Curcumin Decreases Viability and Inhibits Proliferation of Imatinib-Sensitive and Imatinib-Resistant Chronic Myeloid Leukemia Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cultures
2.2. Cell Viability Assay
2.3. Cell Proliferation Assay
2.4. Molecular Docking Study
2.5. Western Blot Analysis
2.6. Statistical Analysis
3. Results
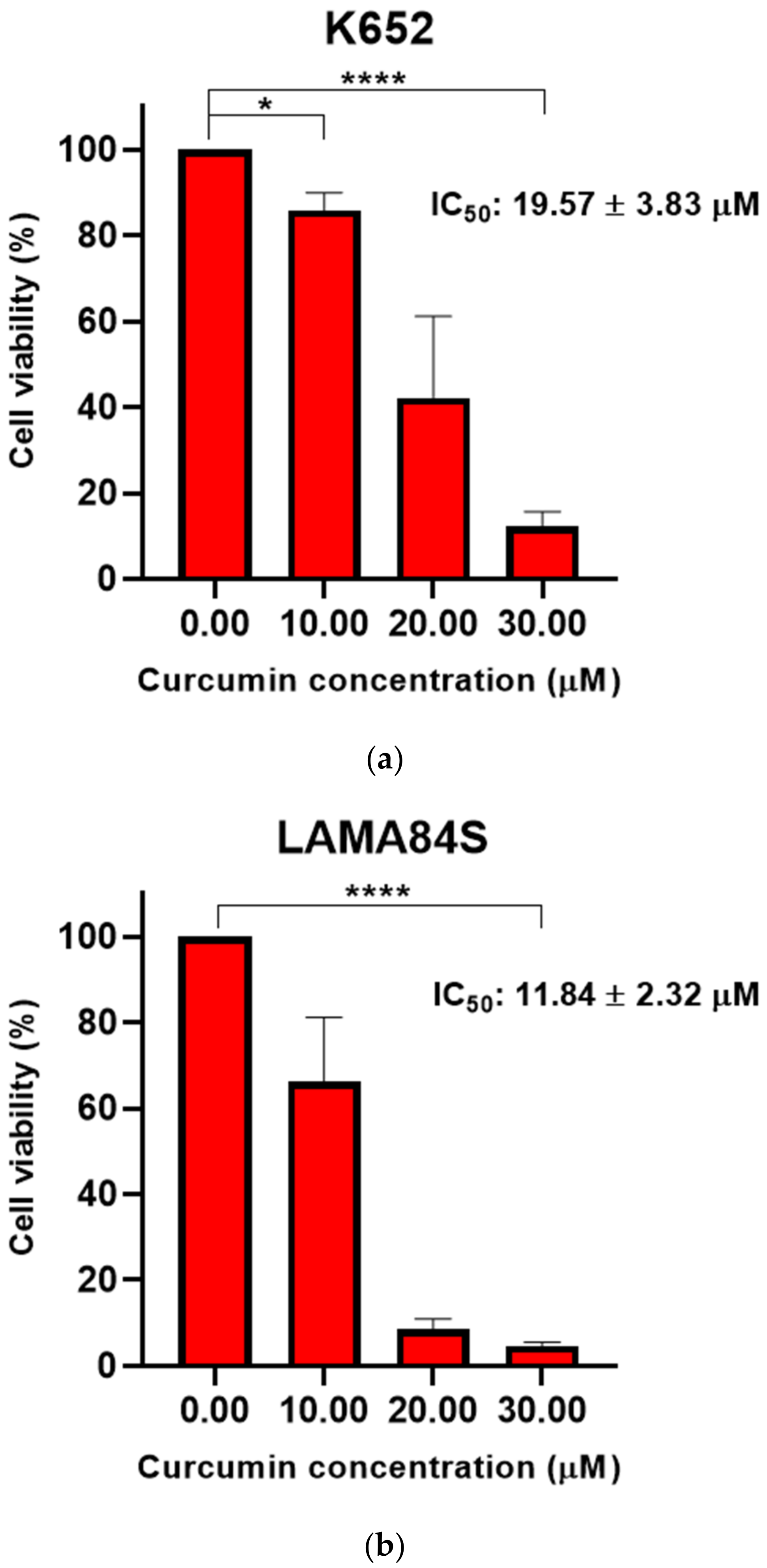
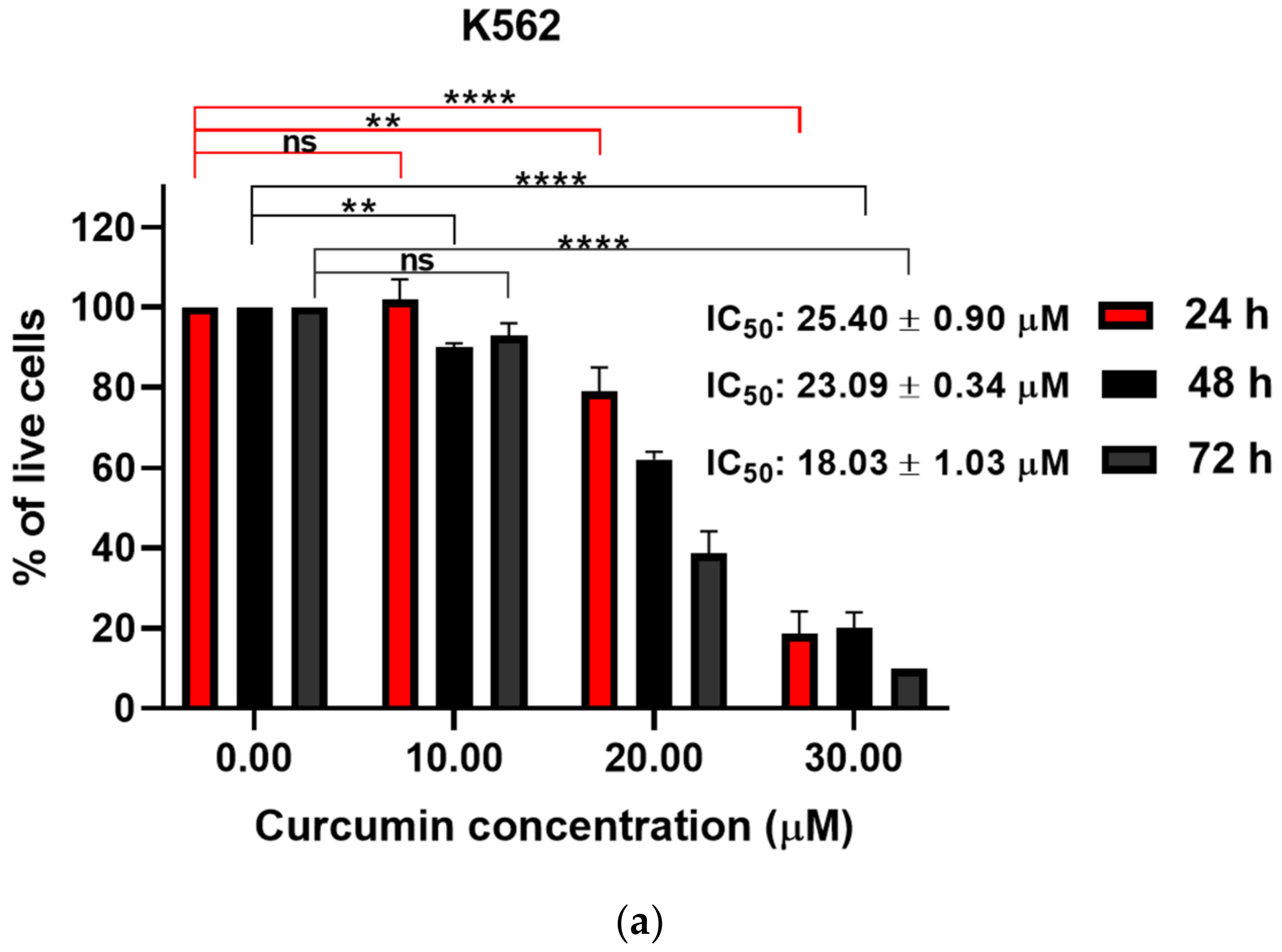
3.1. Curcumin Induced Dose-Dependent Decrease of Cell Viability of Imatinib-Sensitive and Imatinib-Resistant Cell Lines
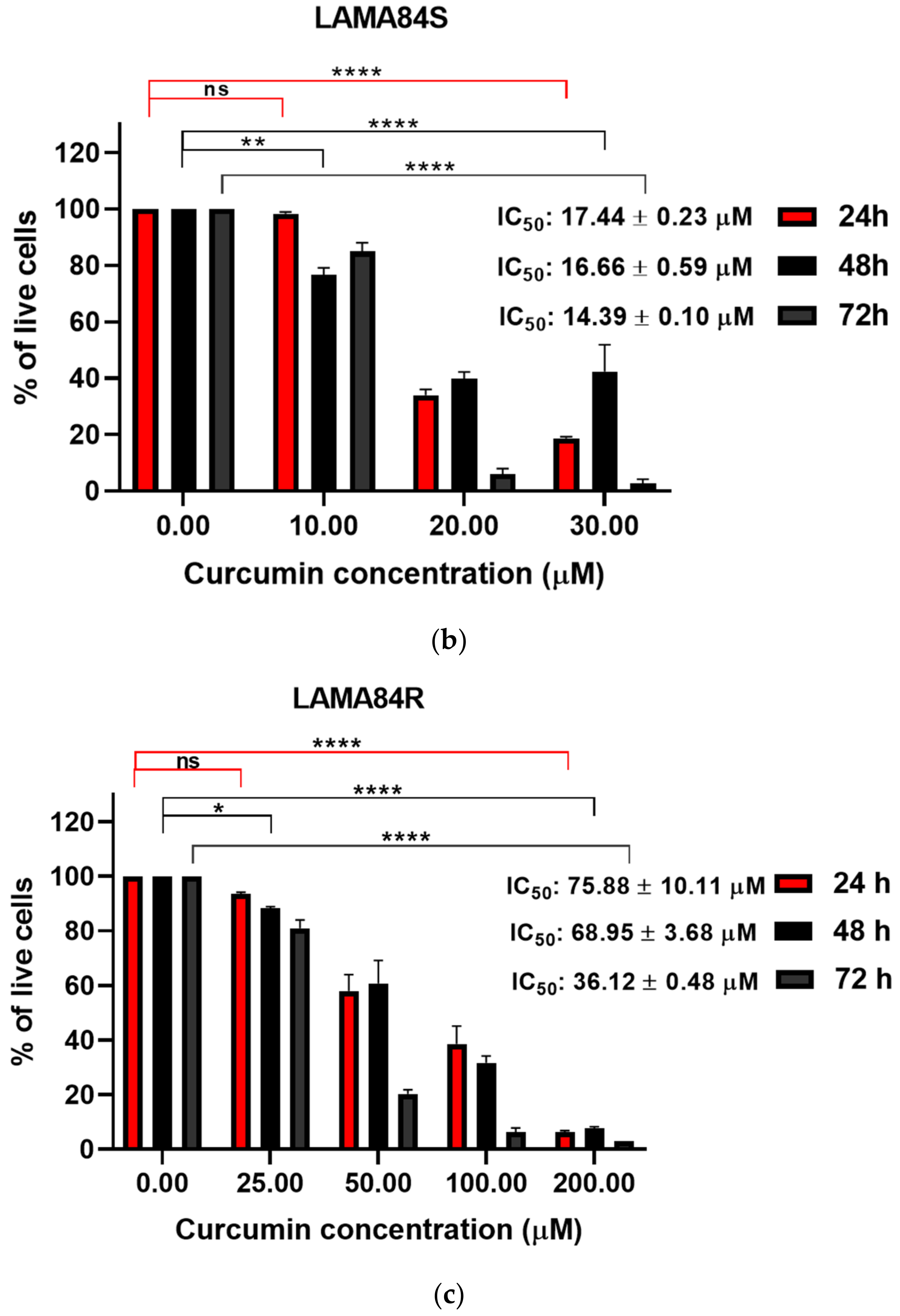
3.2. Curcumin Induced Dose-Dependent Decrease of Cell Proliferation of Imatinib-Sensitive and Imatinib-Resistant Cell Lines
3.3. Molecular Docking Study of Curcumin and Imatinib with Protein Targets
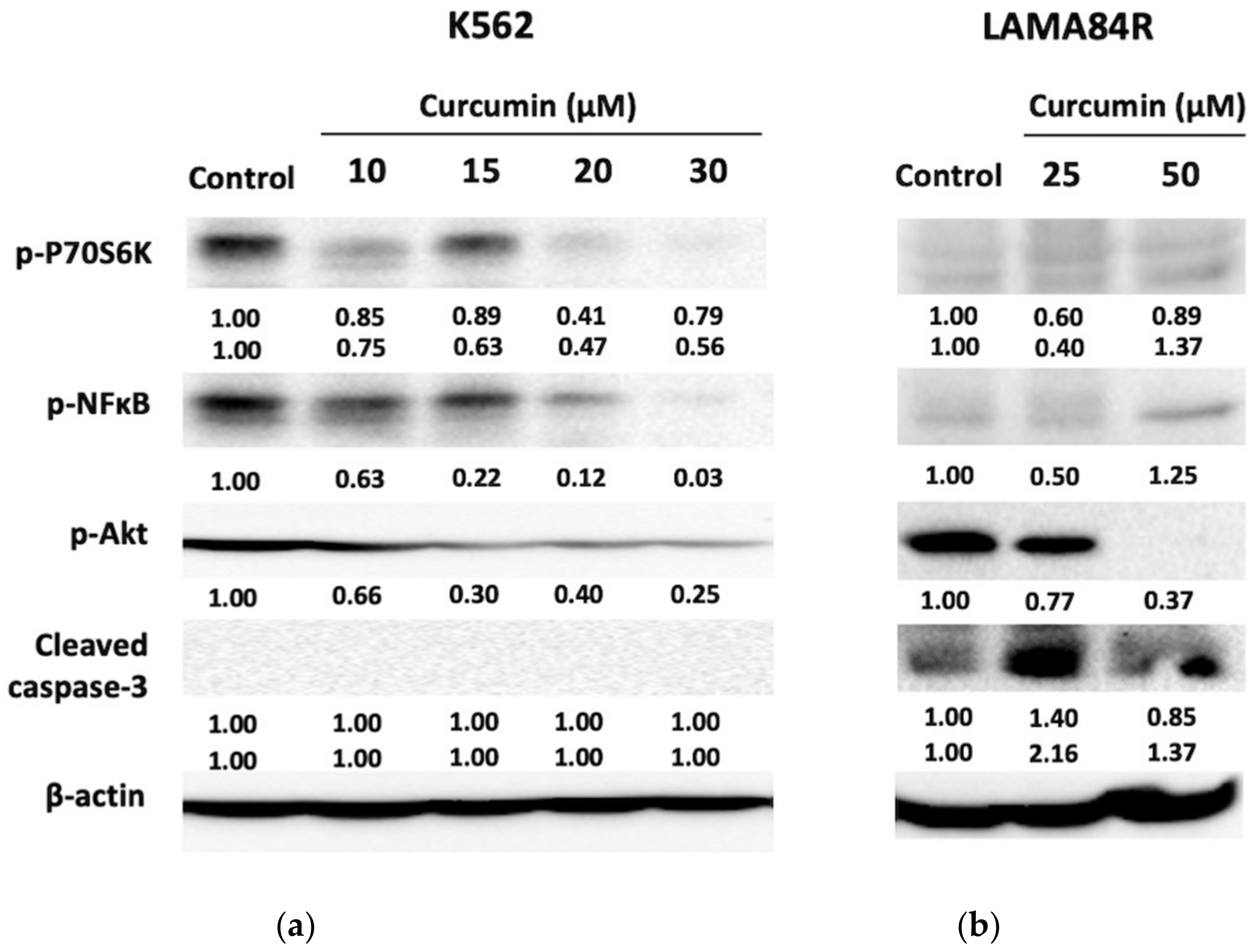
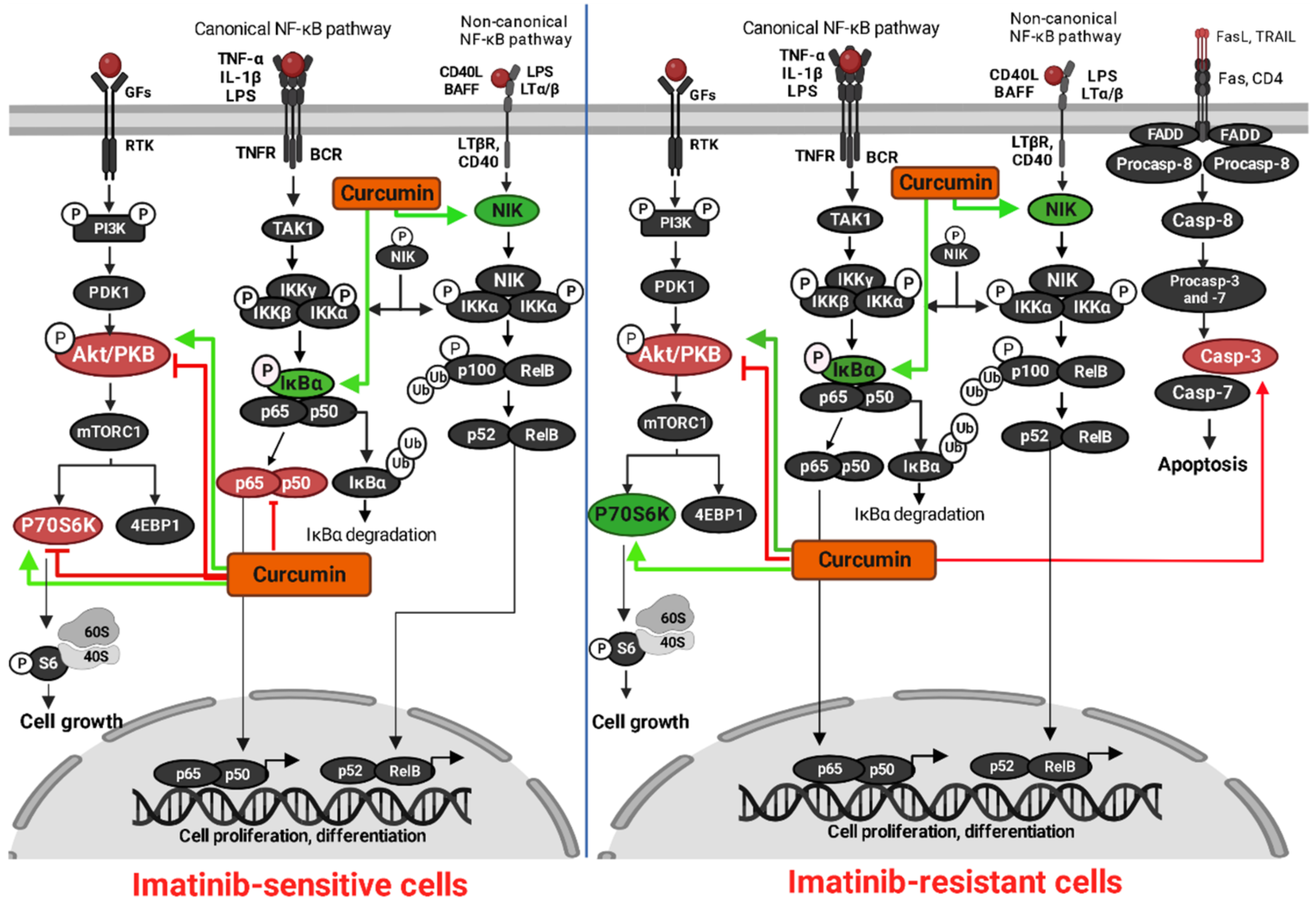
3.4. Analysis of Protein Targets of Curcumin in K562 and LAMA84R Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jabbour, E.; Kantarjian, H. Chronic Myeloid Leukemia: 2018 Update on Diagnosis, Therapy and Monitoring. Am. J. Hematol. 2018, 93, 442–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrigoni, E.; Del Re, M.; Galimberti, S.; Restante, G.; Rofi, E.; Crucitta, S.; Baratè, C.; Petrini, M.; Danesi, R.; Di Paolo, A. Concise Review: Chronic Myeloid Leukemia: Stem Cell Niche and Response to Pharmacologic Treatment: Role of Stem Cells in Chronic Myeloid Leukemia. Stem Cells Transl. Med. 2018, 7, 305–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soverini, S.; Mancini, M.; Bavaro, L.; Cavo, M.; Martinelli, G. Chronic Myeloid Leukemia: The Paradigm of Targeting Oncogenic Tyrosine Kinase Signaling and Counteracting Resistance for Successful Cancer Therapy. Mol. Cancer 2018, 17, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deininger, M.W.N.; Goldman, J.M.; Melo, J.V. The Molecular Biology of Chronic Myeloid Leukemia. Blood 2000, 96, 3343–3356. [Google Scholar] [CrossRef] [PubMed]
- Goldman, J.M.; Melo, J.V. Chronic Myeloid Leukemia—Advances in Biology and New Approaches to Treatment. N. Engl. J. Med. 2003, 349, 1451–1464. [Google Scholar] [CrossRef] [Green Version]
- Flis, S.; Chojnacki, T. Chronic Myelogenous Leukemia, a Still Unsolved Problem: Pitfalls and New Therapeutic Possibilities. Drug Des. Dev. Ther. 2019, 13, 825–843. [Google Scholar] [CrossRef] [Green Version]
