Maternal High-Fat Diet Programs White and Brown Adipose Tissues In Vivo in Mice, with Different Metabolic and Microbiota Patterns in Obesity-Susceptible or Obesity-Resistant Offspring
Abstract
:1. Introduction
2. Results
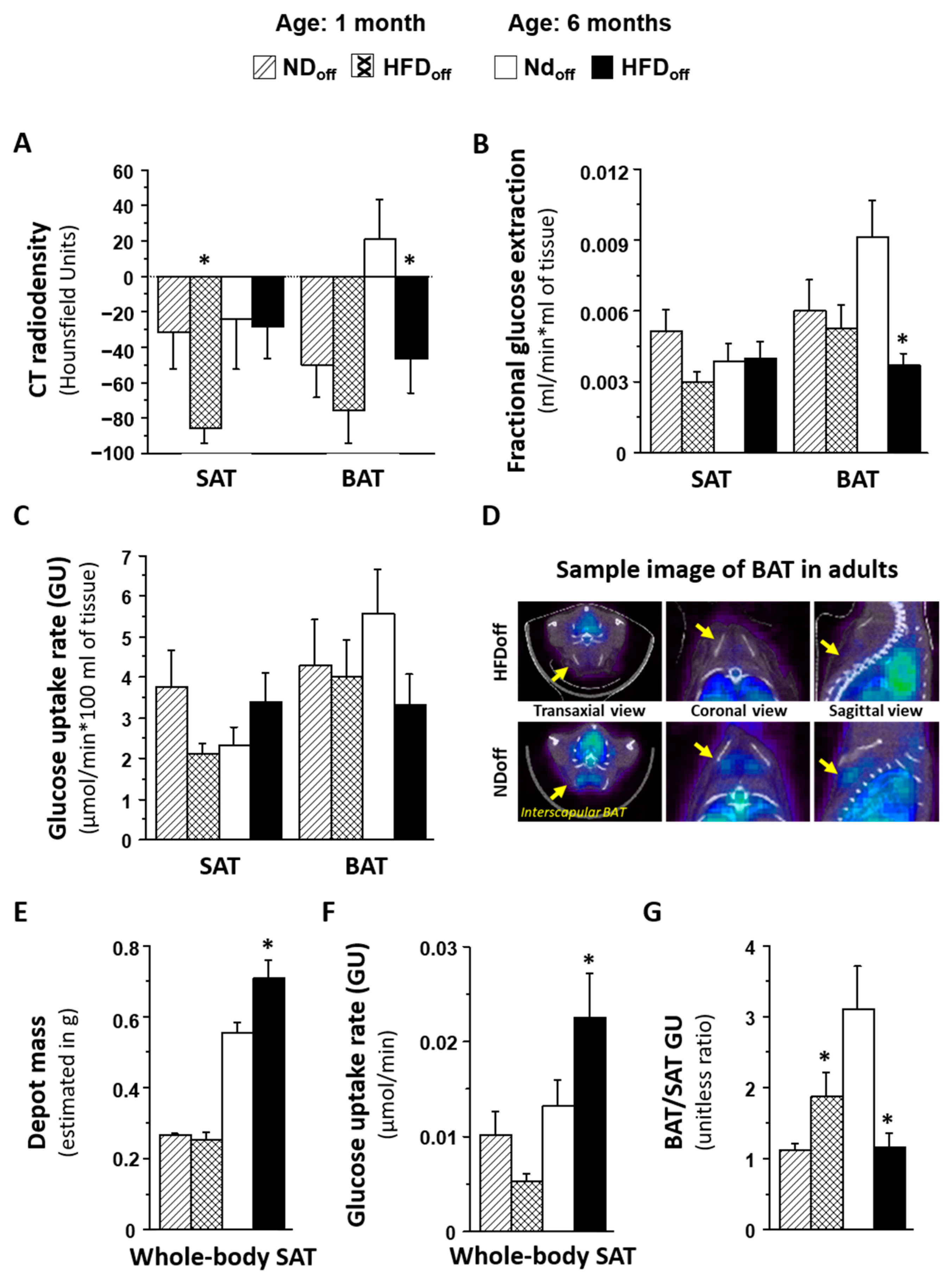
2.1. Metabolic Characteristics
| 1 Month Old | 6 Months Old | |||
|---|---|---|---|---|
| Female | NDoff | HFDoff | NDoff | HFDoff |
| Body weight (g) | 17 ± 1 | 13 ± 1 ^ | 26 ± 2 | 27 ± 0 |
| SAT mass (g) | 0.26 ± 0.02 | 0.19 ± 0.01 ^ | 0.51 ± 0.04 | 0.54 ± 0.00 |
| F-Glycemia (mmol/L) | 6.3 ± 0.2 | 8.5 ± 1.2 * | 5.3 ± 0.3 | 10.5 ± 0.3 * |
| Scan glycemia (mmol/L) | 4.6 ± 0.6 | 6.7 ± 1.2 | 5.6 ± 0.9 | 14.5 ± 3.2 * |
| Triglycerides (mmol/L) | 0.794 ± 0.004 | 0.805 ± 0.015 | 0.790 ± 0.000 | 1.123 ± 0.333 |
| SAT CT (HU) | −34 ± 48 | −94 ± 8 | −2.2 ± 31 | −19 ± 3 |
| BAT CT (HU) | −56 ± 22 | −78 ± 31 | 61 ± 20 | −22 ± 43 ^ |
| SAT GU (µmol/min*100 g) | 1.9 ± 0.7 | 1.7 ± 0.3 | 2.1 ± 0.3 | 5.1 ± 2.0 * |
| Whole SAT GU (µmol/min) | 0.005 ± 0.002 | 0.003 ± 0.002 | 0.011 ± 0.002 | 0.027 ± 0.011 * |
| BAT GU (µmol/min*100 g) | 2.0 ± 1.0 | 3.5 ± 0.8 | 4.9 ± 1.0 | 6.4 ± 2.2 |
| BAT/SAT GU | 1.0 ± 1.8 | 2.1 ± 0.4 | 2.6 ± 0.3 | 1.3 ± 0.1 * |
| Male | NDoff | HFDoff | NDoff | HFDoff |
| Body weight (g) | 18 ± 0 | 19 ± 2 | 31 ± 1 | 38 ± 2 * |
| SAT mass (g) | 0.27 ± 0.01 | 0.29 ± 0.02 | 0.63 ± 0.02 | 0.76 ± 0.05 * |
| F-glycemia (mmol(L) | 8.3 ± 0.6 | 7.8 ± 0.4 | 6.4 ± 0.7 | 7.2 ± 0.8 |
| Scan glycemia (mmol/L) | 7.7 ± 0.9 | 8.1 ± 0.7 | 7.2 ± 1.0 | 7.7 ± 1.1 |
| Triglycerides (mmol/L) | 0.819 ± 0.028 | 0.931 ± 0.141 | 0.790 ± 0.000 | 1.155 ± 0.101 * |
| SAT CT (HU) | −30 ± 21 | −81 ± 13 | −51 ± 53 | −31 ± 23 |
| BAT CT (HU) | −46 ± 28 | −74 ± 26 | −39 ± 27 | −53 ± 23 |
| SAT GU (µmol/min*100 g) | 5.0 ± 1.3 | 2.3 ± 0.4 ^ | 2.5 ± 1.0 | 2.9 ± 0.8 |
| Whole SAT GU (µmol/min) | 0.014 ± 0.004 | 0.007 ± 0.001 | 0.015 ± 0.006 | 0.021 ± 0.005 |
| BAT GU (µmol/min*100 g) | 5.8 ± 1.5 | 4.3 ± 1.4 | 6.6 ± 2.4 | 2.4 ± 0.5 * |
| BAT/SAT GU | 1.2 ± 0.1 | 1.8 ± 0.5 | 3.8 ± 1.3 | 1.1 ± 0.3 * |
2.2. Relationship with Microbiota and Metabolic Pathways
3. Discussion

4. Materials and Methods
4.1. Animal Model and Study Design
4.2. PET-CT Imaging
4.3. Biochemical Analyses
4.4. Gut Bacteria 16SrRNA Gene Sequencing
