Biomarker Discovery for Hepatocellular Carcinoma in Patients with Liver Cirrhosis Using Untargeted Metabolomics and Lipidomics Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Study Cohort and Sample Collection
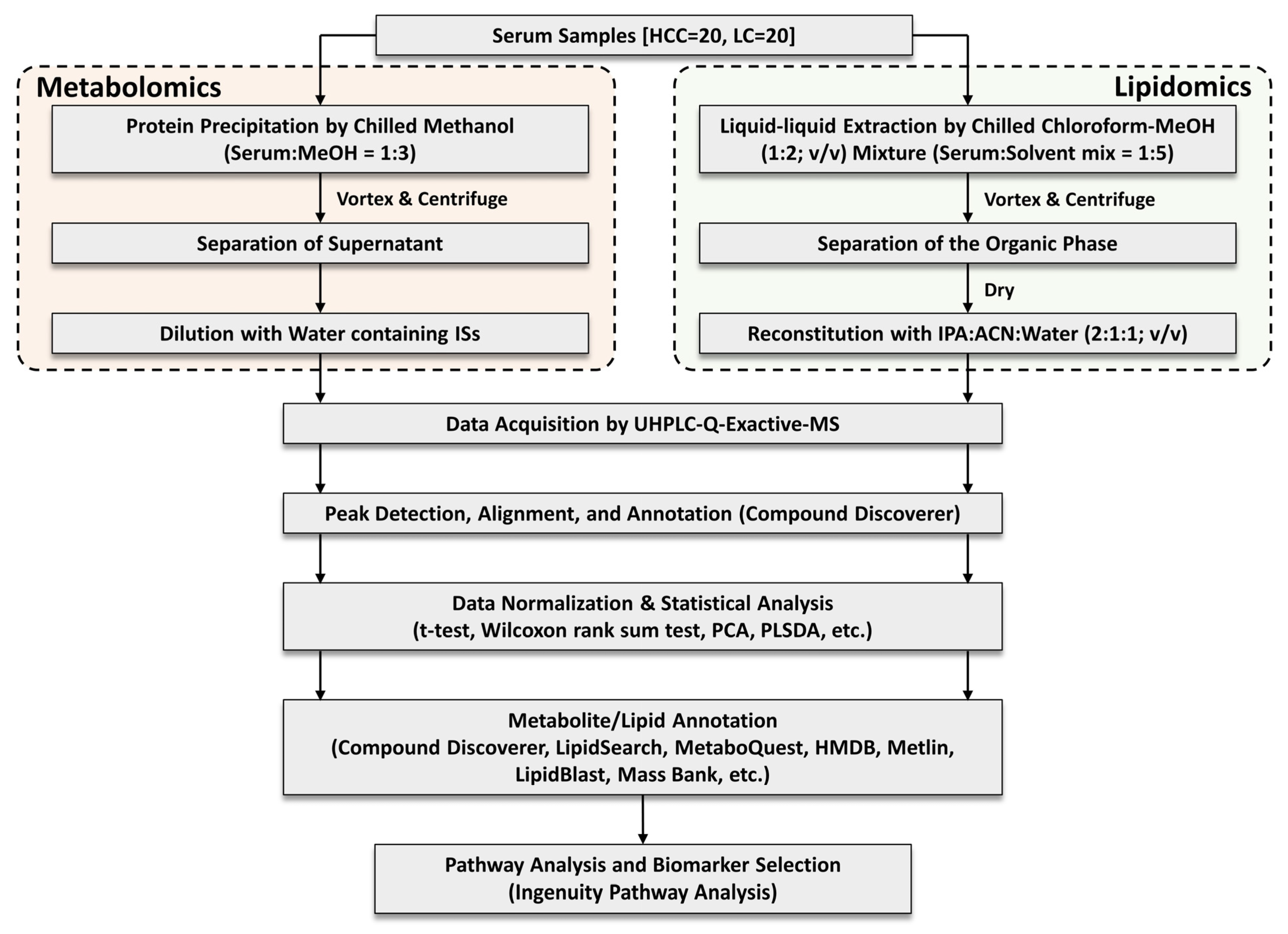
2.3. Serum Sample Preparation
2.4. Instrumental Conditions
2.5. Data Processing and Statistical Analysis
2.6. Metabolite Annotation
2.7. Network and Pathway Analyses
2.8. Receiver Operating Characteristic (ROC) Curve Analysis
3. Results
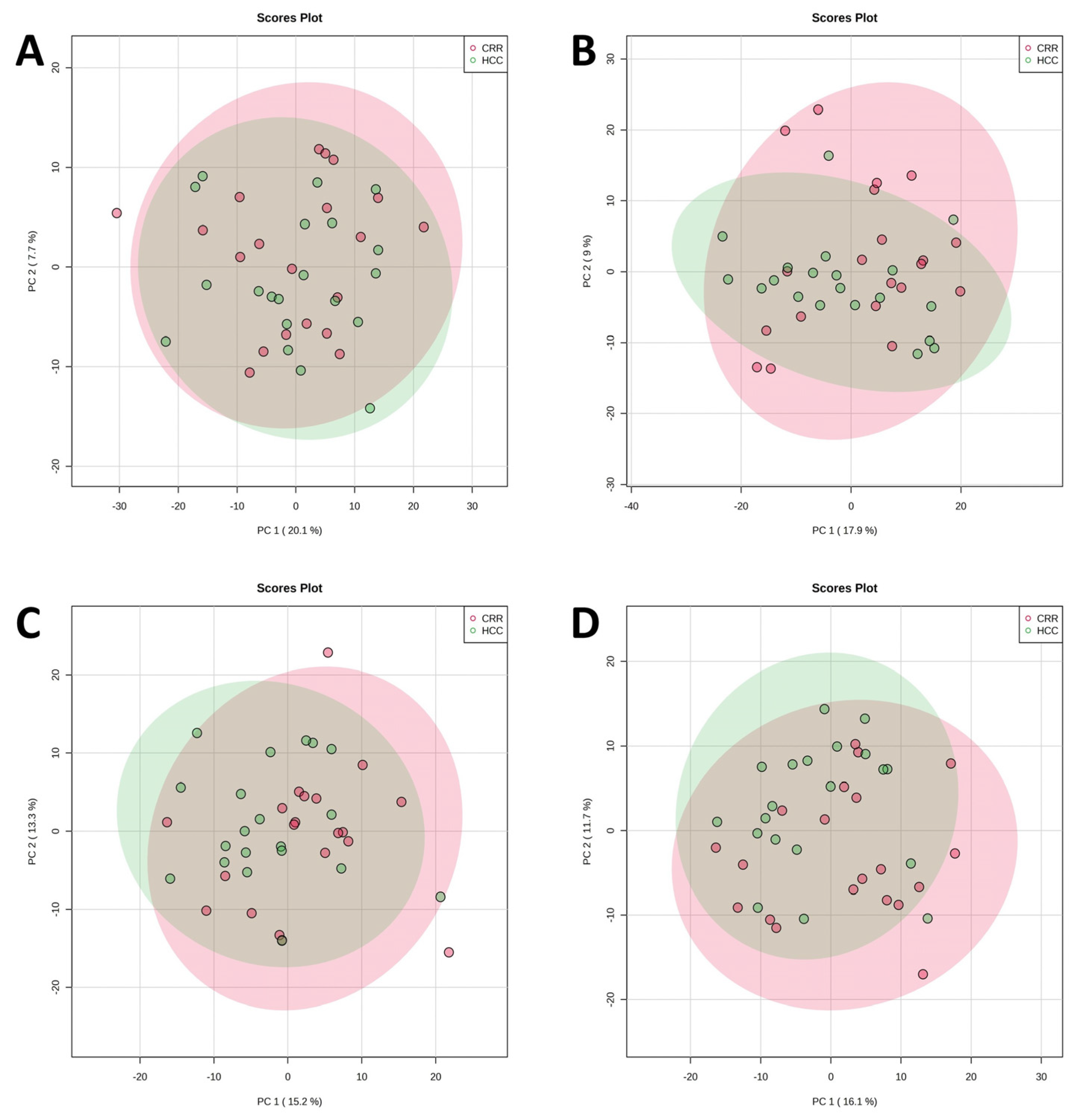
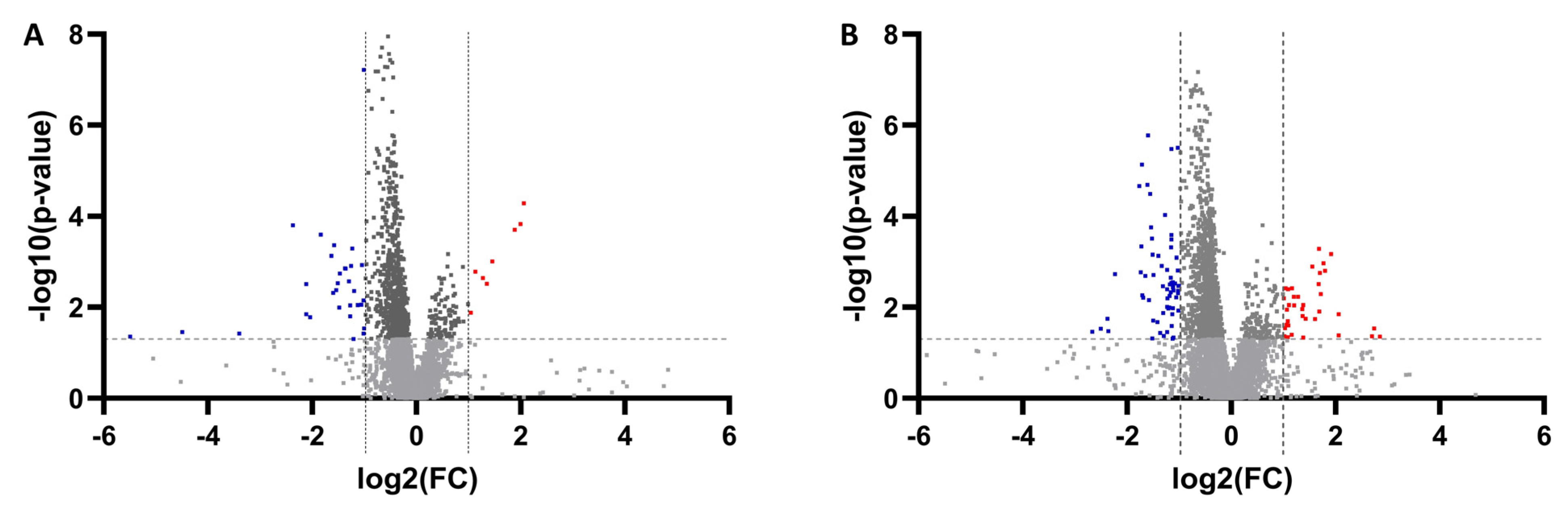
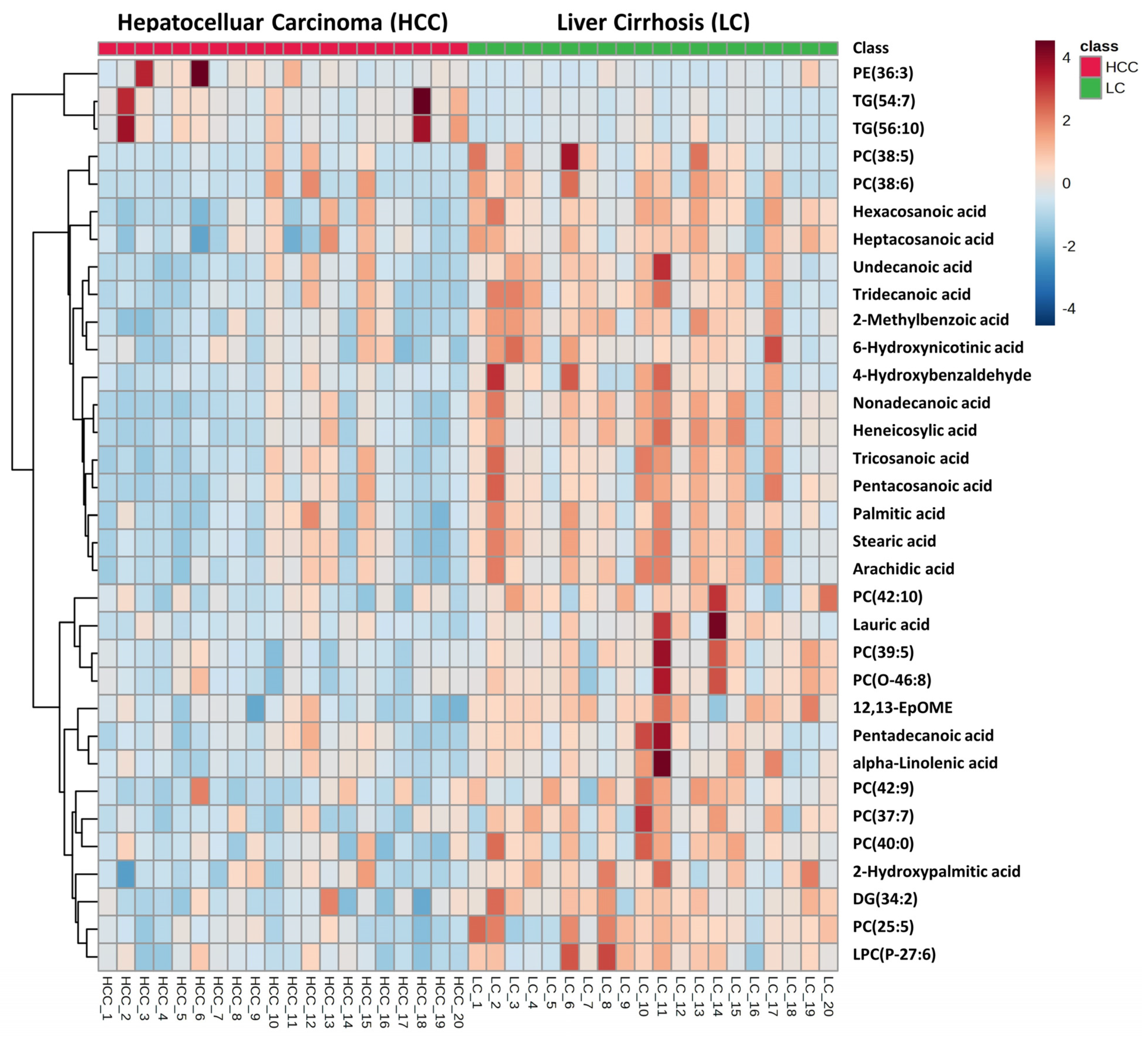
3.1. Untargeted Metabolomics and Lipidomics Analysis of HCC vs. LC
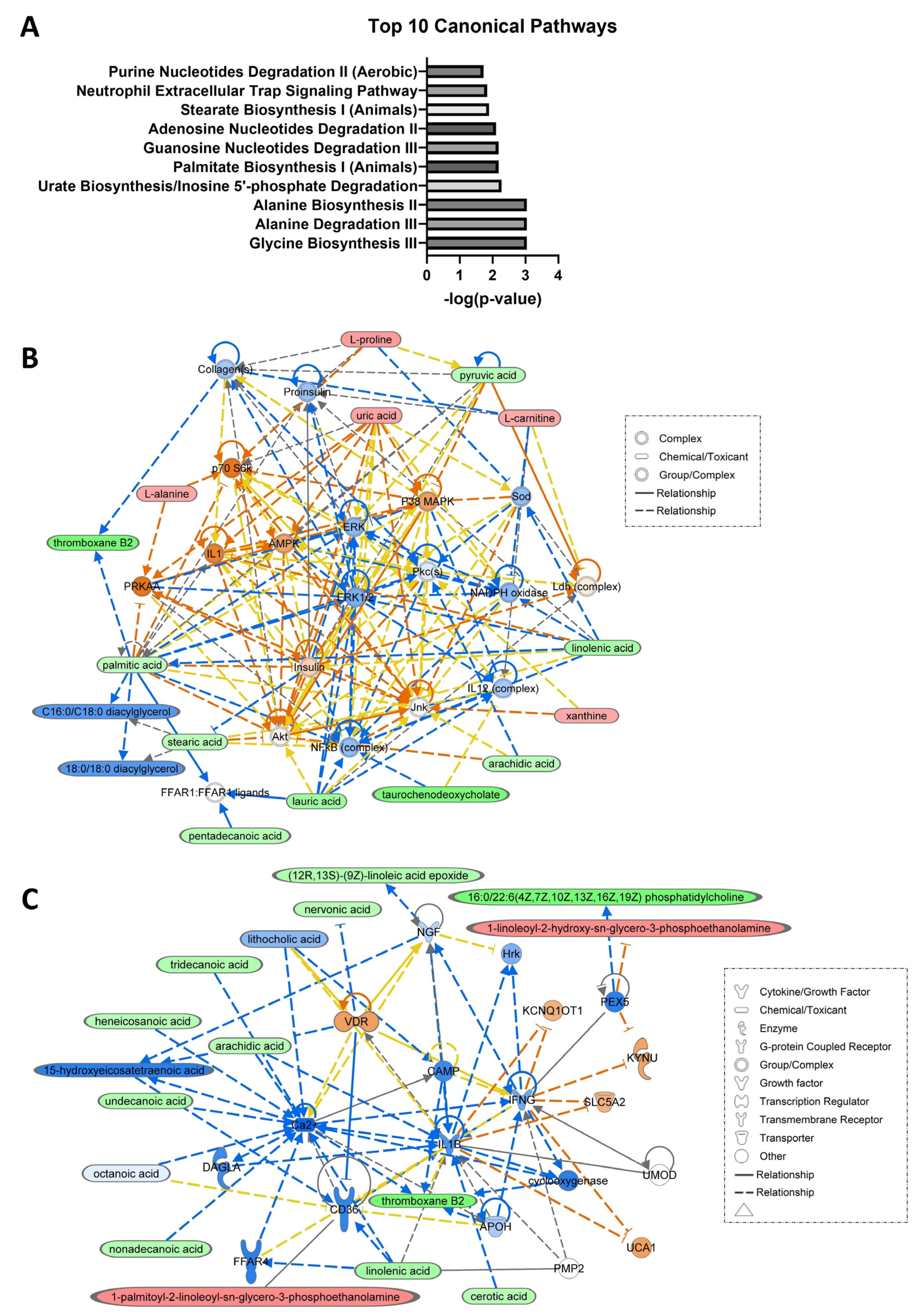
3.2. Network and Pathway Analyses
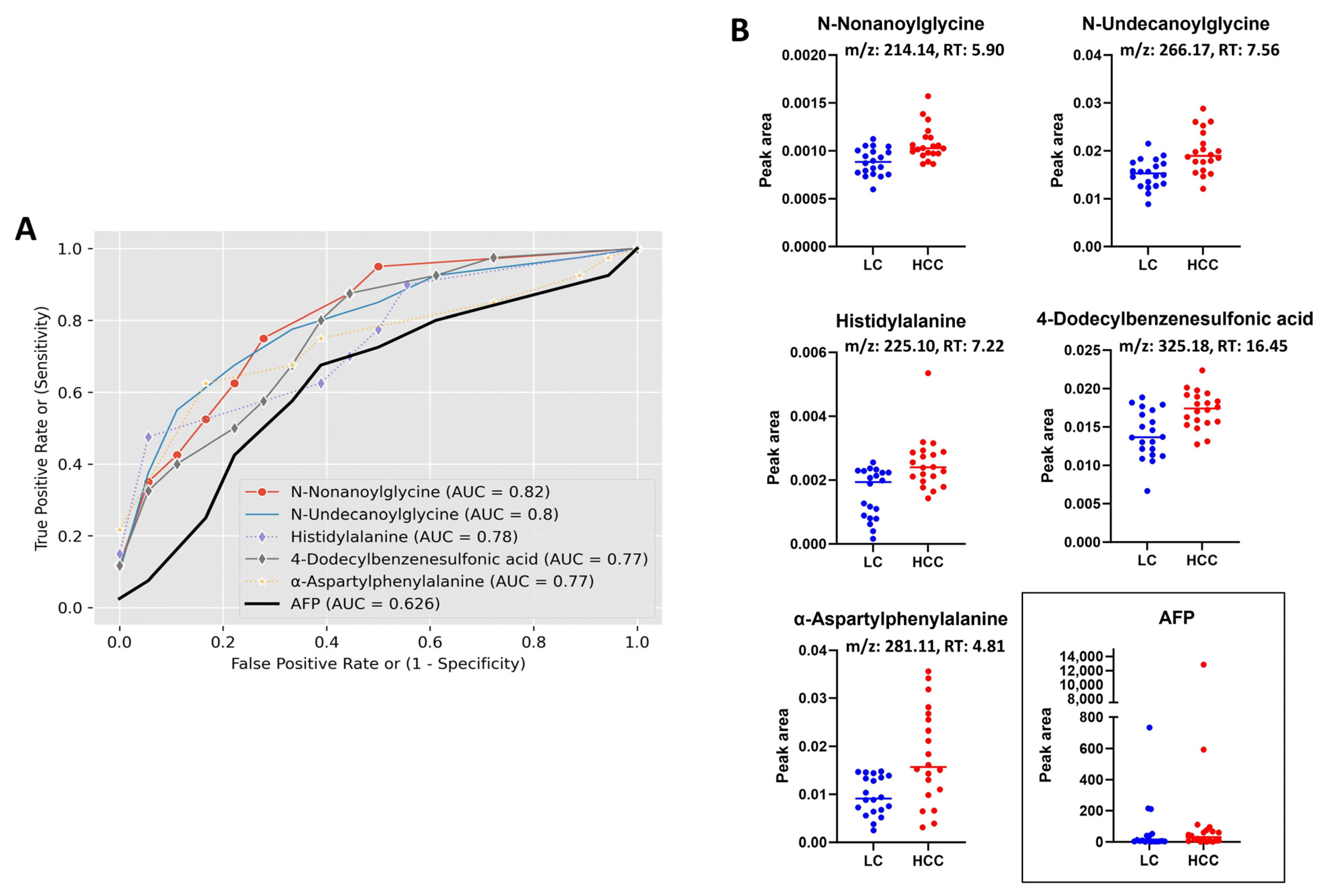
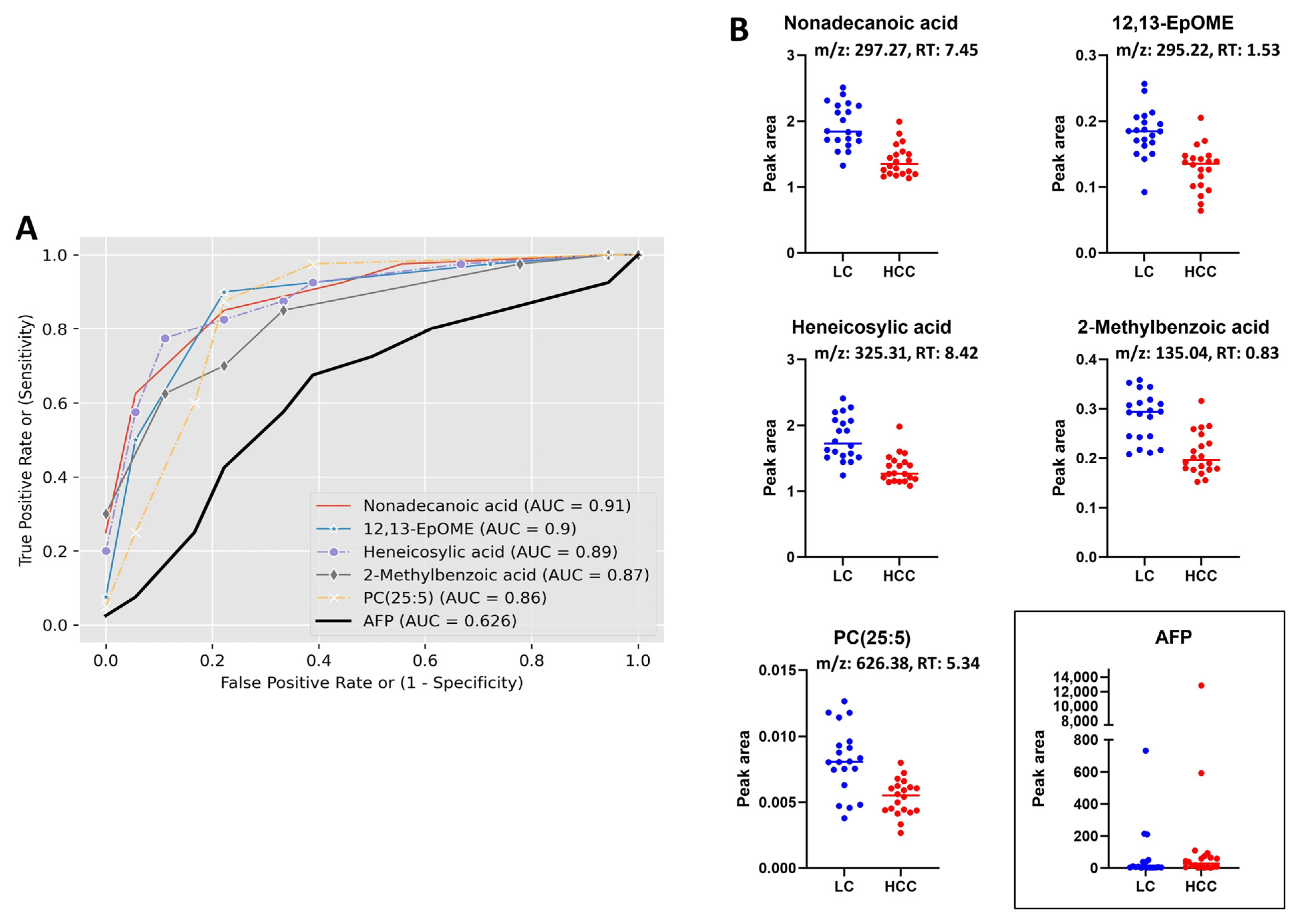
3.3. Receiver Operating Characteristic (ROC) Curve Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khalil, A.; Elfert, A.; Ghanem, S.; Helal, M.; Abdelsattar, S.; Elgedawy, G.; Obada, M.; Abdel-Samiee, M.; El-Said, H. The role of metabolomics in hepatocellular carcinoma. Egypt. Liver J. 2021, 11, 41. [Google Scholar] [CrossRef]
- Morine, Y.; Utsunomiya, T.; Yamanaka-Okumura, H.; Saito, Y.; Yamada, S.; Ikemoto, T.; Imura, S.; Kinoshita, S.; Hirayama, A.; Tanaka, Y.; et al. Essential amino acids as diagnostic biomarkers of hepatocellular carcinoma based on metabolic analysis. Oncotarget 2022, 13, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B.; Davila, J.A. Surveillance for hepatocellular carcinoma: In whom and how? Ther. Adv. Gastroenterol. 2011, 4, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, H.; He, C.; Yang, M.; Chen, H.; Yang, X.; Zhuo, J.; Shen, W.; Hu, Z.; Pan, L.; et al. Metabolomic biomarkers for the diagnosis and post-transplant outcomes of AFP negative hepatocellular carcinoma. Front. Oncol. 2023, 13, 1072775. [Google Scholar] [CrossRef]
- Wong, R.J.; Ahmed, A.; Gish, R.G. Elevated alpha-fetoprotein: Differential diagnosis—Hepatocellular carcinoma and other disorders. Clin. Liver Dis. 2015, 19, 309–323. [Google Scholar] [CrossRef]
- Luo, P.; Yin, P.; Hua, R.; Tan, Y.; Li, Z.; Qiu, G.; Yin, Z.; Xie, X.; Wang, X.; Chen, W.; et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology 2018, 67, 662–675. [Google Scholar] [CrossRef]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Z.; Luo, L.; Shu, D.; Wang, K. Metabolomics in hepatocellular carcinoma: From biomarker discovery to precision medicine. Front. Med. Technol. 2022, 4, 1065506. [Google Scholar] [CrossRef]
- Rashid, M.; Lee, H.; Jung, B.H. Evaluation of the antitumor effects of PP242 in a colon cancer xenograft mouse model using comprehensive metabolomics and lipidomics. Sci. Rep. 2020, 10, 17523. [Google Scholar] [CrossRef]
