–198. [Google Scholar]
- Hayat, M.T.; Xu, J.; Ding, N.; Mahmood, T. Dynamic behavior of persistent organic pollutants in soil and their interaction with organic matter. In Molecular Environmental Soil Science at the Interfaces in the Earth’s Critical Zone; Springer: Berlin/Heidelberg, Germany, 2010; pp. 217–222. [Google Scholar]
- Strauch, G.; Möder, M.; Wennrich, R.; Osenbrück, K.; Gläser, H.R.; Schladitz, T.; Muller, C.; Schirmer, K.; Reinstorf, F.; Schirmer, M. Indicators for assessing anthropogenic impact on urban surface and groundwater. J. Soils Sediments 2008, 8, 23–33. [Google Scholar] [CrossRef]
- Mishra, V.K.; Singh, G.; Shukla, R. Impact of Xenobiotics under a Changing Climate Scenario in Climate Change and Agricultural Ecosystems; Woodhead Publishing: Sawston, UK, 2019; pp. 133–151. [Google Scholar]
- Essumang, D.K. Distribution, levels, and risk assessment of polycyclic aromatic hydrocarbons (PAHs) in some water bodies along the coastal belt of Ghana. Sci. World J. 2010, 10, 972–985. [Google Scholar] [CrossRef]
- Ibor, O.R.; Adeogun, A.O.; Regoli, F.; Arukwe, A. Xenobiotic biotransformation, oxidative stress and obesogenic molecular biomarker responses in Tilapia guineensis from Eleyele Lake, Nigeria. Ecotoxicol. Environ. Saf. 2019, 169, 255–265. [Google Scholar] [CrossRef]
- Burgos-Aceves, M.A.; Cohen, A.; Smith, Y.; Faggio, C. MicroRNAs and their role on fish oxidative stress during xenobiotic environmental exposures. Ecotoxicol. Environ. Saf. 2018, 148, 995–1000. [Google Scholar] [CrossRef]
- Verma, A.; Singh, S.N. Biochemical and ultrastructural changes in plant foliage exposed to auto-pollution. Environ. Monit. Assess. 2006, 120, 585–602. [Google Scholar] [CrossRef]
- Ramel, F.; Sulmon, C.; Serra, A.A.; Gouesbet, G.; Couée, I. Xenobiotic sensing and signalling in higher plants. J. Exp. Bot. 2012, 63, 3999–4014. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Aery, N.C. Impact, Metabolism, and Toxicity of Heavy Metals in Plants. In Plant Responses to Xenobiotics; Singh, A., Prasad, S., Singh, R., Eds.; Springer: Singapore, 2016. [Google Scholar]
- Pande, V.; Pandey, S.C.; Sati, D.; Bhatt, P.; Samant, M. Microbial interventions in bioremediation of heavy metal contaminants in agroecosystem. Front. Microbiol. 2022, 13, 824084. [Google Scholar] [CrossRef]
- Arya, P.; Haq, S.A. Effects of xenobiotics and their biodegradation in marine life. In Smart Bioremediation Technologies; Academic Press: Cambridge, MA, USA, 2019; pp. 63–81. [Google Scholar]
- Abdelkader, E.; Nadjia, L.; Ahmed, B. Degradation study of phenazin neutral red from aqueous suspension by paper sludge. J Chem. Eng. Process. Technol. 2011, 2, 109–114. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, S.; Yan, Y. Isomerization and biodegradation of beta-cypermethrin by Pseudomonas aeruginosa CH7 with biosurfactant production. Bioresour. Technol. 2011, 102, 7139–7146. [Google Scholar] [CrossRef] [PubMed]
- Perkins, S.E.; Hankenson, F.C. Non-experimental xenobiotics: Unintended consequences of intentionally administered substances in terrestrial animal models. ILAR J. 2020, 61, 103. [Google Scholar] [CrossRef]
- Macdonald, N.; Gledhill, A. Potential impact of ABCB1 (glycoprotein) polymorphisms on ivermectin toxicity in humans. Arch. Toxicol. 2007, 81, 553–563. [Google Scholar] [CrossRef]
- Khan, A.M.; Rampal, S. Effects of repeated oral administration of pazufloxacin mesylate and meloxicam on the antioxidant status in rabbits. J. Am. Assoc. Lab. Anim. Sci. 2014, 53, 399–403. [Google Scholar]
- Kobzev, K.; Kobzeva, N.; Chegge, V.; Balinskaya, M.; Bozhenko, E.; Saakian, S.; Sarkisian, D. Ecological problems in the Russian Federation. Impact on the health of people and the country’s economy. E3S Web Conf. 2020, 217, 11001. [Google Scholar] [CrossRef]
