In Vitro Evaluation of Intestinal Transport and High-Density Fermentation of Lactobacillus acidophilus
Abstract
1. Introduction
2. Material and Methods
2.1. Test Strain
2.2. Methods
2.2.1. Preparation of Bacterial Strains
2.2.2. Preliminary Selection of L. acidophilus
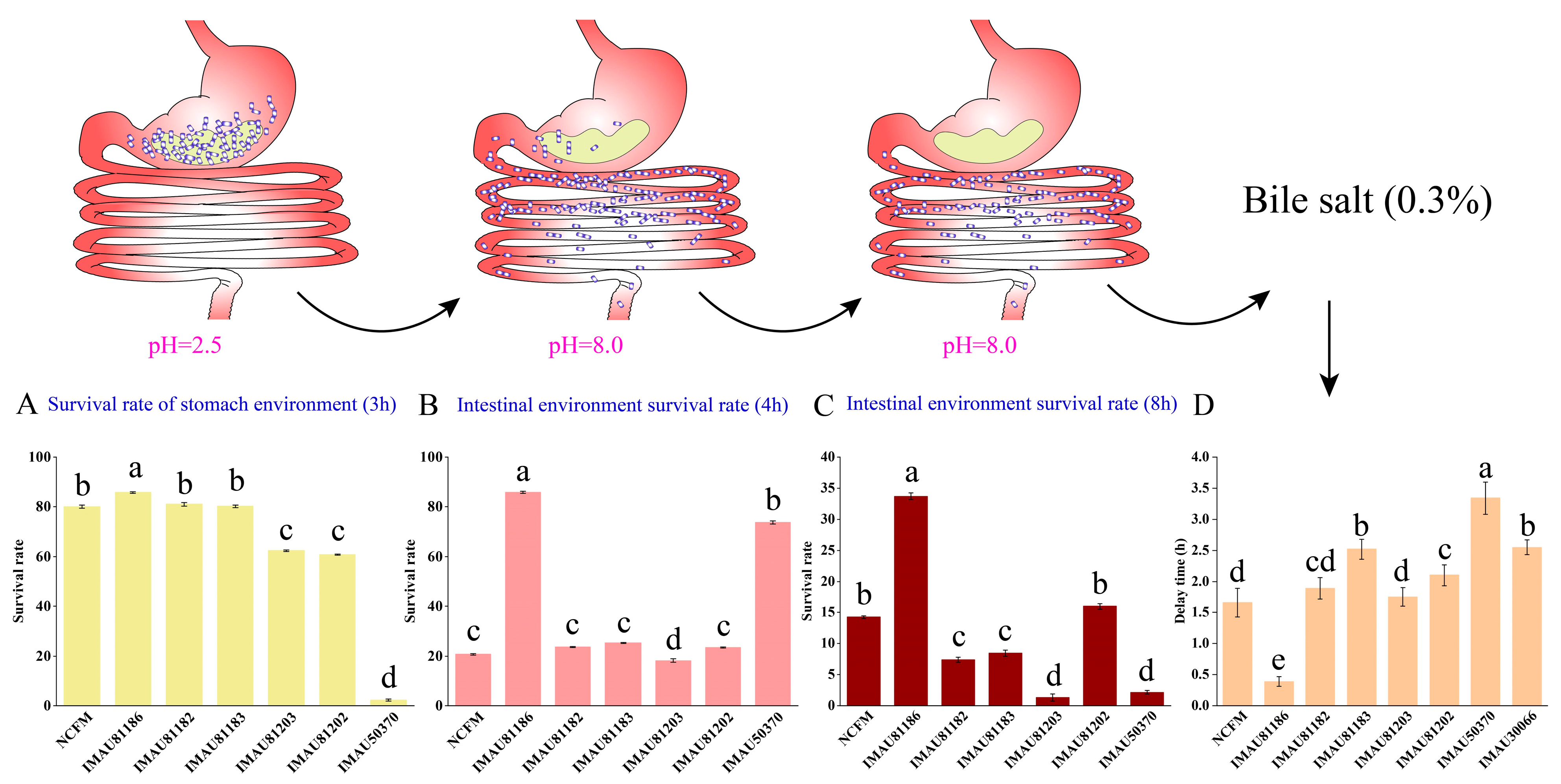
Transit Tolerance in Gastrointestinal Juices
Bile Salt Tolerance of L. acidophilus
2.2.3. High Density Culture
Optimization of Static Culture Conditions
Optimization of Culture Medium Composition
Optimization of High-Density Fermentation Process
Growth Curve of L. acidophilus
2.2.4. Transcriptional Studies at Different Growth Stages
RNA Extraction
Transcriptome Analyses
2.2.5. Statistical Analysis
3. Results and Discussion
3.1. Gastrointestinal Fluid Tolerance and Bile Salt Tolerance
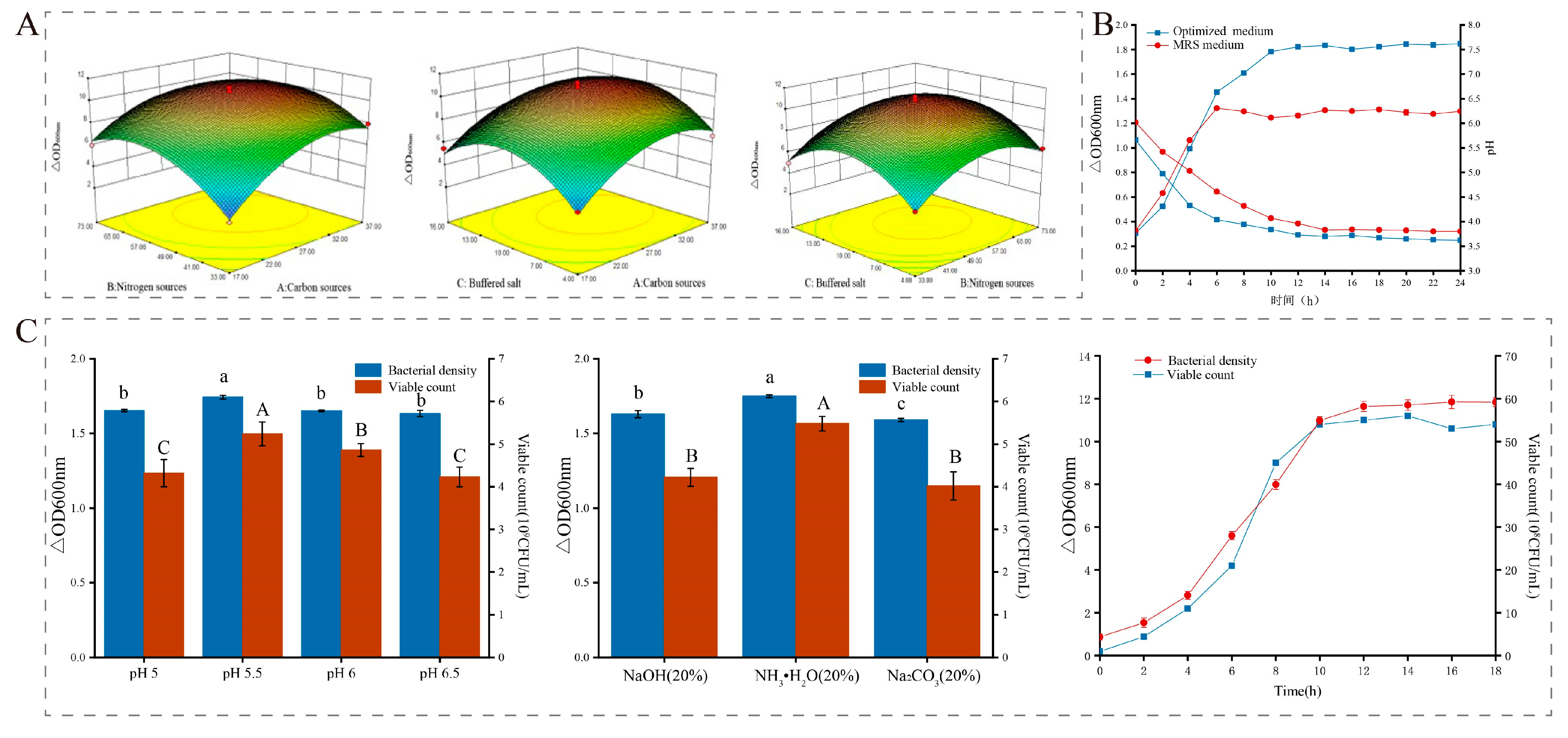
3.2. Optimization of Culture Conditions
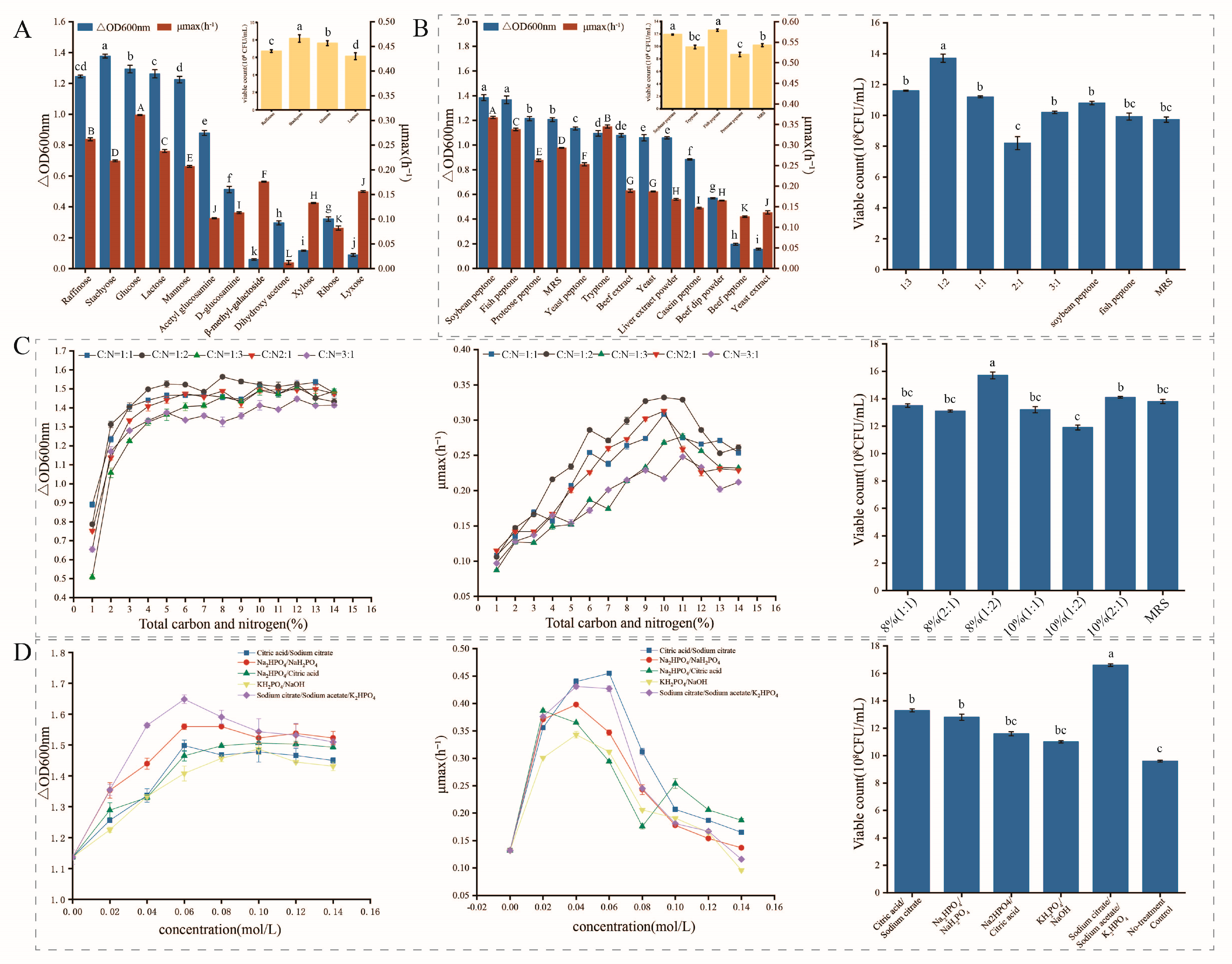
3.3. Optimization of Media
3.3.1. Carbon and Nitrogen Source Optimization
3.3.2. Buffer Salt Optimization
3.3.3. Growth Factor Optimization
3.3.4. Response Surface Optimization
3.4. High-Density Fermentation Technology
3.5. Transcriptional Analysis
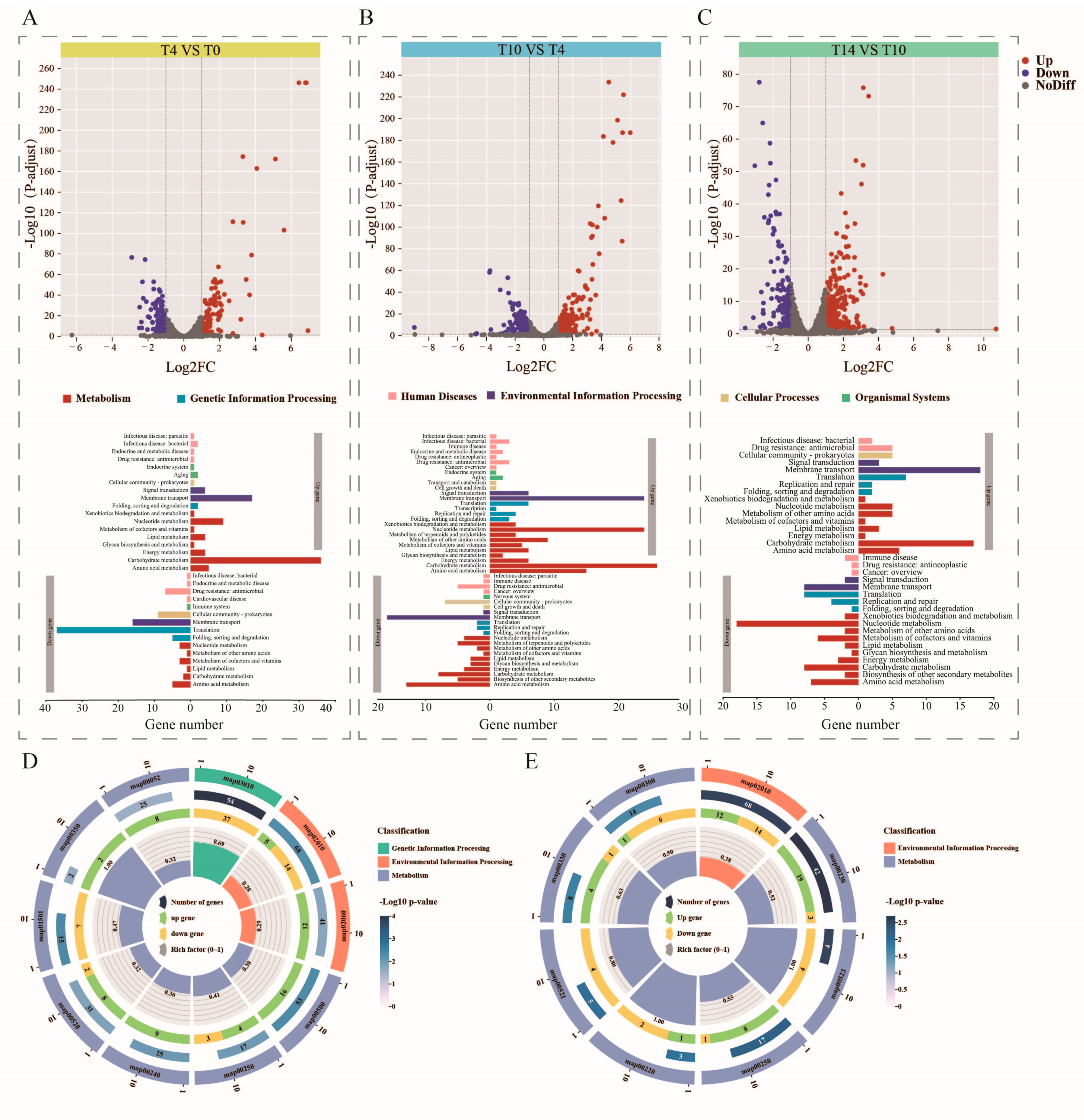
3.5.1. Identification of Differentially Expressed Genes at Various Growth Stages
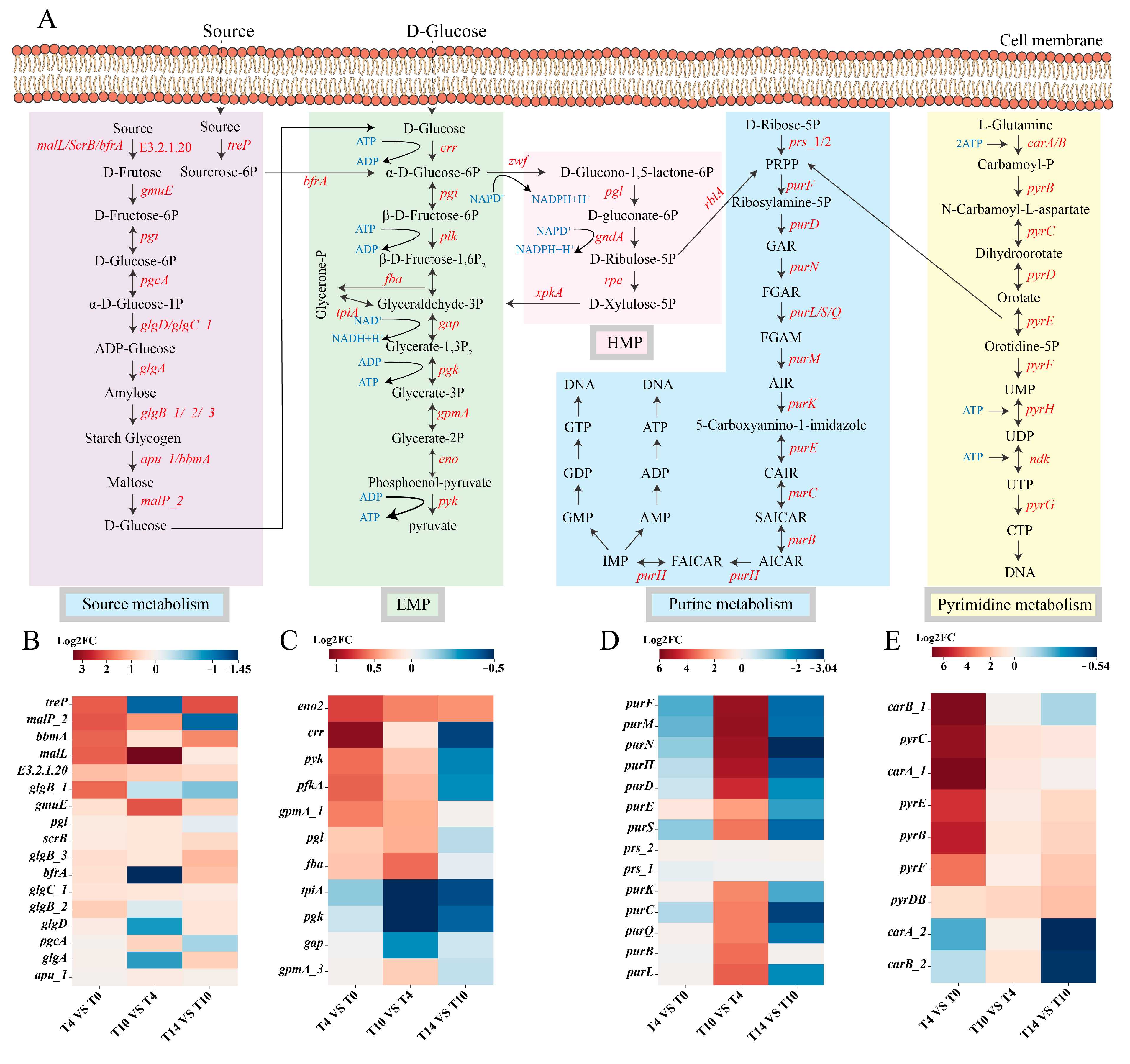
3.5.2. Characterization of Functional Genes in L. acidophilus at Different Growth Phase by KEGG Pathway Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farré-Maduell, E.; Casals-Pascual, C. The origins of gut microbiome research in Europe: From Escherich to Nissle. Hum. Microbiome J. 2019, 14, 100065. [Google Scholar] [CrossRef]