- Liu, X.N.; Cui, D.-N.; Li, Y.-F.; Liu, Y.-H.; Liu, G.; Liu, L. Multiple “Omics” data-based biomarker screening for hepatocellular carcinoma diagnosis. World J. Gastroenterol. 2019, 25, 4199–4212. [Google Scholar] [CrossRef] [PubMed]
- Cotte, A.K.; Cottet, V.; Aires, V.; Mouillot, T.; Rizk, M.; Vinault, S.; Binquet, C.; de Barros, J.-P.P.; Hillon, P.; Delmas, D. Phospholipid profiles and hepatocellular carcinoma risk and prognosis in cirrhotic patients. Oncotarget 2019, 10, 2161–2172. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Cho, E.J.; Yu, K.-S.; Jang, I.-J.; Yoon, J.-H.; Park, T.; Cho, J.-Y. Comprehensive Metabolomic Search for Biomarkers to Differentiate Early Stage Hepatocellular Carcinoma from Cirrhosis. Cancers 2019, 11, 1497. [Google Scholar] [CrossRef] [PubMed]
- Safaei, A.; Oskouie, A.A.; Mohebbi, S.R.; Rezaei-Tavirani, M.; Mahboubi, M.; Peyvandi, M.; Okhovatian, F.; Zamanian-Azodi, M. Metabolomic analysis of human cirrhosis, hepatocellular carcinoma, non-alcoholic fatty liver disease and non-alcoholic steatohepatitis diseases. Gastroenterol. Hepatol. Bed Bench 2016, 9, 158–173. [Google Scholar]
- Feng, N.; Yu, F.; Yu, F.; Feng, Y.; Zhu, X.; Xie, Z.; Zhai, Y. Metabolomic biomarkers for hepatocellular carcinoma: A systematic review. Medicine 2022, 101, e28510. [Google Scholar] [CrossRef]
- Rashid, M.; Lee, H.; Park, J.; Jung, B.H. Comparative metabolomics and lipidomics study to evaluate the metabolic differences between first- and second-generation mammalian or mechanistic target of rapamycin inhibitors. Biomed. Chromatogr. 2021, 35, e5190. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef]
- Yilmaz, H. Uric Acid & Cancer. Eurasian J. Med. Investig. 2022, 6, 141–146. [Google Scholar]
- Yiu, A.; Van Hemelrijck, M.; Garmo, H.; Holmberg, L.; Malmström, H.; Lambe, M.; Hammar, N.; Walldius, G.; Jungner, I.; Wulaningsih, W. Circulating uric acid levels and subsequent development of cancer in 493,281 individuals: Findings from the AMORIS Study. Oncotarget 2017, 8, 42332–42342. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Wang, J.; Chen, L.; Huang, H.; Xiong, Y.; Xie, B. Quantification of serum purine metabolites for distinguishing patients with hepatitis B from hepatocellular carcinoma. Bioanalysis 2019, 11, 1003–1013. [Google Scholar] [CrossRef]
- Wu, L.; Yang, W.; Zhang, Y.; Du, X.; Jin, N.; Chen, W.; Li, H.; Zhang, S.; Xie, B. Elevated Serum Uric Acid is Associated with Poor Survival in Advanced HCC Patients and Febuxostat Improves Prognosis in HCC Rats. Front. Pharmacol. 2021, 12, 778890. [Google Scholar] [CrossRef]
- Thomas, C.E.; Luu, H.N.; Wang, R.; Xie, G.; Adams-Haduch, J.; Jin, A.; Koh, W.-P.; Jia, W.; Behari, J.; Yuan, J.-M. Association between Pre-Diagnostic Serum Bile Acids and Hepatocellular Carcinoma: The Singapore Chinese Health Study. Cancers 2021, 13, 2648. [Google Scholar] [CrossRef]