4.5. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vidal-Puig, A. Adipose tissue expandability, lipotoxicity and the metabolic syndrome. Endocrinol. Nutr. 2013, 60 (Suppl. S1), 39–43. [Google Scholar] [CrossRef]
- Virtanen, K.A.; Iozzo, P.; Hällsten, K.; Huupponen, R.; Parkkola, R.; Janatuinen, T.; Lönnqvist, F.; Viljanen, T.; Rönnemaa, T.; Lönnroth, P.; et al. Increased fat mass compensates for insulin resistance in abdominal obesity and type 2 diabetes: A positron-emitting tomography study. Diabetes 2005, 54, 2720–2726. [Google Scholar] [CrossRef]
- Iozzo, P. Viewpoints on the way to the consensus session: Where does insulin resistance start? The adipose tissue. Diabetes Care 2009, 32 (Suppl. 2), S168–S173. [Google Scholar] [CrossRef]
- Dadson, P.; Hannukainen, J.C.; Din, M.U.; Lahesmaa, M.; Kalliokoski, K.K.; Iozzo, P.; Pihlajamäki, J.; Karlsson, H.K.; Parkkola, R.; Salminen, P.; et al. Brown adipose tissue lipid metabolism in morbid obesity: Effect of bariatric surgery-induced weight loss. Diabetes Obes. Metab. 2018, 20, 1280–1288. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.; Mao, X.; Du, M. Maternal Obesity Impairs Fetal Mitochondriogenesis and Brown Adipose Tissue Development Partially via Upregulation of MiR-204-5p. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2706–2715. [Google Scholar] [CrossRef]
- Raipuria, M.; Bahari, H.; Morris, M. Effects of Maternal Diet and Exercise during Pregnancy on Glucose Metabolism in Skeletal Muscle and Fat of Weanling Rats. PLoS ONE 2015, 10, e0120980. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Goran, M.; Kaur, H.; Nollen, N.; Ahluwalia, J. Developmental Trajectories of Overweight During Childhood: Role of Early Life Factors. Obesity 2007, 15, 760–771. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.G.; Sandboge, S.; Salonen, M.K.; Kajantie, E.; Osmond, C. Long-term consequences of maternal overweight in pregnancy on offspring later health: Findings from the Helsinki Birth Cohort Study. Ann. Med. 2014, 46, 434–438. [Google Scholar] [CrossRef]
- Orsso, C.E.; Colin-Ramirez, E.; Field, C.J.; Madsen, K.L.; Prado, C.M.; Haqq, A.M. Adipose Tissue Development and Expansion from the Womb to Adolescence: An Overview. Nutrients 2020, 12, 2735. [Google Scholar] [CrossRef]
- Chu, D.M.; Antony, K.M.; Ma, J. The Early Infant Gut Microbiome Varies in Association with a Maternal High-Fat Diet. Genome Med. 2016, 8, 77. [Google Scholar] [CrossRef] [Green Version]