- Klotz, C.; Goh, Y.J.; O’Flaherty, S.; Barrangou, R. S-layer associated proteins contribute to the adhesive and immunomodulatory properties of Lactobacillus acidophilus NCFM. BMC Microbiol. 2020, 20, 248. [Google Scholar]
- Lopes, L.A.A.; Pimentel, T.C.; Carvalho, R.D.S.F.; Madruga, M.S.; de Sousa Galvão, M.; Bezerra, T.K.A.; Barão, C.E.; Magnani, M.; Stamford, T.C.M. Spreadable goat Ricotta cheese added with Lactobacillus acidophilus La-05: Can microencapsulation improve the probiotic survival and the quality parameters? Food Chem. 2021, 346, 128769. [Google Scholar] [CrossRef] [PubMed]
- Amiri, S.; Rezazadeh Bari, M.; Alizadeh Khaledabad, M.; Rezaei Mokarram, R.; Sowti Khiabani, M. Co-production of parabiotic metabolites by Lactobacillus acidophilus LA5 and Bifidobacterium animalis subsp. lactis BB12 in dairy effluents. Chem. Rev. Lett. 2021, 4, 66–76. [Google Scholar]
- Jameel, A.A.; Abdullah, A.R.; Hussein, S.I. The antibacterial and antibiofilm activity of Lactobacillus acidophilus isolated from vagina against methicillin-resistant Staphylococcus aureus. Egypt. Pharm. J. 2022, 21, 84. [Google Scholar]
- Huang, Z.; Zhou, X.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Comparative genomics and specific functional characteristics analysis of Lactobacillus acidophilus. Microorganisms 2021, 9, 1992. [Google Scholar] [CrossRef]
- Kim, H.; Jeong, Y.; Kim, J.-E.; Kim, Y.; Paek, N.-S.; Kang, C.-H. Anti-obesity potential of Lactobacillus spp. isolated from infant feces. Biotechnol. Bioprocess Eng. 2021, 26, 575–585. [Google Scholar] [CrossRef]