- Han, J.; Qin, W.-X.; Li, Z.-L.; Xu, A.-J.; Xing, H.; Wu, H.; Zhang, H.; Wang, M.-D.; Li, C.; Liang, L.; et al. Tissue and serum metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin. Chim. Acta 2019, 488, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Wan, D.; Zhao, C.; Chen, J.; Zhao, X.; Wang, W.; Lu, X.; Yang, S.; Gu, J.; Xu, G. A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol. Biosyst. 2009, 5, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Ressom, H.W.; Xiao, J.F.; Tuli, L.; Varghese, R.S.; Zhou, B.; Tsai, T.-H.; Ranjbar, M.R.N.; Zhao, Y.; Wang, J.; Di Poto, C.; et al. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal. Chim. Acta 2012, 743, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Foglia, B.; Beltrà, M.; Sutti, S.; Cannito, S. Metabolic Reprogramming of HCC: A New Microenvironment for Immune Responses. Int. J. Mol. Sci. 2023, 24, 7463. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-A.; Wu, Q.; Chen, Z.; Zhang, W.; Zhou, Y.; Mao, K.; Li, J.; Li, Y.; Chen, J.; Huang, Y.; et al. High-resolution metabolomic biomarkers for lung cancer diagnosis and prognosis. Sci. Rep. 2021, 11, 11805. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Guo, Y.; Li, Z.-W.; Shui, G.; Tian, H.; Li, B.-W.; Kadeerhan, G.; Li, Z.-X.; Li, X.; Zhang, Y.; et al. Identification and Validation of Plasma Metabolomic Signatures in Precancerous Gastric Lesions That Progress to Cancer. JAMA Netw. Open 2021, 4, e2114186. [Google Scholar] [CrossRef]
- Patterson, A.D.; Maurhofer, O.; Beyoglu, D.; Lanz, C.; Krausz, K.W.; Pabst, T.; Gonzalez, F.J.; Dufour, J.-F.; Idle, J.R. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011, 71, 6590–6600. [Google Scholar] [CrossRef]
- Vance, J.E.; Tasseva, G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2013, 1831, 543–554. [Google Scholar] [CrossRef]
- Calzada, E.; Onguka, O.; Claypool, S.M. Phosphatidylethanolamine Metabolism in Health and Disease. Int. Rev. Cell Mol. Biol. 2016, 321, 29–88. [Google Scholar] [PubMed]
- Pandey, S.K.; Paul, A.; Shteinfer-Kuzmine, A.; Zalk, R.; Bunz, U.; Shoshan-Barmatz, V. SMAC/Diablo controls proliferation of cancer cells by regulating phosphatidylethanolamine synthesis. Mol. Oncol. 2021, 15, 3037–3061. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Sakurai, T.; Chen, Z.; Inoue, N.; Chiba, H.; Hui, S.-P. Lysophosphatidylethanolamine Affects Lipid Accumulation and Metabolism in a Human Liver-Derived Cell Line. Nutrients 2022, 14, 579. [Google Scholar] [CrossRef]
- Szlasa, W.; Zendran, I.; Zalesińska, A.; Tarek, M.; Kulbacka, J. Lipid composition of the cancer cell membrane. J. Bioenerg. Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef]