- Stagno, F.; Stella, S.; Spitaleri, A.; Pennisi, M.S.; Di Raimondo, F.; Vigneri, P. Imatinib Mesylate in Chronic Myeloid Leukemia: Frontline Treatment and Long-Term Outcomes. Expert Rev. Anticancer. Ther. 2016, 16, 273–278. [Google Scholar] [CrossRef]
- Kantarjian, H.; Sawyers, C.; Hochhaus, A.; Guilhot, F.; Schiffer, C.; Gambacorti-Passerini, C.; Niederwieser, D.; Resta, D.; Capdeville, R.; Zoellner, U.; et al. Hematologic and Cytogenetic Responses to Imatinib Mesylate in Chronic Myelogenous Leukemia. N. Engl. J. Med. 2002, 346, 645–652. [Google Scholar] [CrossRef]
- Iqbal, N.; Iqbal, N. Imatinib: A Breakthrough of Targeted Therapy in Cancer. Chemother. Res. Pract. 2014, 2014, 357027. [Google Scholar] [CrossRef]
- Marcucci, G.; Perrotti, D.; Caligiuri, M.A. Understanding the Molecular Basis of Imatinib Mesylate Therapy in Chronic Myelogenous Leukemia and the Related Mechanisms of Resistance: Commentary Re: A. N. Mohamed et al., The Effect of Imatinib Mesylate on Patients with Philadelphia Chromosome-Positive Chronic Myeloid Leukemia with Secondary Chromosomal Aberrations. Clin. Cancer Res. 2003, 9, 1248–1252. [Google Scholar]
- Bhamidipati, P.K.; Kantarjian, H.; Cortes, J.; Cornelison, A.M.; Jabbour, E. Management of Imatinib-Resistant Patients with Chronic Myeloid Leukemia. Ther. Adv. Hematol. 2013, 4, 103–117. [Google Scholar] [CrossRef] [Green Version]
- Milojkovic, D.; Apperley, J. Mechanisms of Resistance to Imatinib and Second-Generation Tyrosine Inhibitors in Chronic Myeloid Leukemia. Clin. Cancer Res. 2009, 15, 7519–7527. [Google Scholar] [CrossRef] [Green Version]
- Breccia, M.; Alimena, G. Second-generation tyrosine kinase inhibitors (TKI) as salvage therapy for resistant or intolerant patients to prior TKIs. Mediterr J. Hematol Infect. Dis 2014, 6, e2014003. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H.; Cortes, J. Chronic Myeloid Leukemia and Second-Generation Tyrosine Kinase Inhibitors: When, How, and Which One? Semin. Hematol. 2010, 47, 344–353. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H.; Cortes, J. Use of Second- and Third-Generation Tyrosine Kinase Inhibitors in the Treatment of Chronic Myeloid Leukemia: An Evolving Treatment Paradigm. Clin. Lymphoma Myeloma Leuk. 2015, 15, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Bitencourt, R.; Zalcberg, I.; Louro, I.D. Imatinib Resistance: A Review of Alternative Inhibitors in Chronic Myeloid Leukemia. Rev. Bras. Hematol. Hemoter. 2011, 33, 470–475. [Google Scholar] [CrossRef]