- Korrapati, M.C.; Mehendale, H.M. Xenobiotics. In Encyclopedia of Toxicology, 2nd ed.; Wexler, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 469–470. [Google Scholar]
- Kucherenko, S.V.; Ovcharenko, A.M.; Pushenko, S.L. Xenobiotics: A Threat to the Health of Living Organisms. E3S Web Conf. EDP Sci. 2021, 285, 03006. [Google Scholar] [CrossRef]
- Nagata, Y. Special Issue: Microbial Degradation of Xenobiotics. Microorganisms 2020, 8, 487. [Google Scholar] [CrossRef]
- Nag, M.; Lahiri, D.; Dutta, B.; Jadav, G.; Ray, R.R. Biodegradation of used polyethene bags by a new marine strain of Alcaligenes faecalis LNDR-1. Environ. Sci. Pollut. Res. 2021, 28, 41365–41379. [Google Scholar] [CrossRef] [PubMed]
- Tirkey, S.R.; Ram, S.; Mitra, M.; Mishra, S. Performance analysis of Pseudomonas sp. strain SA3 in naphthalene degradation using phytotoxicity and microcosm studies. Biodegradation 2022, 33, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ma, Y.; Yao, T.; Ma, L.; Zhang, J.; Li, C. Biodegradation Pathway and Detoxification of β-cyfluthrin by the bacterial consortium and its bacterial community structure. J. Agric. Food Chem. 2022, 70, 7626–7635. [Google Scholar] [CrossRef] [PubMed]
- Bodor, A.; Bounedjoum, N.; Feigl, G.; Duzs, Á.; Laczi, K.; Szilágyi, Á.; Perei, K. Exploitation of extracellular organic matter from Micrococcus luteus to enhance ex-situ bioremediation of soils polluted with used lubricants. J. Hazard. Mat. 2021, 417, 125996. [Google Scholar] [CrossRef]
- Srinivasan, S.; Sadasivam, S.K. Biodegradation of textile azo dyes by textile effluent non-adapted and adapted Aeromonas hydrophila. Environ. Res. 2021, 194, 110643. [Google Scholar] [CrossRef]
- Feller, F.M.; Eilebrecht, S.; Nedielkov, R.; Yücel, O.; Alvincz, J.; Salinas, G.; Philipp, B. Investigations on the Degradation of the Bile Salt Cholate via the 9, 10-Seco-Pathway Reveals the Formation of a Novel Recalcitrant Steroid Compound by a Side Reaction in Sphingobium sp. Strain Chol11. Microorganisms 2021, 9, 2146. [Google Scholar] [CrossRef]
- Benguenab, A.; Chibani, A. Biodegradation of petroleum hydrocarbons by filamentous fungi (Aspergillus ustus and Purpureocilliumlilacinum) isolated from used engine oil-contaminated soil. Acta Ecol. Sin. 2021, 41, 416–423. [Google Scholar] [CrossRef]
- Correa, L.O.; Bezerra, A.F.M.; Honorato, L.R.S.; Cortêz, A.C.A.; Souza, J.V.B.; Souza, E.S. Amazonian soil fungi are efficient degraders of glyphosate herbicide; novel isolates of Penicillium, Aspergillus, and Trichoderma. Braz. J. Biol. 2021, 83, e242830. [Google Scholar] [CrossRef]
- Benmessaoud, S.; Anissi, J.; Kara, M.; Assouguem, A.; AL-Huqail, A.A.; Germoush, M.O.; Bahhou, J. Isolation and Characterization of Three New Crude Oil Degrading Yeast Strains, Candida parapsilosis SK1, Rhodotorulamucilaginosa SK2 and SK3. Sustainability 2022, 14, 3465. [Google Scholar] [CrossRef]
- Lakshmi, V.V. Genomics Approach to Bioremediation. Bioremediat. Technol. 2010, 7, 206–244. [Google Scholar]
- Rodríguez, A.; Castrejón-Godínez, M.L.; Salazar-Bustamante, E.; Gama-Martínez, Y.; Sánchez-Salinas, E.; Mussali-Galante, P.; Tovar-Sánchez, E.; Ortiz-Hernández, M.L. Omics Approaches to Pesticide Biodegradation. Curr. Microbiol. 2020, 77, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.N.; Perry, B.J.; Bergsveinson, J.; Freeman, C.N.; Sheedy, C.; Nilsson, D.; Yost, C.K. Metagenomic and metatranscriptomic analysis reveals enrichment for xenobiotic-degrading bacterial specialists and xenobiotic-degrading genes in a Canadian Prairie two-cell biobed system. Environ. Microbiol. Rep. 2021, 13, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Fortin, N.; Elias, M.; Wasserscheid, J.; King, T.L.; Lee, K.; Greer, C.W. Metagenomic and metatranscriptomic responses of natural oil-degrading bacteria in the presence of dispersants. Environ. Microbiol. 2019, 21, 2307–2319. [Google Scholar] [CrossRef] [PubMed]