- Di Poto, C.; Ferrarini, A.; Zhao, Y.; Varghese, R.S.; Tu, C.; Zuo, Y.; Wang, M.; Ranjbar, M.R.N.; Luo, Y.; Zhang, C.; et al. Metabolomic Characterization of Hepatocellular Carcinoma in Patients with Liver Cirrhosis for Biomarker Discovery. Cancer Epidemiol. Biomarkers Prev. 2017, 26, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.T.; Elfert, A.; Helal, M.; Salama, I.; El-Said, H.; Fiehn, O. Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma. Cancers 2020, 13, 88. [Google Scholar] [CrossRef]
- Law, S.-H.; Chan, M.-L.; Marathe, G.K.; Parveen, F.; Chen, C.-H.; Ke, L.-Y. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int. J. Mol. Sci. 2019, 20, 1149. [Google Scholar] [CrossRef]
- Wang, B.; Chen, D.; Chen, Y.; Hu, Z.; Cao, M.; Xie, Q.; Chen, Y.; Xu, J.; Zheng, S.; Li, L. Metabonomic Profiles Discriminate Hepatocellular Carcinoma from Liver Cirrhosis by Ultraperformance Liquid Chromatography–Mass Spectrometry. J. Proteome Res. 2012, 11, 1217–1227. [Google Scholar] [CrossRef]
- Paul, B.; Lewinska, M.; Andersen, J.B. Lipid alterations in chronic liver disease and liver cancer. JHEP Rep. 2022, 4, 100479. [Google Scholar] [CrossRef]
- Wan, Y.; Zhang, J.; Chen, M.; Ma, M.; Sheng, B. Elevated serum triglyceride levels may be a key independent predicting factor for gallbladder cancer risk in gallbladder stone disease patients: A case–control study. BMC Endocr. Disord. 2022, 22, 270. [Google Scholar] [CrossRef]
- Van Hemelrijck, M.; Garmo, H.; Holmberg, L.; Walldius, G.; Jungner, I.; Hammar, N.; Lambe, M. Prostate cancer risk in the Swedish AMORIS study: The interplay among triglycerides, total cholesterol, and glucose. Cancer 2011, 117, 2086–2095. [Google Scholar] [CrossRef] [PubMed]
- Vatten, L.J.; Foss, O.P. Total serum cholesterol and triglycerides and risk of breast cancer: A prospective study of 24,329 Norwegian women. Cancer Res. 1990, 50, 2341–2346. [Google Scholar]
- Liu, X.; Li, M.; Wang, X.; Dang, Z.; Jiang, Y.; Wang, X.; Yang, Z. Effect of serum triglyceride level on the prognosis of patients with hepatocellular carcinoma in the absence of cirrhosis. Lipids Health Dis. 2018, 17, 248. [Google Scholar] [CrossRef] [PubMed]
- Borena, W.; Stocks, T.; Jonsson, H.; Strohmaier, S.; Nagel, G.; Bjørge, T.; Manjer, J.; Hallmans, G.; Selmer, R.; Almquist, M.; et al. Serum triglycerides and cancer risk in the metabolic syndrome and cancer (Me-Can) collaborative study. Cancer Causes Control 2011, 22, 291–299. [Google Scholar] [CrossRef]
- Mroweh, M.; Roth, G.; Decaens, T.; Marche, P.N.; Lerat, H.; Jílková, Z.M. Targeting Akt in Hepatocellular Carcinoma and Its Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 1794. [Google Scholar] [CrossRef]
- Lai, K.P.; Cheung, A.; Ho, C.H.; Tam, N.Y.-K.; Li, J.W.; Lin, X.; Chan, T.F.; Lee, N.P.-Y.; Li, R. Transcriptomic analysis reveals the oncogenic role of S6K1 in hepatocellular carcinoma. J. Cancer 2020, 11, 2645–2655. [Google Scholar] [CrossRef]
- Moon, H.; Ro, S.W. MAPK/ERK Signaling Pathway in Hepatocellular Carcinoma. Cancers 2021, 13, 3026. [Google Scholar] [CrossRef]