- Guzzardi, M.A.; Ederveen, T.H.A.; Rizzo, F.; Weisz, A.; Collado, M.C.; Muratori, F.; Gross, G.; Alkema, W.; Iozzo, P. Maternal pre-pregnancy overweight and neonatal gut bacterial colonization are associated with cognitive development and gut microbiota composition in pre-school-age offspring. Brain Behav. Immun. 2022, 100, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Cassidy-Bushrow, A.E.; Burmeister, C.; Havstad, S.; Levin, A.M.; Lynch, S.V.; Ownby, D.R.; Rundle, A.G.; Woodcroft, K.J.; Zoratti, E.M.; Johnson, C.C.; et al. Prenatal Antimicrobial Use and Early-Childhood Body Mass Index. Int. J. Obes. 2018, 42, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.T.; Whyatt, R.; Hoepner, L.; Oberfield, S.; Dominguez-Bello, M.G.; Widen, E.M.; Hassoun, A.; Perera, F.; Rundle, A. Prenatal Exposure to Antibiotics, Cesarean Section and Risk of Childhood Obesity. Int. J. Obes. 2015, 39, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Yamanishi, S.; Cox, L.; Methe, B.A.; Zavadil, J.; Li, K.; Gao, Z.; Mahana, D.; Raju, K.; Teitler, I.; et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012, 488, 621–626. [Google Scholar] [CrossRef]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Yang, Q.; Zhang, L.; Maricelli, J.W.; Rodgers, B.D.; Zhu, M.J.; Du, M. Maternal high-fat diet during lactation impairs thermogenic function of brown adipose tissue in offspring mice. Sci. Rep. 2016, 6, 34345. [Google Scholar] [CrossRef]
- Iozzo, P.; Sanguinetti, E. Early Dietary Patterns and Microbiota Development: Still a Way to Go from Descriptive Interactions to Health-Relevant Solutions. Front. Nutr. 2018, 5, 5. [Google Scholar] [CrossRef]
- Yu, H.; Dilbaz, S.; Coßmann, J.; Hoang, A.C.; Diedrich, V.; Herwig, A.; Harauma, A.; Hoshi, Y.; Moriguchi, T.; Landgraf, K.; et al. Breast Milk Alkylglycerols Sustain Beige Adipocytes through Adipose Tissue Macrophages. J. Clin. Investig. 2019, 129, 2485–2499. [Google Scholar] [CrossRef]
- van den Elsen, L.W.J.; Verhasselt, V. Human Milk Drives the Intimate Interplay Between Gut Immunity and Adipose Tissue for Healthy Growth. Front. Immunol. 2021, 12, 645415. [Google Scholar] [CrossRef]
- Savva, C.; Helguero, L.A.; González-Granillo, M.; Melo, T.; Couto, D.; Buyandelger, B.; Gustafsson, S.; Liu, J.; Domingues, M.R.; Li, X.; et al. Maternal high-fat diet programs white and brown adipose tissue lipidome and transcriptome in offspring in a sex- and tissue-dependent manner in mice. Int. J. Obes. 2022, 46, 831–842. [Google Scholar] [CrossRef]