- Sunu, P.; Sunarti, D.; Mahfudz, L.D.; Yunianto, V.D. Effect of synbiotic from Allium sativum and Lactobacillus acidophilus on hematological indices, antioxidative status and intestinal ecology of broiler chicken. J. Saudi Soc. Agric. Sci. 2021, 20, 103–110. [Google Scholar]
- Dar, A.; Singh, S.; Rahman, J.; Ahmad, S. The effects of probiotic Lactobacillus acidophilus and/or prebiotic mannan oligosaccharides on growth performance, nutrient utilization, blood metabolites, faecal bacteria, and economics of crossbred calves. Iran J. Vet. Res. 2022, 23, 322–330. [Google Scholar]
- Gao, H.; Li, X.; Chen, X.; Hai, D.; Wei, C.; Zhang, L.; Li, P. The functional roles of Lactobacillus acidophilus in different physiological and pathological processes. J. Microbiol. Biotechnol. 2022, 32, 1226. [Google Scholar]
- Ouwehand, A.C.; Nermes, M.; Collado, M.C.; Rautonen, N.; Salminen, S.; Isolauri, E. Specific probiotics alleviate allergic rhinitis during the birch pollen season. World J. Gastroenterol. 2009, 15, 3261. [Google Scholar] [PubMed]
- Zhuo, Q.; Yu, B.; Zhou, J.; Zhang, J.; Zhang, R.; Xie, J.; Wang, Q.; Zhao, S. Lysates of Lactobacillus acidophilus combined with CTLA-4-blocking antibodies enhance antitumor immunity in a mouse colon cancer model. Sci. Rep. 2019, 9, 20128. [Google Scholar] [PubMed]
- Yang, A.; Liao, Y.; Zhu, J.; Zhang, J.; Wu, Z.; Li, X.; Tong, P.; Chen, H.; Wang, S.; Liu, Z. Screening of anti-allergy Lactobacillus and its effect on allergic reactions in BALB/c mice sensitized by soybean protein. J. Funct. Foods 2021, 87, 104858. [Google Scholar]