- Hamad, A.; Sahli, Z.; El Sabban, M.; Mouteirik, M.; Nasr, R. Emerging Therapeutic Strategies for Targeting Chronic Myeloid Leukemia Stem Cells. Stem Cells Int. 2013, 2013, 724360. [Google Scholar] [CrossRef] [Green Version]
- Abruzzese, E.; Breccia, M.; Latagliata, R. Second-Generation Tyrosine Kinase Inhibitors in First-Line Treatment of Chronic Myeloid Leukaemia (CML). BioDrugs 2014, 28, 17–26. [Google Scholar] [CrossRef]
- Kenzik, K.M.; Bhatia, R.; Bhatia, S. Expenditures for First- and Second-Generation Tyrosine Kinase Inhibitors Before and After Transition of Imatinib to Generic Status. JAMA Oncol. 2020, 6, 542. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Sundaram, C.; Malani, N.; Ichikawa, H. Curcumin: The Indian Solid Gold. In The Molecular Targets and Therapeutic Uses of Curcumin in Health and Disease; Aggarwal, B.B., Surh, Y.-J., Shishodia, S., Eds.; Springer: Boston, MA, USA, 2007; Volume 595, pp. 1–75. ISBN 978-0-387-46400-8. [Google Scholar]
- Hosseini, A.; Ghorbani, A. Cancer Therapy with Phytochemicals: Evidence from Clinical Studies. Avicenna J. Phytomed. 2015, 5, 84–97. [Google Scholar]
- Noorafshan, A.; Ashkani-Esfahani, S. A Review of Therapeutic Effects of Curcumin. Curr. Pharm. Des. 2013, 19, 2032–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Jiang, S.; Zhou, L.; Yu, F.; Ding, H.; Li, P.; Zhou, M.; Wang, K. Potential Mechanisms of Action of Curcumin for Cancer Prevention: Focus on Cellular Signaling Pathways and MiRNAs. Int. J. Biol. Sci. 2019, 15, 1200–1214. [Google Scholar] [CrossRef] [PubMed]
- Priyadarsini, K. The Chemistry of Curcumin: From Extraction to Therapeutic Agent. Molecules 2014, 19, 20091–20112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocaadam, B.; Şanlier, N. Curcumin, an Active Component of Turmeric (Curcuma Longa), and Its Effects on Health. Crit. Rev. Food Sci. Nutr. 2017, 57, 2889–2895. [Google Scholar] [CrossRef] [PubMed]
- Ghalaut, V.S.; Sangwan, L.; Dahiya, K.; Ghalaut, P.; Dhankhar, R.; Saharan, R. Effect of Imatinib Therapy with and without Turmeric Powder on Nitric Oxide Levels in Chronic Myeloid Leukemia. J. Oncol. Pharm. Pract. 2012, 18, 186–190. [Google Scholar] [CrossRef]
- Song, X.; Zhang, M.; Dai, E.; Luo, Y. Molecular Targets of Curcumin in Breast Cancer (Review). Mol. Med. Report 2018, 19, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Wan Mohd Tajuddin, W.N.B.; Lajis, N.H.; Abas, F.; Othman, I.; Naidu, R. Mechanistic Understanding of Curcumin’s Therapeutic Effects in Lung Cancer. Nutrients 2019, 11, 2989. [Google Scholar] [CrossRef] [Green Version]