- Baba, S.; Jacene, H.A.; Engles, J.M.; Honda, H.; Wahl, R.L. CT Hounsfield units of brown adipose tissue increase with activation: Preclinical and clinical studies. J. Nucl. Med. 2010, 51, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Din, M.U.; Raiko, J.; Saari, T.; Saunavaara, V.; Kudomi, N.; Solin, O.; Parkkola, R.; Nuutila, P.; Virtanen, K.A. Human Brown Fat Radiodensity Indicates Underlying Tissue Composition and Systemic Metabolic Health. J. Clin. Endocrinol. Metab. 2017, 102, 2258–2267. [Google Scholar] [CrossRef] [PubMed]
- Torriani, M.; Oliveira, A.L.; Azevedo, D.C.; Bredella, M.A.; Yu, E.W. Effects of roux-en-Y gastric bypass surgery on visceral and subcutaneous fat density by computed tomography. Obes. Surg. 2015, 25, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, K.J.; Pedley, A.; Massaro, J.M.; Therkelsen, K.E.; Murabito, J.M.; Hoffmann, U.; Fox, C.S. Visceral and subcutaneous fat quality and cardiometabolic risk. JACC Cardiovasc. Imag. 2013, 6, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.H.; Chung, S.A.; Nayak, K.S.; Jackson, H.A.; Gilsanz, V. Differential computed tomographic attenuation of metabolically active and inactive adipose tissues: Preliminary findings. J. Comput. Assist. Tomogr. 2011, 35, 65e71. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, P. Metabolic imaging in obesity: Underlying mechanisms and consequences in the whole body. Ann. NY Acad. Sci. 2015, 1353, 21–40. [Google Scholar] [CrossRef]
- Summerfield, M.; Zhou, Y.; Zhou, T.; Wu, C.; Alpini, G.; Zhang, K.K.; Xie, L. A long-term maternal diet transition from high-fat diet to normal fat diet during pre-pregnancy avoids adipose tissue inflammation in next generation. PLoS ONE 2018, 13, e0209053. [Google Scholar] [CrossRef]
- Guo, S.; Zhao, H.; Ma, Z.; Zhang, S.; Li, M.; Zheng, Z.; Ren, X.; Ho, C.T.; Bai, N. Anti-Obesity and Gut Microbiota Modulation Effect of Secoiridoid-Enriched Extract from Fraxinus mandshurica Seeds on High-Fat Diet-Fed Mice. Molecules 2020, 25, 4001. [Google Scholar] [CrossRef]
- Karvonen, A.M.; Sordillo, J.E.; Gold, D.R.; Bacharier, L.B.; O’Connor, G.T.; Zeiger, R.S.; Beigelman, A.; Weiss, S.T.; Litonjua, A.A. Gut microbiota and overweight in 3-year old children. Int. J. Obes. 2019, 43, 713–723. [Google Scholar] [CrossRef]