- Lokapirnasari, W.P.; Sahidu, A.M.; Maslachah, L.; Yulianto, A.; Najwan, R. The effect of combination Bifidobacterium sp. and Lactobacillus acidophilus probiotic on egg yolk cholesterol, HDL, and LDL. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2020; p. 012049. [Google Scholar]
- Shoaib, A.; Xin, L.; Xin, Y. Oral administration of Lactobacillus acidophilus alleviates exacerbations in Pseudomonas aeruginosa and Staphylococcus aureus pulmonary infections. Pak. J. Pharm. Sci. 2019, 32, 1621–1630. [Google Scholar] [PubMed]
- Hosoda, M.; Hashimoto, H.; He, F.; Morita, H.; Hosono, A. Effect of administration of milk fermented with Lactobacillus acidophilus LA-2 on fecal mutagenicity and microflora in the human intestine. J. Dairy Sci. 1996, 79, 745–749. [Google Scholar] [CrossRef]
- Hove, H.; Nørgaard, H.; Brøbech Mortensen, P. Lactic acid bacteria and the human gastrointestinal tract. Eur. J. Clin. Nutr. 1999, 53, 339–350. [Google Scholar] [CrossRef]
- Bull, M.; Plummer, S.; Marchesi, J.; Mahenthiralingam, E. The life history of Lactobacillus acidophilus as a probiotic: A tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol. Lett. 2013, 349, 77–87. [Google Scholar] [CrossRef]
- Papizadeh, M.; Rohani, M.; Nahrevanian, H.; Hosseini, S.N.; Shojaosadati, S.A.; Pourshafie, M.R. Using various approaches of design of experiments for high cell density production of the functionally probiotic Lactobacillus plantarum strain RPR42 in a cane molasses-based medium. Curr. Microbiol. 2020, 77, 1756–1766. [Google Scholar]
- Osborne, R. Manufacture of starters: Production of frozen concentrated cheese starters by diffusion culture. Int. J. Dairy Technol. 1977, 30, 40–44. [Google Scholar] [CrossRef]