- Siddique, A.B.; Albrectsen, B.R.; Ilbi, H.; Siddique, A.B. Optimization of Protocol for Construction of Fungal ITS Amplicon Library for High-Throughput Illumina Sequencing to Study the Mycobiome of Aspen Leaves. Appl. Sci. 2022, 12, 1136. [Google Scholar] [CrossRef]
- Jeffries, T.C.; Rayu, S.; Nielsen, U.N.; Lai, K.; Ijaz, A.; Nazaries, L.; Singh, B.K. Metagenomic Functional Potential Predicts Degradation Rates of a Model Organophosphorus Xenobiotic in Pesticide Contaminated Soils. Front. Microbiol. 2018, 9, 147. [Google Scholar] [CrossRef]
- Billet, L.; Devers-Lamrani, M.; Serre, R.F.; Julia, E.; Vandecasteele, C.; Rouard, N.; Spor, A. Complete genome sequences of four atrazine-degrading bacterial strains, Pseudomonas sp. strain ADPe, Arthrobacter sp. strain TES, Variovorax sp. strain 38R, and Chelatobacter sp. strain SR38. Microbiol. Resour. Announc. 2021, 10, e01080-20. [Google Scholar] [CrossRef]
- Delegan, Y.A.; Valentowich, L.N.; Shafieva, S.M.; Ganbarov, K.G.; Filonov, A.E.; Vainstein, M.B. Characterization and genomic analysis of highly efficient thermotolerant oil-degrading bacterium Gordonia sp. 1D. Folia Microbiol. 2019, 64, 41–48. [Google Scholar] [CrossRef]
- Eyers, L.; George, I.; Schuler, L.; Stenuit, B.; Agathos, S.N.; ElFantroussi, S. Environmental genomics: Exploring the unmined richness of microbes to degrade xenobiotics. Appl. Microbiol. Biotechnol. 2004, 66, 123–130. [Google Scholar] [CrossRef] [Green Version]
- Lam, K.N.; Cheng, J.; Engel, K.; Neufeld, J.D.; Charles, T.C. Current and future resources for functional metagenomics. Front. Microbiol. 2015, 6, 1196. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, D.; Pandya, L.; Pandit, P.R.; Patel, Z.; Bhairappanavar, S.; Das, J. Gene-targeted metagenomics approach for the degradation of organic pollutants. In Emerging Technologies in Environmental Bioremediation; Shah, M.P., Rodriguez-Couto, S., Sengor, S.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 257–273. [Google Scholar]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, W.; Bhatt, P.; Chen, S. Recent Advanced Technologies for the Characterization of Xenobiotic-Degrading Microorganisms and Microbial Communities. Front. Bioeng. Biotechnol. 2021, 9, 632059. [Google Scholar] [CrossRef]
- Singh, O.V.; Nagaraj, N.S. Transcriptomics, proteomics and interactomics: Unique approaches to track the insights of Bioremediation. Brief. Funct. Genom. Proteomic 2006, 4, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Dharmadi, Y.; Gonzalez, R.D.N.A. microarrays: Experimental issues, data analysis, and application to bacterial systems. Biotechnol. Prog. 2004, 20, 1309–1324. [Google Scholar] [CrossRef] [PubMed]
- Roh, S.W.; Abell, G.; Kim, K.H.; Nam, Y.D.; Bae, J.W. Comparing microarray and next-generation sequencing technologies for microbial ecology research. Trends Biotechnol. 2010, 28, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Davolos, D.; Russo, F.; Canfora, L.; Malusà, E.; Tartanus, M.; Furmanczyk, E.M.; Persiani, A.M. A Genomic and Transcriptomic Study on the DDT-Resistant Trichoderma hamatum FBL 587: First Genetic Data into Mycoremediation Strategies for DDT-Polluted Sites. Microorganisms 2021, 9, 1680. [Google Scholar] [CrossRef] [PubMed]
- Lima-Morales, D.; Jáuregui, R.; Camarinha-Silva, A.; Geffers, R.; Pieper, D.H.; Vilchez-Vargas, R. Linking microbial community and catabolic gene structures during the adaptation of three contaminated soils under continuous long-term pollutant stress. Appl. Environ. Microbiol. 2016, 82, 2227–2237. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Wang, R.; Lin, C.Y.; Chen, S.H.; Chuang, C.H.; Chou, T.H.; Shih, Y.H. The degradation mechanisms of Rhodopseudomonas palustris toward hexabromocyclododecane by time-course transcriptome analysis. Chem. Eng. J. 2021, 425, 130489. [Google Scholar] [CrossRef]