- Companys, J.; Gosalbes, M.J.; Pla-Pagà, L.; Calderón-Pérez, L.; Llauradó, E.; Pedret, A.; Valls, R.M.; Jiménez-Hernández, N.; Sandoval-Ramirez, B.A.; Del Bas, J.M.; et al. Gut Microbiota Profile and Its Association with Clinical Variables and Dietary Intake in Overweight/Obese and Lean Subjects: A Cross-Sectional Study. Nutrients 2021, 13, 2032. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469.e5. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Zhang, Y.; Li, M.; Guo, C.; Mi, S. RBUD: A New Functional Potential Analysis Approach for Whole Microbial Genome Shotgun Sequencing. Microorganisms 2020, 8, 1563. [Google Scholar] [CrossRef] [PubMed]
- Valeri, F.; Endres, K. How biological sex of the host shapes its gut microbiota. Front. Neuroendocrinol. 2021, 61, 100912. [Google Scholar] [CrossRef] [PubMed]
- Guzzardi, M.A.; La Rosa, F.; Campani, D.; Cacciato Insilla, A.; De Sena, V.; Panetta, D.; Brunetto, M.R.; Bonino, F.; Collado, M.C.; Iozzo, P. Maturation of the Visceral (Gut-Adipose-Liver) Network in Response to the Weaning Reaction versus Adult Age and Impact of Maternal High-Fat Diet. Nutrients 2021, 13, 3438. [Google Scholar] [CrossRef] [PubMed]
- Guzzardi, M.A.; Guiducci, L.; Campani, D.; La Rosa, F.; Cacciato Insilla, F.; Bartoli, A.; Cabiati, M.; De Sena, V.; Del Ry, S.; Burchielli, S.; et al. Leptin resistance before and after obesity: Evidence that tissue glucose uptake underlies adipocyte enlargement and liver steatosis/steatohepatitis in Zucker rats from early-life stages. Int. J. Obes. 2022, 46, 50–58. [Google Scholar] [CrossRef]
- Sanguinetti, E.; Guzzardi, M.A.; Tripodi, M.; Panetta, D.; Selma-Royo, M.; Zega, A.; Telleschi, M.; Collado, M.C.; Iozzo, P. Microbiota signatures relating to reduced memory and exploratory behaviour in the offspring of overweight mothers in a murine model. Sci. Rep. 2019, 9, 12609. [Google Scholar] [CrossRef]
- Tekus, E.; Miko, A.; Furedi, N.; Rostas, I.; Tenk, J.; Kiss, T.; Szitter, I.; Balasko, M.; Helyes, Z.; Wilhelm, M.; et al. Body fat of rats of different age groups and nutritional states: Assessment by micro-CT and skinfold thickness. J. Appl. Physiol. (1985) 2018, 124, 268–275. [Google Scholar] [CrossRef] [Green Version]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar]

| Parameter | Level | Bacteria Taxa | R | p | p FDR-Corr | Mean Abund |
|---|---|---|---|---|---|---|