- Mbese, Z.; Khwaza, V.; Aderibigbe, B.A. Curcumin and Its Derivatives as Potential Therapeutic Agents in Prostate, Colon and Breast Cancers. Molecules 2019, 24, 4386. [Google Scholar] [CrossRef] [Green Version]
- Kouhpeikar, H.; Butler, A.E.; Bamian, F.; Barreto, G.E.; Majeed, M.; Sahebkar, A. Curcumin as a Therapeutic Agent in Leukemia. J. Cell Physiol. 2019, 234, 12404–12414. [Google Scholar] [CrossRef]
- Kelkel, M.; Jacob, C.; Dicato, M.; Diederich, M. Potential of the Dietary Antioxidants Resveratrol and Curcumin in Prevention and Treatment of Hematologic Malignancies. Molecules 2010, 15, 7035–7074. [Google Scholar] [CrossRef] [Green Version]
- Giordano, A. Tommonaro Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Castillo, M.; Bonilla-Moreno, R.; Aleman-Lazarini, L.; Meraz-Rios, M.A.; Orozco, L.; Cedillo-Barron, L.; Cordova, E.J.; Villegas-Sepulveda, N. A Subpopulation of the K562 Cells Are Killed by Curcumin Treatment after G2/M Arrest and Mitotic Catastrophe. PLoS ONE 2016, 11, e0165971. [Google Scholar] [CrossRef]
- Taverna, S.; Giallombardo, M.; Pucci, M.; Flugy, A.; Manno, M.; Raccosta, S.; Rolfo, C.; De Leo, G.; Alessandro, R. Curcumin Inhibits in Vitro and in Vivo Chronic Myelogenous Leukemia Cells Growth: A Possible Role for Exosomal Disposal of MiR-21. Oncotarget 2015, 6, 21918–21933. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-J.; Choi, Y.-H.; Kim, C.U.; Bae, E.K.; Kim, B.-S.; Ahn, K.-S.; Lee, J.-S.; Park, S.; Kim, B.K.; Yoon, S.-S. Effect of the Combination of Imatinib Mesylate (Glivec) and Curcumin in Chronic Myeloid Leukemia Cell Line. Blood 2004, 104, 4685. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New Ways to Boost Molecular Dynamics Simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular Graphics for All Devices—From Smartphones to Workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, M.D.; Harrison, S.C. Structure of an IkappaBalpha/NF-KappaB Complex. Cell 1998, 95, 749–758. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.; Lin, J.; Wu, W.-I.; Ballard, J.; Lee, B.B.; Gloor, S.L.; Vigers, G.P.A.; Morales, T.H.; Friedman, L.S.; Skelton, N.; et al. An ATP-Site On-Off Switch That Restricts Phosphatase Accessibility of Akt. Sci. Signal. 2012, 5, ra37. [Google Scholar] [CrossRef]