- Meng, D.; Zhang, L.; Meng, J.; Tian, Q.; Zhai, L.; Hao, Z.; Guan, Z.; Cai, Y.; Liao, X. Evaluation of the Strain Bacillus amyloliquefaciens YP6 in Phoxim degradation via transcriptomic data and product analysis. Molecules 2019, 24, 3997. [Google Scholar] [CrossRef]
- Kleiner, M. Metaproteomics: Much more than measuring gene expression in microbial communities. mSystems 2019, 4, e00115–e00119. [Google Scholar] [CrossRef] [Green Version]
- Castrejón-Godínez, M.L.; Tovar-Sánchez, E.; Ortiz-Hernández, M.L.; Encarnación-Guevara, S.; Batallar, Á.G.M.; Ortiz, M.H.; Mussali-Galante, P. Proteomic analysis of Burkholderia zhejiangensis CEIB S4–3 during the methyl parathion degradation process. Pestic. Biochem. Physiol. 2022, 187, 105197. [Google Scholar] [CrossRef]
- An, X.; Chen, Y.; Chen, G.; Feng, L.; Zhang, Q. Integrated metagenomic and metaproteomic analyses reveal potential degradation mechanism of azo dye-Direct Black G by thermophilic microflora. Ecotoxicol. Environ. Saf. 2020, 196, 110557. [Google Scholar] [CrossRef]
- Dangi, A.K.; Sharma, B.; Hill, R.T.; Shukla, P. Bioremediation through microbes: Systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 2019, 39, 79–98. [Google Scholar] [CrossRef]
- Bedia, C. Metabolomics in environmental toxicology: Applications and challenges. Trends Environ. Anal. C 2022, 34, e00161. [Google Scholar] [CrossRef]
- Koppel, N.; Bisanz, J.E.; Pandelia, M.E.; Turnbaugh, P.J.; Balskus, E.P. Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. Elife 2018, 7, e33953. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Kato, H.; Ohtsubo, Y.; Tsuda, M. Lessons from the genomes of lindane-degrading sphingomonads. Environ. Microbiol. Rep. 2019, 11, 630–644. [Google Scholar] [CrossRef]
- Singh, D.P.; Prabha, R.; Gupta, V.K.; Verma, M.K. Metatranscriptome Analysis Deciphers Multifunctional Genes and Enzymes Linked with the Degradation of Aromatic Compounds and Pesticides in the Wheat Rhizosphere. Front. Microbiol. 2018, 9, 1331. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Keum, Y.S.; Li, Q.X. Metabolomic and proteomic insights into carbaryl catabolism by Burkholderia sp. C3 and degradation of ten N-methylcarbamate. Biodegradation 2013, 24, 795–811. [Google Scholar] [CrossRef]
- Wang, T.; Hu, C.; Zhang, R.; Sun, A.; Li, D.; Shi, X. Mechanism study of cyfluthrin biodegradation by Photobacterium ganghwense with comparative metabolomics. Appl. Microbiol. Biotechnol. 2019, 103, 473–488. [Google Scholar] [CrossRef]
- Jones, O.A.H.; Sdepanian, S.; Lofts, S.; Svendsen, C.; Spurgeon, D.J.; Maguire, M.L.; Griffin, J.L. Metabolomic analysis of soil communities can be used for pollution assessment. Environ. Toxicol. Chem. 2018, 33, 61–64. [Google Scholar] [CrossRef] [Green Version]
- Wright, R.; Bosch, R.; Gibson, M.I.; Christie-Oleza, J. Plasticizer degradation by a marine bacterial isolates: A proteogenomic and metabolomic characterization. Environ. Sci. Technol. 2020, 54, 2244–2256. [Google Scholar] [CrossRef]
- D’Errico, G.; Aloj, V.; Flematti, G.R.; Sivasithamparam, K.; Worth, C.M. Metabolites of Drechslera sp. endophyte with potential as biocontrol and bioremediation agent. Nat. Prod. Res. 2020, 35, 4508–4516. [Google Scholar] [CrossRef]
- Gao, M.; Gu, X.; Satterlee, T.; Duke, M.V.; Scheffler, B.E.; Gold, S.E.; Glenn, A.E. Transcriptomic Responses of Fusarium verticillioides to Lactam and Lactone Xenobiotics. Front. Fungal Biol. 2022, 3, 923112. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Song, Q.; Yang, J.; Xie, L.; Yu, S.; Zheng, L. Polycyclic aromatic hydrocarbon and n-alkane pollution characteristics and structural and functional perturbations to the microbial community: A case-study of historically petroleum-contaminated soil. Environ. Sci. Pollut. Res. Int. 2021, 28, 10589–10602. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Rajput, V.; Dharne, M. Functional metagenomic landscape of polluted river reveals potential genes involved in degradation of xenobiotic pollutants. Environ. Res. 2021, 192, 110332. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Stenuit, B.; Tremblay, J.; Yu, K.; Tringe, S.G.; Alvarez-Cohen, L. (Structural dynamics and transcriptomic analysis of Dehalococcoidesmccartyi within a TCE-Dechlorinating community in a completely mixed flow reactor. Water Res. 2019, 158, 146–156. [Google Scholar] [CrossRef]