- Luo, X.; Zheng, E.; Wei, L.; Zeng, H.; Qin, H.; Zhang, X.; Liao, M.; Chen, L.; Zhao, L.; Ruan, X.Z.; et al. The fatty acid receptor CD36 promotes HCC progression through activating Src/PI3K/AKT axis-dependent aerobic glycolysis. Cell Death Dis. 2021, 12, 328. [Google Scholar] [CrossRef] [PubMed]

| Variables | HCC (n = 20) | LC (n = 20) | p-Value | |
|---|---|---|---|---|
| Age | Mean (SD) | 59 (6) | 58 (6) | 0.4870 |
| Gender | Male | 12 | 13 | 1.0000 |
| Race | AA | 10 | 7 | 0.5231 |
| EA | 10 | 13 | ||
| BMI | Mean (SD) | 30 (7.2) | 30 (4.8) | 0.9663 |
| Etiology | HCV | 17 | 16 | 1.0000 |
| Alcohol | 6 | 7 | 1.0000 | |
| HCV Serology | HCV Ab+ | 16 | 15 | 0.6948 |
| HBV Serology | anti HBC+ | 9 | 7 | 0.7475 |
| HBs Ag+ | 1 | 0 | 1.0000 | |
| Smoking | Current | 5 | 5 | 1.0000 |
| Former | 11 | 10 | ||
| None | 4 | 5 | ||
| Alcohol | Current | 5 | 4 | 1.0000 |
| Former | 11 | 12 | ||
| None | 4 | 4 | ||
| AFP | Median (IQR) | 29.1 (60.8) | 7.3 (35.4) | 0.3389 |
| AST | Median (IQR) | 107.5 (83.2) | 94 (70) | 0.1903 |
| ALT | Median (IQR) | 98.5 (53.2) | 53.2 (47) | 0.0695 |
| MELD | Median (IQR) | 10.5 (5.2) | 13.5 (9.3) | 0.0474 |
| Child Pugh score | Mean (SD) | 6.8 (1.8) | 9.1 (2.8) | 0.0116 |
| Median (IQR) | 6 (2.5) | 9 (5) | ||
| Child Pugh Class | A | 9 | 3 | |
| B | 7 | 8 | ||
| C | 3 | 5 | ||
| HCC Stage | Stage I | 6 | None | |
| Stage II | 13 | |||
| Stage III | 1 | |||
| Metabolite Name | Class | ID | Exact Mass | m/z | RT | Adduct | HCC vs. LC | Fold Change (HCC/LC) | Trend | |
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | FDR | |||||||||
| Pyruvic acid | Keto acids and derivatives | HMDB0000243 | 88.0148 | 87.0070 | 0.91 | [M − H]− | 0.04081 | 0.70955 | −1.29 | ↓ |
| L-Alanine | Carboxylic acids and derivatives | HMDB0000161 | 89.0475 | 90.0547 | 0.82 | [M + H]+ | 0.00740 | 0.88660 | 1.27 | ↑ |
| Proline | Carboxylic acids and derivatives | HMDB0251528 | 115.0631 | 116.0703 | 1.42 | [M + H]+ | 0.04101 | 0.97773 | 1.41 | ↑ |
| N-Undecanoylglycine | Carboxylic acids and derivatives | HMDB0013286 | 243.1828 | 266.1720 | 7.56 | [M + Na]+ | 0.00022 | 0.37484 | 1.30 | ↑ |
| N-Nonanoylglycine | Carboxylic acids and derivatives | HMDB0013279 | 215.1522 | 214.1444 | 5.90 | [M − H]− | 0.00062 | 0.50963 | 1.22 | ↑ |
| Histidylalanine | Carboxylic acids and derivatives | HMDB0028878 | 226.1166 | 225.1088 | 7.22 | [M − H]− | 0.00064 | 0.50963 | 1.60 | ↑ |
| α-Aspartylphenylalanine | Carboxylic acids and derivatives | HMDB0000706 | 280.1052 | 281.1125 | 4.81 | [M + H]+ | 0.00073 | 0.49771 | 1.85 | ↑ |
| L-Carnitine | Organonitrogen compounds | HMDB0000062 | 161.1048 | 162.1120 | 0.82 | [M + H]+ | 0.00813 | 0.88660 | 1.24 | ↑ |
| 4-Dodecylbenzenesulfonic acid | Benzene and substituted derivatives | HMDB0251568 | 326.1919 | 325.1841 | 16.45 | [M – H]− | 0.00071 | 0.50963 | 1.23 | ↑ |