- De Leon-Boenig, G.; Bowman, K.K.; Feng, J.A.; Crawford, T.; Everett, C.; Franke, Y.; Oh, A.; Stanley, M.; Staben, S.T.; Starovasnik, M.A.; et al. The Crystal Structure of the Catalytic Domain of the NF-ΚB Inducing Kinase Reveals a Narrow but Flexible Active Site. Structure 2012, 20, 1704–1714. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Cron, P.; Thompson, V.; Good, V.M.; Hess, D.; Hemmings, B.A.; Barford, D. Molecular Mechanism for the Regulation of Protein Kinase B/Akt by Hydrophobic Motif Phosphorylation. Molecular Cell 2002, 9, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Sunami, T.; Byrne, N.; Diehl, R.E.; Funabashi, K.; Hall, D.L.; Ikuta, M.; Patel, S.B.; Shipman, J.M.; Smith, R.F.; Takahashi, I.; et al. Structural Basis of Human P70 Ribosomal S6 Kinase-1 Regulation by Activation Loop Phosphorylation. J. Biol. Chem. 2010, 285, 4587–4594. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A Point-Charge Force Field for Molecular Mechanics Simulations of Proteins Based on Condensed-Phase Quantum Mechanical Calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Molnar, L.F.; Jung, Y.; Kussmann, J.; Ochsenfeld, C.; Brown, S.T.; Gilbert, A.T.B.; Slipchenko, L.V.; Levchenko, S.V.; O’Neill, D.P.; et al. Advances in Methods and Algorithms in a Modern Quantum Chemistry Program Package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [Google Scholar] [CrossRef]
- Konagurthu, A.S.; Whisstock, J.C.; Stuckey, P.J.; Lesk, A.M. MUSTANG: A Multiple Structural Alignment Algorithm. Proteins 2006, 64, 559–574. [Google Scholar] [CrossRef]
- Jang, S.-K.; Hong, S.-E.; Lee, D.-H.; Kim, J.Y.; Kim, J.-Y.; Hong, J.; Park, I.-C.; Jin, H.-O. Inhibition of AKT Enhances the Sensitivity of NSCLC Cells to Metformin. Anticancer Res. 2021, 41, 3481–3487. [Google Scholar] [CrossRef]
- Mu, H.; Zhu, X.; Jia, H.; Zhou, L.; Liu, H. Combination Therapies in Chronic Myeloid Leukemia for Potential Treatment-Free Remission: Focus on Leukemia Stem Cells and Immune Modulation. Front. Oncol. 2021, 11, 643382. [Google Scholar] [CrossRef]
- Bhaskar, A.; Raturi, K.; Dang, S.; Gabrani, R. Current Perspectives on the Therapeutic Aspects of Chronic Myelogenous Leukemia. Expert Opin. Ther. Pat. 2014, 24, 1117–1127. [Google Scholar] [CrossRef]
- Glamoclija, U.; Mahmutovic, L.; Bilajac, E.; Soljic, V.; Vukojevic, K.; Suljagic, M. Metformin and Thymoquinone Synergistically Inhibit Proliferation of Imatinib-Resistant Human Leukemic Cells. Front. Pharmacol. 2022, 13, 867133. [Google Scholar] [CrossRef]