- Sengupta, K.; Swain, M.T.; Livingstone, P.G.; Whitworth, D.E.; Saha, P. Genome sequencing and comparative transcriptomics provide a holistic view of 4-nitrophenol degradation and concurrent fatty acid catabolism by Rhodococcus sp. strain BUPNP1. Front. Microbiol. 2019, 9, 3209. [Google Scholar] [CrossRef]
- Di Canito, A.; Zampolli, J.; Orro, A.; D’Ursi, P.; Milanesi, L.; Sello, G.; Di Gennaro, P. Genome-based analysis for the identification of genes involved in o-xylene degradation in Rhodococcusopacus R7. BMC Genom. 2018, 19, 587. [Google Scholar] [CrossRef]
- Kim, H.M.; Kang, J.S. Metabolomic Studies for the Evaluation of Toxicity Induced by Environmental Toxicants on Model Organisms. Metabolites 2021, 11, 485. [Google Scholar] [CrossRef]
- Plumb, J.A.; Finn, P.W.; Williams, R.J.; Bandara, M.J.; Romero, M.R.; Watkins, C.J.; La Thangue, N.B.; Brown, R. Pharmacodynamic response and inhibition of growth of human tumor xenografts by the novel histone deacetylase inhibitor PXD101. Mol. Cancer Ther. 2003, 2, 721–728. [Google Scholar]
- Chen, C.; Kim, S. LC-MS based metabolomics of xenobiotic-induced toxicities. Comput. Struct. Biotechnol. J. 2013, 4, e201301008. [Google Scholar] [CrossRef] [Green Version]
- Percival, B.C.; Grootveld, M.; Gibson, M.; Osman, Y.; Molinari, M.; Jafari, F.; Sahota, T.; Martin, M.; Casanova, F.; Mather, M.L. Low-field, benchtop NMR spectroscopy as a potential tool for point-of-care diagnostics of metabolic conditions: Validation, protocols and computational models. High-Throughput 2019, 8, 2. [Google Scholar] [CrossRef]
- Fraga-Corral, M.; Carpena, M.; Garcia-Oliveira, P.; Pereira, A.; Prieto, M.; Simal-Gandara, J. Analytical metabolomics and applications in health, environmental and food science. Crit. Rev. Anal. Chem. 2020, 52, 712–734. [Google Scholar] [CrossRef]
- Zu, L.; Li, G.; An, T.; Wong, P.K. Biodegradation kinetics and mechanism of 2,4,6-tribromophenol by Bacillus sp. GZT: A phenomenon of xenobiotic methylation during debromination. Bioresour. Technol. 2012, 110, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Robledo, V.; Vendrell, A.; García-Cifuentes, A.; Villaseca-González, N.; Guiberteau-Cabanillas, C.; Martínez, L.L.; Garde, J.J.; Fernández-Santos, M.R. Determination of atrazine and propazine metabolites deemed endocrine disruptors in human seminal plasma by LC–ESI-MS/MS. Chem. Biol Technol. Agric. 2022, 9, 22. [Google Scholar] [CrossRef]
- Lee, S.Y.; Sekhon, S.S.; Ban, Y.H.; Ahn, J.Y.; Ko, J.H.; Lee, L.; Kim, S.Y.; Kim, Y.C.; Kim, Y.H. Proteomic analysis of polycyclic aromatic hydrocarbons (PAHs) degradation and detoxification in Sphingobiumchungbukense DJ77. J. Microbiol. Biotechnol. 2016, 26, 1943–1950. [Google Scholar] [CrossRef]
- Facey, S.J.; Nebel, B.A.; Kontny, L.; Allgaier, M.; Hauer, B. Rapid and complete degradation of diclofenac by native soil microorganisms. Environ. Technol. Innov. 2018, 10, 55–61. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Poi, R.; Sen, M.B.; Hazra, D.K.; Ghosh, R.; Mandal, S.; Karmakar, R. Establishment of modified QuEChERS-GC–MS-LC–MS/MS method for simultaneous screening of multi-class multi-pesticide residues in betelvine and consumer risk assessment. Microchem. J. 2022, 179, 107444. [Google Scholar] [CrossRef]
- Tuzimski, T.; Sherma, J. Determination of Targe Xenobiotics and Unknown Compound Residues in Food, Environmental, and Biological Samples; Taylor & Francis Group: Boca Raton, FL, USA, 2019. [Google Scholar]