- Krzywonos, M.; Eberhard, T. High density process to cultivate Lactobacillus plantarum biomass using wheat stillage and sugar beet molasses. Electron. J. Biotechnol. 2011, 14, 6. [Google Scholar] [CrossRef]
- Elsayed, E.A.; Othman, N.Z.; Malek, R.; Tang, T.; El Enshasy, H. Improvement of cell mass production of Lactobacillus delbrueckii sp. bulgaricus WICC-B-02: A newly isolated probiotic strain from mother’s milk. J. Appl. Pharm. Sci. 2014, 4, 008–014. [Google Scholar]
- Liu, M.; Yang, L.; Cai, M.; Feng, C.; Zhao, Z.; Yang, D.; Ding, P. Transcriptome analysis reveals important candidate gene families related to oligosaccharides biosynthesis in Morinda officinalis. Plant Physiol. Biochem. 2021, 167, 1061–1071. [Google Scholar] [PubMed]
- Yang, L.; Tang, J.; Chen, J.-J.; Peng, A.-Y.; Wang, Q.-M.; Rao, L.-Q.; Yang, H.; Zhang, X.-W.; Yang, H.-Z.; Zhang, C. Transcriptome analysis of three cultivars of Poria cocos reveals genes related to the biosynthesis of polysaccharides. J. Asian Nat. Prod. Res. 2019, 21, 462–475. [Google Scholar] [CrossRef]
- Li, X.; Hu, D.; Tian, Y.; Song, Y.; Hou, Y.; Sun, L.; Zhang, Y.; Man, C.; Zhang, W.; Jiang, Y. Protective effects of a novel Lactobacillus rhamnosus strain with probiotic characteristics against lipopolysaccharide-induced intestinal inflammation in vitro and in vivo. Food Funct. 2020, 11, 5799–5814. [Google Scholar] [CrossRef]
- Pacheco, K.C.; del Toro, G.V.; Martínez, F.R.; Durán-Páramo, E. Viability of LACTOBACILLUS delbrueckii under human gastrointestinal conditions simulated in vitro. Am. J. Agric. Biol. Sci. 2010, 5, 37–42. [Google Scholar] [CrossRef]