- Mutlu Altundağ, E.; Yılmaz, A.M.; Koçtürk, S.; Taga, Y.; Yalçın, A.S. Synergistic Induction of Apoptosis by Quercetin and Curcumin in Chronic Myeloid Leukemia (K562) Cells. Nutr. Cancer 2018, 70, 97–108. [Google Scholar] [CrossRef]
- Mukherjee, A.; Sarkar, R.; Mukherjee, S.; Biswas, J.; Roy, M. Curcumin Boosts up the Efficacy of Imatinib Mesylate in Chronic Myelogenic Leukemia Cell Line K-562 by Modulation of Various Markers. Int. J. Curr. Microbiol. App. Sci. 2016, 5, 240–255. [Google Scholar] [CrossRef] [Green Version]
- Lopes-Rodrigues, V.; Oliveira, A.; Correia-da-Silva, M.; Pinto, M.; Lima, R.T.; Sousa, E.; Vasconcelos, M.H. A Novel Curcumin Derivative Which Inhibits P-Glycoprotein, Arrests Cell Cycle and Induces Apoptosis in Multidrug Resistance Cells. Bioorganic Med. Chem. 2017, 25, 581–596. [Google Scholar] [CrossRef]
- Wu, L.; Wu, Y.; Chen, R.; Liu, Y.; Huang, L.; Lou, L.; Zheng, Z.; Chen, Y.; Xu, J. Curcumin Derivative C817 Inhibits Proliferation of Imatinib-Resistant Chronic Myeloid Leukemia Cells with Wild-Type or Mutant Bcr-Abl in Vitro. Acta Pharmacol. Sin. 2014, 35, 401–409. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Zhou, Y.; Zhang, L.; Lin, Q.; Gao, P.; Cai, F.; Zhu, L.; Liu, B.; Xu, J. C1206, a Novel Curcumin Derivative, Potently Inhibits Hsp90 and Human Chronic Myeloid Leukemia Cells in Vitro. Acta Pharmacol. Sin. 2018, 39, 649–658. [Google Scholar] [CrossRef]
- Ustun Alkan, F.; Anlas, C.; Cinar, S.; Yildirim, F.; Ustuner, O.; Bakirel, T.; Gurel, A. Effects of Curcumin in Combination with Cyclophosphamide on Canine Mammary Tumour Cell Lines. Vet. Med. 2014, 59, 553–572. [Google Scholar] [CrossRef] [Green Version]
- Khazaei Koohpar, Z.; Entezari, M.; Movafagh, A.; Hashemi, M. Anticancer Activity of Curcumin on Human Breast Adenocarcinoma: Role of Mcl-1 Gene. Iran. J. Cancer Preven. 2015, 8, e2331. [Google Scholar] [CrossRef] [Green Version]
- Calaf, G.; Ponce-Cusi, R.; Carrión, F. Curcumin and Paclitaxel Induce Cell Death in Breast Cancer Cell Lines. Oncol. Rep. 2018, 40, 2381–2388. [Google Scholar] [CrossRef]
- Wolanin, K. Curcumin Affects Components of the Chromosomal Passenger Complex and Induces Mitotic Catastrophe in Apoptosis-Resistant Bcr-Abl-Expressing Cells. Mol. Cancer Res. 2006, 4, 457–469. [Google Scholar] [CrossRef] [Green Version]