- Kim, L.; Lee, D.; Cho, H.K.; Choi, S.D. Review of the QuEChERS method for the analysis of organic pollutants: Persistent organic pollutants, polycyclic aromatic hydrocarbons, and pharmaceuticals. Trends Environ. Anal. Chem. 2019, 22, e00063. [Google Scholar] [CrossRef]
- Azzouz, A.; Kumar Kailasa, S.; Soo Lee, S.; Rascón, A.J.; Ballesteros, E.; Zhang, M.; Kim, K.H. Review of nanomaterials as sorbents in solid-phase extraction for environmental samples. Trends Anal. Chem. 2018, 108, 347–369. [Google Scholar] [CrossRef]
- Lashgari, M.; Singh, V.; Pawliszyn, J. A critical review on regulatory sample preparation methods: Validating solid-phase microextraction techniques. Trends Anal. Chem. 2019, 119, 115618. [Google Scholar] [CrossRef]
- David, F.; Sandra, P. Stir bar sorptive extraction for trace analysis. J. Chromatogr. A 2007, 1152, 54–69. [Google Scholar] [CrossRef]
- Khan, W.A.; Arain, M.B.; Yamini, Y.; Shah, N.; Kazi, T.G.; Pedersen-Bjergaard, S.; Tajik, M. Hollow fiber-based liquid phase microextraction followed by analytical instrumental techniques for quantitative analysis of heavy metal ions and pharmaceuticals. J. Pharm. Anal. 2020, 10, 109–122. [Google Scholar] [CrossRef]
- Zgoła-Grześkowiak, A.; Kaczorek, E. Isolation, preconcentration and determination of rhamnolipids in aqueous samples by dispersive liquid-liquid microextraction and liquid chromatography with tandem mass spectrometry. Talanta 2011, 83, 744–750. [Google Scholar] [CrossRef]
- Errekatxo, A.; Prieto, A.; Zuloaga, O.; Usobiaga, A.; Etxebarria, N.; Fernández, L.A. Simultaneous extraction of several persistent organic pollutants in sediment using focused ultrasonic solid-liquid extraction. Anal. Bioanal. Chem. 2008, 392, 1471–1478. [Google Scholar] [CrossRef]
- Godoy, P.; Reina, R.; Calderón, A.; Wittich, R.M.; García-Romera, I.; Aranda, E. Exploring the potential of fungi isolated from PAH-polluted soil. as a source of xenobiotics-degrading fungi. Environ. Sci. Pollut. Res. 2016, 23, 20985–20996. [Google Scholar] [CrossRef]
- Ijoma, G.N.; Tekere, M. Potential microbial applications of co-cultures involving ligninolytic fungi in the Bioremediation of recalcitrant xenobiotic compounds. Int. J. Environ. Sci. Technol. 2017, 14, 1787–1806. [Google Scholar] [CrossRef]
- Arora, P.K. Bacilli-Mediated Degradation of Xenobiotic Compounds and Heavy Metals. Front. Bioeng. Biotechnol. 2020, 8, 570307. [Google Scholar] [CrossRef]
- Gangola, S.; Joshi, S.; Kumar, S.; Pandey, S.C. Comparative analysis of fungal and bacterial enzymes in biodegradation of xenobiotic compounds. In Smart Bioremediation Technologies; Academic Press: Cambridge, MA, USA, 2016; pp. 169–189. [Google Scholar]
- Abbasian, F.; Palanisami, T.; Megharaj, M.; Naidu, R.; Lockington, R.; Ramadass, K. Microbial diversity and hydrocarbon degrading gene capacity of a crude oil field soil as determined by metagenomics analysis. Biotechnol. Prog. 2016, 32, 638–648. [Google Scholar] [CrossRef]
- Okolafor, F.; Ekhaise, F.O. Microbial Enzyme Remediation of Poly-Aromatic Hydrocarbon (PAH’s): A review. J. Int. Environ. Appl. Sci. 2022, 17, 10–21. [Google Scholar]
- Janssen, D.B.; Dinkla, I.J.T.; Poelarends, G.J.; Terpstra, P. Bacterial degradation of xenobiotic compounds: Evolution and distribution of novel enzyme activities. Environ. Microbiol. 2005, 7, 1868–1882. [Google Scholar] [CrossRef]
- Kulig, J.K.; Spandolf, C.; Hyde, R.; Ruzzini, A.C.; Eltis, L.D.; Grönberg, G.; Grogan, G. A P450 fusion library of heme domains from Rhodococcusjostii RHA1 and its evaluation for the biotransformation of drug molecules. Bioorg. Med. Chem. 2015, 23, 5603–5609. [Google Scholar] [CrossRef]