- Dong, Z.; Gu, L.; Zhang, J.; Wang, M.; Du, G.; Chen, J.; Li, H. Optimisation for high cell density cultivation of Lactobacillus salivarius BBE 09-18 with response surface methodology. Int. Dairy J. 2014, 34, 230–236. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Differential analysis of count data–the DESeq2 package. Genome Biol. 2014, 15, 10–1186. [Google Scholar]
- Kim, Y.; Whang, J.Y.; Whang, K.Y.; Oh, S.; Kim, S.H. Characterization of the cholesterol-reducing activity in a cell-free supernatant of Lactobacillus acidophilus ATCC 43121. Biosci. Biotechnol. Biochem. 2008, 72, 1483–1490. [Google Scholar] [CrossRef]
- Goldin, B.R.; Gorbach, S.L. The effect of milk and lactobacillus feeding on human intestinal bacterial enzyme activity. Am. J. Clin. Nutr. 1984, 39, 756–761. [Google Scholar] [CrossRef]
- Cook, M.T.; Tzortzis, G.; Charalampopoulos, D.; Khutoryanskiy, V.V. Production and evaluation of dry alginate-chitosan microcapsules as an enteric delivery vehicle for probiotic bacteria. Biomacromolecules 2011, 12, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- Conway, P.; Gorbach, S.; Goldin, B. Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J. Dairy Sci. 1987, 70, 1–12. [Google Scholar] [CrossRef]
- Pedram, N.; Ataei, S. Optimization of a modified gS medium for a probiotic strain (L. acidophilus ATCC4356). Appl. Food Biotechnol. 2014, 1, 25–29. [Google Scholar]