- Eryanti, Y.; Zamri, A.; Frimayanti, N.; Teruna, H.Y.; Supratmman, U.; Herlina, T.; Shiono, Y. Synthesis, Structure-Activity Relationship, Docking and Molecular Dynamic Simulation of Curcumin Analogues Against HL-60 for Anti Cancer Agents (Leukemia). Orient. J. Chem. 2017, 33, 2164–2172. [Google Scholar] [CrossRef]
- Tong, Y.; Xiang, Y.; Li, B.; Bao, S.; Zhou, Y.; Yuan, W.; Ling, Y.; Hao, D.; Zhu, H.; Sun, Z. Association between TERT Gene Polymorphisms and Acute Myeloid Leukemia Susceptibility in a Chinese Population: A Case–Control Study. Cancer Cell Int. 2020, 20, 313. [Google Scholar] [CrossRef]
- Kumar, A.; Bora, U. Molecular Docking Studies of Curcumin Natural Derivatives with DNA Topoisomerase I and II-DNA Complexes. Interdiscip. Sci. Comput. Life Sci. 2014, 6, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, A. Curcumin Targets in Inflammation and Cancer. EMIDDT 2015, 15, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Charlet, J.; Juncker, T.; Teiten, M.-H.; Dicato, M.; Diederich, M. Effect of Curcumin on Nuclear Factor ΚB Signaling Pathways in Human Chronic Myelogenous K562 Leukemia Cells. Ann. N. Y. Acad. Sci. 2009, 1171, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Castillo, M.; Villegas-Sepúveda, N.; Meraz-Rios, M.; Hernández-Zavala, A.; Berumen, J.; Coleman, M.; Orozco, L.; Cordova, E. Curcumin Differentially Affects Cell Cycle and Cell Death in Acute and Chronic Myeloid Leukemia Cells. Oncol. Lett. 2018, 15, 6777–6783. [Google Scholar] [CrossRef]
- Jia, Y.-L.; Li, J.; Qin, Z.-H.; Liang, Z.-Q. Autophagic and Apoptotic Mechanisms of Curcumin-Induced Death in K562 Cells. J. Asian Nat. Prod. Res. 2009, 11, 918–928. [Google Scholar] [CrossRef]
- Speciale, A.; Chirafisi, J.; Saija, A.; Cimino, F. Nutritional Antioxidants and Adaptive Cell Responses: An Update. Curr. Mol. Med. 2011, 11, 770–789. [Google Scholar] [CrossRef]


| IC50 | ||||||
|---|---|---|---|---|---|---|
| Cell Line | K562 | LAMA84S | LAMA84R | |||
| Assay (48 h) | WST-8 | Trypan blue | WST-8 | Trypan blue | WST-8 | Trypan blue |
| 20.11 ± 3.10 | 21.62 ± 0.37 | 12.60 ± 2.16 | 16.66 ± 0.59 | 71.34 ± 6.26 | 68.95 ± 3.68 | |
| Protein (PDB ID)—Ligand Complex | Binding Energy (kcal/mol) | Dissociation Constant (μM) | Contacting Amino Acid Residues (H-Bonds) |
|---|---|---|---|
| I-Kappa-B-Alpha/NF-Kappa-B (PDB ID: 1NFI)—curcumin | −10.45 | 0.022 | Gln 111 |