| CAECUM: Age 1 month | ||||||
| SAT GE | Genus | Dorea | −0.7 | 0.0006 | 0.023 | 1.01 |
| CAECUM: Age 6 months | ||||||
| BAT GE | Order | Bacillales | −0.67 | 0.0024 | 0.038 | 2.97 |
| Family | Enterococcaceae | −0.78 | 0.0001 | 0.005 | 1.55 | |
| Family | Bacillaceae | −0.72 | 0.0008 | 0.013 | 1.53 | |
| Family | Streptococcaceae | −0.63 | 0.0054 | 0.042 | 0.03 | |
| Family | Peptostreptococcaceae | 0.62 | 0.0061 | 0.042 | 2.02 | |
| Family | Aerococcaceae | −0.62 | 0.0066 | 0.042 | 0.42 | |
| Genus | Unclassified.Enterococcaceae | −0.79 | 0.0001 | 0.004 | 1.54 | |
| Genus | Bacillus | −0.72 | 0.0008 | 0.020 | 1.53 | |
| BAT GU | Family | Enterococcaceae | −0.77 | 0.0002 | 0.006 | 1.55 |
| Family | Streptococcaceae | −0.66 | 0.0027 | 0.041 | 0.03 | |
| Family | Staphylococcaceae | −0.64 | 0.0039 | 0.041 | 0.98 | |
| Genus | Unclassified.Enterococcaceae | −0.79 | 0.0001 | 0.004 | 1.54 | |
| BAT/SAT GU | Phylum | Proteobacteria | −0.67 | 0.0034 | 0.024 | 3.42 |
| Family | Rikenellaceae | −0.73 | 0.0009 | 0.014 | 0.20 | |
| Family | Unclassified.Clostridiales | 0.71 | 0.0014 | 0.015 | 16.01 | |
| Genus | Bacillus | −0.79 | 0.0002 | 0.009 | 1.53 | |
| Genus | Unclassified.Rikenellaceae | −0.73 | 0.0009 | 0.021 | 0.20 | |
| Genus | Unclassified.Clostridiales | 0.71 | 0.0014 | 0.022 | 16.01 | |
| Whole SAT GU | Family | Ruminococcaceae | −0.8 | 0.0001 | 0.004 | 6.67 |
| Family | Peptococcaceae | −0.72 | 0.0011 | 0.013 | 0.38 | |
| Family | Christensenellaceae | −0.72 | 0.0012 | 0.013 | 0.14 | |
| Family | Unclassified.Clostridiales | −0.69 | 0.0021 | 0.017 | 16.01 | |
| Family | Dehalobacteriaceae | −0.64 | 0.0054 | 0.035 | 0.07 | |
| Genus | rc44 | −0.72 | 0.0011 | 0.017 | 0.38 | |
| Genus | Unclassified.Christensenellaceae | −0.72 | 0.0012 | 0.017 | 0.14 | |
| Genus | Unclassified.Ruminococcaceae | −0.71 | 0.0013 | 0.017 | 3.85 | |
| Genus | Coprococcus | −0.71 | 0.0015 | 0.017 | 0.62 | |
| Genus | Anaerotruncus | −0.7 | 0.0018 | 0.017 | 0.02 | |
| Genus | Unclassified.Clostridiales | −0.69 | 0.0021 | 0.017 | 16.01 | |
| Genus | Oscillospira | −0.65 | 0.0044 | 0.030 | 2.01 | |
| Genus | Dehalobacterium | −0.64 | 0.0054 | 0.032 | 0.07 | |
| Genus | Unclassified.Erysipelotrichaceae | −0.61 | 0.0093 | 0.050 | 1.14 | |
| Parameter | Pathway | R | p | p FDR-corr |
|---|---|---|---|---|
| CAECUM: Age 1 month | ||||