- Wang, L.; Wu, L.; Chen, Q.; Li, S.; Zhu, Y.; Wu, J.; Chu, J.; Wu, S. Development of sugarcane resource for efficient fermentation of exopolysaccharide by using a novel strain of Kosakonia cowanii LT-1. Bioresour. Technol. 2019, 280, 247–254. [Google Scholar] [CrossRef]
- Xu, X.; Nie, Z.; Zheng, Z.; Zhu, L.; Zhan, X. Production and rheological properties of welan gum produced by Sphingomonas sp. ATCC 31555 with different nitrogen sources. Microbiol. Physlol. 2017, 27, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Safari, R.; Motamedzadegan, A.; Ovissipour, M.; Regenstein, J.M.; Gildberg, A.; Rasco, B. Use of hydrolysates from yellowfin tuna (Thunnus albacares) heads as a complex nitrogen source for lactic acid bacteria. Food Bioprocess Technol. 2012, 5, 73–79. [Google Scholar] [CrossRef]
- Wu, P.; Li, J.-Z.; Wang, Y.-L.; Liu, X.-S.; Du, C.; Tong, Q.-Y.; Li, N. Promoting the growth of Rubrivivax gelatinosus in sewage purification by the addition of magnesium ions. Biochem. Eng. J. 2014, 91, 66–71. [Google Scholar] [CrossRef]
- Amouzou, K.; Prevost, H.; Divies, C. Effects of milk magnesium supplementation on lactic acid fermentation by Streptococcus lactis and Streptococcus thermophilus. Lait 1985, 65, 21–34. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Li, Y.; Li, S.; Li, Q.; Fan, W.; Kiatoukosin, L.; Chen, J. Extracellular polysaccharides of endophytic fungus Alternaria tenuissima F1 from Angelica sinensis: Production conditions, purification, and antioxidant properties. Int. J. Biol. Macromol. 2019, 133, 172–183. [Google Scholar]
- Rettger, L.F.; Cheplin, H.A. Therapeutic application of Bacillus acidophilus. Proc. Soc. Exp. Biol. Med. 1921, 19, 72–76. [Google Scholar] [CrossRef]