| Xanthine | Imidazopyrimidines | HMDB0000292 | 152.0324 | 151.0246 | 1.19 | [M − H]− | 0.02981 | 0.70955 | 1.25 | ↑ |
| Uric acid | Imidazopyrimidines | HMDB0000289 | 168.0280 | 169.0353 | 0.89 | [M + H]+ | 0.04447 | 0.98349 | 1.32 | ↑ |
| Indoleacrylic acid | Indoles and derivatives | HMDB0000734 | 187.0630 | 188.0703 | 6.36 | [M + H]+ | 0.01981 | 0.90379 | −2.22 | ↓ |
| Sebacic acid | Fatty acyls | HMDB0000792 | 202.1200 | 201.1122 | 7.06 | [M − H]− | 0.03118 | 0.70955 | 1.34 | ↑ |
| 2-Hydroxymyristic acid | Fatty acyls | HMDB0002261 | 244.2039 | 243.1961 | 11.32 | [M − H]− | 0.01980 | 0.70955 | −1.18 | ↓ |
| Prostaglandin A2 | Fatty acyls | HMDB0002752 | 334.2128 | 333.2050 | 10.17 | [M − H]− | 0.03417 | 0.70955 | −1.44 | ↓ |
| Nervonic acid | Fatty acyls | HMDB0002368 | 366.3502 | 365.3424 | 15.99 | [M − H]− | 0.02158 | 0.70955 | −1.35 | ↓ |
| Thromboxane B2 | Fatty acyls | HMDB0003252 | 370.2361 | 369.2283 | 7.67 | [M − H]− | 0.03649 | 0.70955 | −2.51 | ↓ |
| 2-Linoleoyl Glycerol | Fatty Acyls | HMDB0245187 | 354.2760 | 355.2833 | 12.52 | [M + H]+ | 0.04812 | 0.99111 | 2.59 | ↑ |
| Glycodeoxycholic acid (GDCA) | Steroids and steroid derivatives | HMDB0000631 | 449.3141 | 432.3098 | 8.01 | [M + H − H2O]+ | 0.01730 | 0.90379 | −3.97 | ↓ |
| Glycoursodeoxycholic acid (GUDCA) | Steroids and steroid derivatives | HMDB0000708 | 449.3149 | 448.3071 | 8.02 | [M − H]− | 0.02246 | 0.70955 | −3.79 | ↓ |
| Taurochenodeoxycholic acid (TCDCA) | Steroids and steroid derivatives | HMDB0000951 | 499.2978 | 498.2900 | 8.11 | [M − H]− | 0.02940 | 0.70955 | −2.40 | ↓ |
| Testosterone sulfate | Steroids and steroid derivatives | HMDB0002833 | 368.1661 | 367.1583 | 7.72 | [M − H]− | 0.02959 | 0.70955 | 1.85 | ↑ |
| LysoPE(18:3/0:0); LPE(18:3) | Glycerophospholipids | HMDB0011509 | 475.2707 | 474.2629 | 10.94 | [M − H]− | 0.02921 | 0.70955 | 1.62 | ↑ |
| LysoPE(18:2/0:0); LPE(18:2) | Glycerophospholipids | HMDB0011507 | 477.2843 | 478.2916 | 11.88 | [M + H]+ | 0.00765 | 0.88660 | 1.62 | ↑ |
| PE(16:0/18:2); PE(34:2) | Glycerophospholipids | HMDB0008928 | 715.5165 | 714.5087 | 16.67 | [M − H]− | 0.01653 | 0.70955 | 1.66 | ↑ |
| PE(18:1/18:2); PE(36:3) | Glycerophospholipids | HMDB0009027 | 741.5324 | 740.5246 | 16.89 | [M − H]− | 0.04691 | 0.70955 | 1.38 | ↑ |
| PE(18:1/20:4); PE(38:5) | Glycerophospholipids | HMDB0009036 | 765.5323 | 764.5245 | 16.65 | [M − H]− | 0.01554 | 0.70955 | 1.60 | ↑ |
| PE(O−16:1/20:4); PE(O-36:5) | Glycerophospholipids | 723.5220 | 722.5142 | 16.55 | [M − H]− | 0.00356 | 0.62660 | 1.80 | ↑ | |
| PE(O-16:1/22:6); PE(O-38:7) | Glycerophospholipids | 747.5222 | 746.5144 | 16.45 | [M − H]− | 0.01257 | 0.70955 | 1.68 | ↑ | |
| PE(O-16:1/18:2); PE(O-34:3) | Glycerophospholipids | 699.5218 | 698.5140 | 17.04 | [M − H]− | 0.01861 | 0.70955 | 1.49 | ↑ | |