- Torres-Farradá, G.; Manzano Leon, A.M.; Rineau, F.; Ledo Alonso, L.L.; Sánchez-López, M.I.; Thijs, S.; Vangronsveld, J. Diversity of ligninolytic enzymes and their genes in strains of the genus Ganoderma: Applicable for biodegradation of xenobiotic compounds? Front. Microbiol. 2017, 8, 898. [Google Scholar] [CrossRef]
- Budeli, P.; Ekwanzala, M.D.; Unuofin, J.O.; Momba, M.N.B. Endocrine disruptive estrogens in wastewater: Revisiting bacterial degradation and zymoremediation. Environ. Tech. Innov. 2021, 21, 101248. [Google Scholar] [CrossRef]
- Misal, S.A.; Gawai, K.R. Azoreductase: A key player of xenobiotic Metabolism. Bioresour. Bioprocess. 2018, 5, 17. [Google Scholar] [CrossRef]
- Xu, M.; Liu, D.; Sun, P.; Li, Y.; Wu, M.; Liu, W.; Guo, L. Degradation of 2, 4, 6-Trinitrotoluene (TNT): Involvement of Protocatechuate 3, 4-Dioxygenase (P34O) in Buttiauxella sp. S19-1. Toxics 2021, 9, 231. [Google Scholar] [CrossRef]
- Mohapatra, B.; Phale, P.S. Microbial degradation of naphthalene and substituted naphthalenes: Metabolic diversity and genomic insight for Bioremediation. Front. Bioeng. Biotechnol. 2021, 9, 602445. [Google Scholar] [CrossRef]
- Dhiman, N.; Jasrotia, T.; Sharma, P.; Negi, S.; Chaudhary, S.; Kumar, R.; Mahnashi, M.H.; Umar, A.; Kumar, R. Immobilization interaction between xenobiotic and Bjerkanderaadusta for the biodegradation of atrazine. Chemosphere 2020, 257, 127060. [Google Scholar] [CrossRef]
- Esparza-Naranjo, S.B.; da Silva, G.F.; Duque-Castaño, D.C.; Araujo, W.L.; Peres, C.K.; Boroski, M.; Bonugli- Santos, R.C. Potential for the Biodegradation of Atrazine Using Leaf Litter Fungi from a Subtropical Protection Area. Curr. Microbiol. 2021, 78, 358–368. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Alkooranee, J.T.; Zhang, X.; Ma, F. Fungal Degradation of Polycyclic Aromatic Hydrocarbons. Int. J. Pure Appl. Biosci. 2018, 6, 8–24. [Google Scholar] [CrossRef]
- AlMatar, M.; Makky, E.A. Cladosporium cladosporioides from the perspectives of medical and biotechnological approaches. 3 Biotech 2016, 6, 4. [Google Scholar] [CrossRef]
- Kantharaj, P.; Boobalan, B.; Sooriamuthu, S.; Mani, R. Lignocellulose Degrading Enzymes from Fungi and Their Industrial Applications. Int. J. Curr. Res. Rev. 2017, 9, 1–12. [Google Scholar]
- Bilal, M.; Bagheri, A.R.; Bhatt, P.; Chen, S. Environmental occurrence, toxicity concerns, and remediation of recalcitrant nitroaromatic compounds. J. Environ. Manag. 2021, 291, 112685. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, B.; Rajesh, B.; Janardhan, A.; Kumar, A.P.; Narasimha, G. Fungal Laccases and Their Applications in Bioremediation. Enzyme Res. 2014, 2014, 163242. [Google Scholar] [CrossRef]
- González-Abradelo, D.; Pérez-Llano, Y.; Peidro-Guzmán, H.; del Rayo Sánchez-Carbente, M.; Folch-Mallol, J.L.; Aranda, E.; Vaidyanathan, V.K.; Cabana, H.; Gunde-Cimerman, N.; Batista-García, R.A. First demonstration that the ascomycetous halophilic fungi Aspergillus sydowii and Aspergillus destruens are useful in xenobiotic mycoremediation under high salinity conditions. Bioresour. Technol. 2019, 279, 287–296. [Google Scholar] [CrossRef]
- Peidro-Guzmán, H.; Pérez-Llano, Y.; González-Abradelo, D.; Fernández-López, M.G.; Dávila-Ramos, S. Transcriptomic analysis of polyaromatic hydrocarbon degradation by the halophilic fungus Aspergillus sydowii at hypersaline conditions. Environ. Microbiol. 2021, 23, 3435–3459. [Google Scholar] [CrossRef]
- Patrick, F.D.; Samuel, K.S. Biologically Pure Culture of Yeast Strain Used for the Microbial Detoxification of Xenobiotics. United States Patent US4968620A, 6 November 1990. [Google Scholar]
- Michael, S.P.; Juan Matías, S.S.; Francisca Acevedo, C. PCB-Degrading Recombinant Bacterium, Product for the Bioremediation and Method of Bioremediation. United States Patent US 7,989,194B2, 2011. Available online: https://patentimages.storage.googleapis.com/ae/90/40/4afda38f65437b/US7989194.pdf (accessed on 1 May 2022).