| I-Kappa-B-Alpha/NF-Kappa-B (PDB ID: 1NFI)—imatinib | −10.67 | 0.015 | Tyr 181, Lys 326 |
| Akt1 (binding pocket of AMP-PNP) (PDB ID: 4EKK)—curcumin | −11.30 | 0.005 | Lys 158, Glu 234, Asp 439 |
| Akt1 (binding pocket of AMP-PNP) (PDB ID: 4EKK)—imatinib | −9.81 | 0.064 | Lys 179, Asp 439 |
| NF-kappaB inducing kinase (PDB ID: 4G3D)—curcumin | −10.12 | 0.038 | Leu 445, Gly 446 |
| NF-kappaB inducing kinase (PDB ID: 4G3D)—imatinib | −8.52 | 0.569 | Gly 446 |
| NF-kappaB inducing kinase (binding pocket) (PDB ID: 4G3D)—curcumin | −9.81 | 0.064 | Glu 440, Cys 444, Gln 479 |
| NF-kappaB inducing kinase (binding pocket) (PDB ID: 4G3D)—imatinib | −5.05 | 200.06 | Gln 479, Phe 535 |
| Protein kinase B unphosphorylated (PDB ID: 1GZO)—curcumin | −11.67 | 0.003 | Thr 313, Gly 335 |
| Protein kinase B unphosphorylated (PDB ID: 1GZO)—imatinib | −11.16 | 0.007 | Trp 334 |
| Phosphorylated p70S6K1 (binding pocket) (PDB ID: 3A62)—curcumin | −9.31 | 0.150 | Lys 123, Glu 143 |
| Phosphorylated p70S6K1 (binding pocket) (PDB ID: 3A62)—imatinib | −9.10 | 0.212 | Leu 175 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bilajac, E.; Mahmutović, L.; Glamočlija, U.; Osmanović, A.; Hromić-Jahjefendić, A.; Tambuwala, M.M.; Suljagić, M. Curcumin Decreases Viability and Inhibits Proliferation of Imatinib-Sensitive and Imatinib-Resistant Chronic Myeloid Leukemia Cell Lines. Metabolites 2023, 13, 58. https://doi.org/10.3390/metabo13010058
Bilajac E, Mahmutović L, Glamočlija U, Osmanović A, Hromić-Jahjefendić A, Tambuwala MM, Suljagić M. Curcumin Decreases Viability and Inhibits Proliferation of Imatinib-Sensitive and Imatinib-Resistant Chronic Myeloid Leukemia Cell Lines. Metabolites. 2023; 13(1):58. https://doi.org/10.3390/metabo13010058
Chicago/Turabian StyleBilajac, Esma, Lejla Mahmutović, Una Glamočlija, Amar Osmanović, Altijana Hromić-Jahjefendić, Murtaza M. Tambuwala, and Mirza Suljagić. 2023. "Curcumin Decreases Viability and Inhibits Proliferation of Imatinib-Sensitive and Imatinib-Resistant Chronic Myeloid Leukemia Cell Lines" Metabolites 13, no. 1: 58. https://doi.org/10.3390/metabo13010058
APA StyleBilajac, E., Mahmutović, L., Glamočlija, U., Osmanović, A., Hromić-Jahjefendić, A., Tambuwala, M. M., & Suljagić, M. (2023). Curcumin Decreases Viability and Inhibits Proliferation of Imatinib-Sensitive and Imatinib-Resistant Chronic Myeloid Leukemia Cell Lines. Metabolites, 13(1), 58. https://doi.org/10.3390/metabo13010058