- Meena, G.S.; Kumar, N.; Majumdar, G.C.; Banerjee, R.; Meena, P.K.; Yadav, V. Growth characteristics modeling of Lactobacillus acidophilus using RSM and ANN. Braz. Arch. Biol. Technol. 2014, 57, 15–22. [Google Scholar] [CrossRef]
- Weisberger, D.; Johnson, F.G. Nutritional requirements for an oral strain of Lactobacillus acidophilus. J. Dent. Res. 1946, 25, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Choi, J.W.; Jhun, J.; Kwon, J.Y.; Lee, B.-I.; Yang, C.W.; Park, S.-H.; Cho, M.-L. Lactobacillus acidophilus improves intestinal inflammation in an acute colitis mouse model by regulation of Th17 and Treg cell balance and fibrosis development. J. Med. Food 2018, 21, 215–224. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, X.; Menghe, B.; Zhang, H.; Liu, W. In Vitro Evaluation of Intestinal Transport and High-Density Fermentation of Lactobacillus acidophilus. Metabolites 2023, 13, 1077. https://doi.org/10.3390/metabo13101077
Su X, Menghe B, Zhang H, Liu W. In Vitro Evaluation of Intestinal Transport and High-Density Fermentation of Lactobacillus acidophilus. Metabolites. 2023; 13(10):1077. https://doi.org/10.3390/metabo13101077
Chicago/Turabian StyleSu, Xin, Bilige Menghe, Heping Zhang, and Wenjun Liu. 2023. "In Vitro Evaluation of Intestinal Transport and High-Density Fermentation of Lactobacillus acidophilus" Metabolites 13, no. 10: 1077. https://doi.org/10.3390/metabo13101077
APA StyleSu, X., Menghe, B., Zhang, H., & Liu, W. (2023). In Vitro Evaluation of Intestinal Transport and High-Density Fermentation of Lactobacillus acidophilus. Metabolites, 13(10), 1077. https://doi.org/10.3390/metabo13101077