| SAT CT | Inositol.phosphate.metabolism | 0.84 | 1.59 × 10−5 | 0.003 |
| Replication.recombination.and.repair.proteins | −0.78 | 1.49 × 10−4 | 0.009 | |
| Tetracycline.biosynthesis | −0.77 | 1.74 × 10−4 | 0.009 | |
| Fatty.acid.biosynthesis | −0.77 | 1.75 × 10−4 | 0.009 | |
| Chromosome | −0.71 | 1.06 × 10−3 | 0.045 | |
| SAT GE | Nitrotoluene.degradation | −0.82 | 3.42 × 10−5 | 0.007 |
| CAECUM: Age 6 months | ||||
| BAT/SAT GU | Bacterial.secretion.system | −0.8 | 0.0001 | 0.024 |
| Whole SAT GU | beta.Lactam.resistance | 0.78 | 0.0002 | 0.023 |
| Flagellar.assembly | −0.77 | 0.0003 | 0.023 | |
| Transcription.related.proteins | 0.77 | 0.0003 | 0.023 | |
| Bacterial.chemotaxis | −0.75 | 0.0005 | 0.026 | |
| Bacterial.motility.proteins | −0.74 | 0.0006 | 0.026 | |
| Chaperones.and.folding.catalysts | 0.74 | 0.0007 | 0.026 | |
| Sulfur.relay.system | 0.71 | 0.0013 | 0.039 | |
| Flavone.and.flavonol.biosynthesis | −0.70 | 0.0018 | 0.048 | |
| Bacterial.secretion.system | 0.69 | 0.0021 | 0.050 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzzardi, M.A.; Collado, M.C.; Panetta, D.; Tripodi, M.; Iozzo, P. Maternal High-Fat Diet Programs White and Brown Adipose Tissues In Vivo in Mice, with Different Metabolic and Microbiota Patterns in Obesity-Susceptible or Obesity-Resistant Offspring. Metabolites 2022, 12, 828. https://doi.org/10.3390/metabo12090828
Guzzardi MA, Collado MC, Panetta D, Tripodi M, Iozzo P. Maternal High-Fat Diet Programs White and Brown Adipose Tissues In Vivo in Mice, with Different Metabolic and Microbiota Patterns in Obesity-Susceptible or Obesity-Resistant Offspring. Metabolites. 2022; 12(9):828. https://doi.org/10.3390/metabo12090828
Chicago/Turabian StyleGuzzardi, Maria Angela, Maria Carmen Collado, Daniele Panetta, Maria Tripodi, and Patricia Iozzo. 2022. "Maternal High-Fat Diet Programs White and Brown Adipose Tissues In Vivo in Mice, with Different Metabolic and Microbiota Patterns in Obesity-Susceptible or Obesity-Resistant Offspring" Metabolites 12, no. 9: 828. https://doi.org/10.3390/metabo12090828
APA StyleGuzzardi, M. A., Collado, M. C., Panetta, D., Tripodi, M., & Iozzo, P. (2022). Maternal High-Fat Diet Programs White and Brown Adipose Tissues In Vivo in Mice, with Different Metabolic and Microbiota Patterns in Obesity-Susceptible or Obesity-Resistant Offspring. Metabolites, 12(9), 828. https://doi.org/10.3390/metabo12090828