| Cer(d18:1/16:0); Cer(d34:1) | Sphingolipids | 583.5179 | 582.5101 | 16.44 | [M + HCOO] | 0.03797 | 0.70955 | 1.50 | ↑ | |
| Metabolite Name | Class | ID | Exact Mass | m/z | RT | Adduct | HCC vs. LC | Fold Change (HCC/LC) | Trend | |
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | FDR | |||||||||
| Undecanoic acid | Fatty acyls | HMDB0000947 | 186.1609 | 185.1531 | 2.17 | [M − H]− | 0.00009 | 0.00700 | −1.46 | ↓ |
| Lauric acid/Dodecanoic acid | Fatty acyls | HMDB0000638 | 200.1767 | 199.1689 | 2.88 | [M − H]− | 0.00034 | 0.01528 | −1.79 | ↓ |
| Tridecanoic acid | Fatty acyls | HMDB0000910 | 214.1925 | 213.1847 | 3.70 | [M − H]− | 0.00010 | 0.00724 | −1.44 | ↓ |
| Pentadecanoic acid | Fatty acyls | HMDB0000826 | 242.2241 | 241.2163 | 5.10 | [M − H]− | 0.00239 | 0.04856 | −1.35 | ↓ |
| Palmitic acid | Fatty acyls | HMDB0000220 | 256.2398 | 255.2320 | 5.65 | [M − H]− | 0.00058 | 0.02039 | −1.18 | ↓ |
| 2-Hydroxypalmitic acid | Fatty acyls | HMDB0031057 | 272.2351 | 271.2273 | 3.27 | [M − H]− | 0.00076 | 0.02357 | −1.37 | ↓ |
| alpha-Linolenic acid | Fatty acyls | HMDB0001388 | 278.2224 | 277.2146 | 5.09 | [M − H]− | 0.00096 | 0.02738 | −1.61 | ↓ |
| Stearic acid | Fatty acyls | HMDB0000827 | 284.2713 | 283.2635 | 6.70 | [M − H]− | 0.00008 | 0.00643 | −1.25 | ↓ |
| 12,13-EpOME | Fatty acyls | HMDB0004702 | 296.2349 | 295.2271 | 1.53 | [M − H]− | 0.00003 | 0.00311 | −1.43 | ↓ |
| Nonadecanoic acid | Fatty acyls | HMDB0000772 | 298.2870 | 297.2792 | 7.45 | [M − H]− | 0.00000 | 0.00025 | −1.38 | ↓ |
| Arachidic acid | Fatty acyls | HMDB0002212 | 312.3028 | 311.2950 | 7.71 | [M − H]− | 0.00018 | 0.01032 | −1.26 | ↓ |
| Heneicosylic acid | Fatty acyls | HMDB0002345 | 326.3182 | 325.3104 | 8.42 | [M − H]− | 0.00000 | 0.00056 | −1.34 | ↓ |
| Tricosanoic acid | Fatty acyls | HMDB0001160 | 354.3496 | 353.3418 | 9.12 | [M − H]− | 0.00045 | 0.01783 | −1.28 | ↓ |
| Pentacosanoic acid | Fatty acyls | HMDB0002361 | 382.3809 | 381.3731 | 9.98 | [M − H]− | 0.00010 | 0.00749 | −1.33 | ↓ |
| Hexacosanoic acid | Fatty acyls | HMDB0002356 | 396.3964 | 395.3886 | 10.38 | [M − H]− | 0.00004 | 0.00352 | −1.32 | ↓ |
| Heptacosanoic acid | Fatty acyls | HMDB0002063 | 410.4122 | 409.4044 | 10.76 | [M − H]− | 0.00056 | 0.02001 | −1.32 | ↓ |
| 4-Hydroxybenzaldehyde | Organooxygen compounds | HMDB0011718 | 122.0354 | 121.0276 | 0.83 | [M − H]− | 0.00138 | 0.03373 | −1.87 | ↓ |
| 2-Methylbenzoic acid | Benzene and substituted derivatives | HMDB0002340 | 136.0511 | 135.0433 | 0.83 | [M − H]− | 0.00000 | 0.00085 | −1.36 | ↓ |
| 6-Hydroxynicotinic acid | Pyridines and derivatives | HMDB0002658 | 139.0256 | 138.0178 | 0.93 | [M − H]− | 0.00072 | 0.02321 | −1.46 | ↓ |
| PC(22:5/3:0); PC(25:5) | Glycerophospholipids | NIST598 | 625.3754 | 626.3833 | 5.34 | [M + H]+ | 0.00010 | 0.00525 | −1.53 | ↓ |