- Pfeiffer, E.; Kommer, A.; Dempe, J.S.; Hildebrand, A.A.; Metzler, M. Absorption and Metabolism of the mycotoxin zearalenone and the growth promotor zeranol in Caco-2 cells in vitro. Mol. Nutr. Food Res. 2011, 55, 560–567. [Google Scholar] [CrossRef]
- Popowska, M.; Boszczyk-Maleszak, H.; Komorowska, I. Composition Of Bacterial Strains, Bioremediation Mixture and Use of This Composition for The Removal of Contaminants from The Soil and A Method for Purifying of The Soil Contaminants. European Patent Specification EP 2 788 512 B1, 2015. Available online: https://patentimages.storage.googleapis.com/5a/82/f0/6108eb476fe385/EP2788512B1.pdf (accessed on 25 April 2022).
- Knowies, C.J.; Wyatt, J.M. Microbial Degradation of Waste. European Patent Application 0 274 856 A1, 20 July 1988. [Google Scholar]
- Karl, S.; Peter, R. Process for Microbial Removal of Xenobiotic Dyes. German Patent DD290004A5, 16 May 1991. [Google Scholar]
- Dowd, P.F.; Shen, S.K. The contribution of symbiotic yeast to toxin resistance of the cigarette beetle (Lasiodermaserricorne). Entomol. Exp. Appl. 1990, 56, 241–248. [Google Scholar] [CrossRef]
- Daugulis, A.J.; Collins, L.D. Two-Phase Partitioning Bioreactor For The Degradation Of A Xenobiotech. Canadian Intellectual Property Office CA2216327A1, 19 March 1999. [Google Scholar]
- Michael, R.H.; Kenneth, W.; Lynda, M.C. Bioremediation of Xenobiotics Including Methyl Tert-Butylether. United States Patent US 6,194,197B1, 27 February 2001. [Google Scholar]
- Razavi-Shirazi, F. Biological Permeable Barrier to Treat Contaminated Groundwater Using Immobilized Cells. Australian Patent Office WO 01/32566 Al, 10 May 2001. [Google Scholar]
- Mihara, C.; Yano, T.; Kozaki, S.; Imamura, T. Novel Microbial Strain, Method for Biodegrading Organic Compounds and Method for Environmental Remediation. European Patent Specification EP 0 822 253 B1, 7 September 2003. [Google Scholar]
- Enamul, H. Microbial Decomposition of Xenobiotics, Useful Especially for Degrading Isoproturon, Uses Mixture of Fungi with oxygenase and Glutathione-S-Transferase Activities. German Patent DE10125365A1, 5 June 2003. [Google Scholar]
- Schmidt, J.E.; Trably, E.; Batstone, J.D.; Angelidaki, I.; Christensen, N. Anaerobic Microbial Degradation of Phthalic acid Esters. World Intellectual Property Organization WO2006136173A2, 28 December 2006. [Google Scholar]
- Soljan, V. Method for Simultaneous Biological Removal of Nitrogen Compounds and Xenobiotics of Wastewaters. World Intellectual Property Organization WO2013166611, 14 November 2013. [Google Scholar]
- Bazhanuofu, D.; Li, H.; Li, C.; Li, J.; Yang, H. Atrazine Degrading Bacterium and Application Thereof. Chinese Patent CN104762227A, 8 July 2015. [Google Scholar]
- Maddaloni, M.; Pascual, D.W.; Ellis, J. Bioremediation of Xenobiotics in the Honey Bee Hive. United States Patent US2021378263A1, 9 December 2021. [Google Scholar]
- Tramontano, M.; Klünemann, M.; Pruteanu, M.; Maier, L.; Kuhn, M.; Andrejev, S.; Kim, Y.; Bork, P.; Typas, A.; Patil, K.R.; et al. In-Vitro Model of the Human Gut Microbiome and Uses Thereof in the Analysis of the Impact of Xenobiotics. United States Patent Application Publication US20200370005, 26 November 2020. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).