| PC(22:5/16:0); PC(38:5) | Glycerophospholipids | HMDB0008692 | 807.5531 | 830.5635 | 7.77 | [M + Na]+ | 0.00141 | 0.03407 | −2.56 | ↓ |
| PC(16:0/22:6); PC(38:6) | Glycerophospholipids | HMDB0007991 | 805.54666 | 806.5545 | 7.16 | [M + H]+ | 0.00268 | 0.04897 | −2.45 | ↓ |
| PC(15:1/24:4); PC(39:5) | Glycerophospholipids | NIST3518 | 821.6588 | 822.6667 | 10.85 | [M + H]+ | 0.00024 | 0.00988 | −1.59 | ↓ |
| PC(15:1/22:6); PC(37:7) | Glycerophospholipids | NIST2714 | 789.5303 | 790.5382 | 8.94 | [M + H]+ | 0.00054 | 0.01819 | −1.27 | ↓ |
| PC(20:3/22:6); PC(42:9) | Glycerophospholipids | HMDB0008387 | 855.5665 | 856.5743 | 7.77 | [M + H]+ | 0.00187 | 0.03991 | −1.37 | ↓ |
| PC(18:0/22:0); PC(40:0) | Glycerophospholipids | HMDB0008051 | 845.6607 | 846.6685 | 10.01 | [M + H]+ | 0.00190 | 0.03991 | −1.25 | ↓ |
| PC(20:4/22:6); PC(42:10) | Glycerophospholipids | HMDB0008485 | 853.5579 | 854.5657 | 8.70 | [M + H]+ | 0.00189 | 0.03991 | −1.61 | ↓ |
| LPC(P-27:6) | Glycerophospholipids | 621.4174 | 622.4252 | 6.65 | [M + H]+ | 0.00022 | 0.00886 | −1.29 | ↓ | |
| PC(O-46:8) | Glycerophospholipids | 899.6740 | 900.6818 | 10.85 | [M + H]+ | 0.00115 | 0.03006 | −1.52 | ↓ | |
| DG(20:1/14:1/0:0); DG(34:2) | Glycerolipids | HMDB0007386 | 592.5066 | 599.5241 | 8.35 | [M + Li]+ | 0.00012 | 0.00552 | −1.34 | ↓ |
| PE(18:3/18:0); PE(36:3) | Glycerophospholipids | HMDB0009156 | 741.5303 | 740.5225 | 8.65 | [M − H]− | 0.00127 | 0.03220 | 2.93 | ↑ |
| TG(16:0/16:1/22:6); TG(54:7) | Glycerolipids | HMDB0044079 | 876.7207 | 894.7535 | 13.15 | [M + NH4]+ | 0.00005 | 0.00321 | 4.17 | ↑ |
| TG(18:3/18:2/20:5); TG(56:10) | Glycerolipids | HMDB0053065 | 898.7010 | 899.7089 | 13.15 | [M + H]+ | 0.00020 | 0.00833 | 3.70 | ↑ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, M.M.; Varghese, R.S.; Ding, Y.; Ressom, H.W. Biomarker Discovery for Hepatocellular Carcinoma in Patients with Liver Cirrhosis Using Untargeted Metabolomics and Lipidomics Studies. Metabolites 2023, 13, 1047. https://doi.org/10.3390/metabo13101047
Rashid MM, Varghese RS, Ding Y, Ressom HW. Biomarker Discovery for Hepatocellular Carcinoma in Patients with Liver Cirrhosis Using Untargeted Metabolomics and Lipidomics Studies. Metabolites. 2023; 13(10):1047. https://doi.org/10.3390/metabo13101047
Chicago/Turabian StyleRashid, Md Mamunur, Rency S. Varghese, Yuansong Ding, and Habtom W. Ressom. 2023. "Biomarker Discovery for Hepatocellular Carcinoma in Patients with Liver Cirrhosis Using Untargeted Metabolomics and Lipidomics Studies" Metabolites 13, no. 10: 1047. https://doi.org/10.3390/metabo13101047
APA StyleRashid, M. M., Varghese, R. S., Ding, Y., & Ressom, H. W. (2023). Biomarker Discovery for Hepatocellular Carcinoma in Patients with Liver Cirrhosis Using Untargeted Metabolomics and Lipidomics Studies. Metabolites, 13(10), 1047. https://doi.org/10.3390/metabo13101047






