Hibernation-Induced microRNA Expression Promotes Signaling Pathways and Cell Cycle Dysregulation in Ictidomys tridecemlineatus Cardiac Tissue
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Collection
2.2. RNA Extraction
2.3. Small RNA Sequencing
2.4. Read Processing
2.5. Differential Expression Analysis and Clustering
2.6. Gene Set Analysis
2.7. Statistical Analysis and Visualization
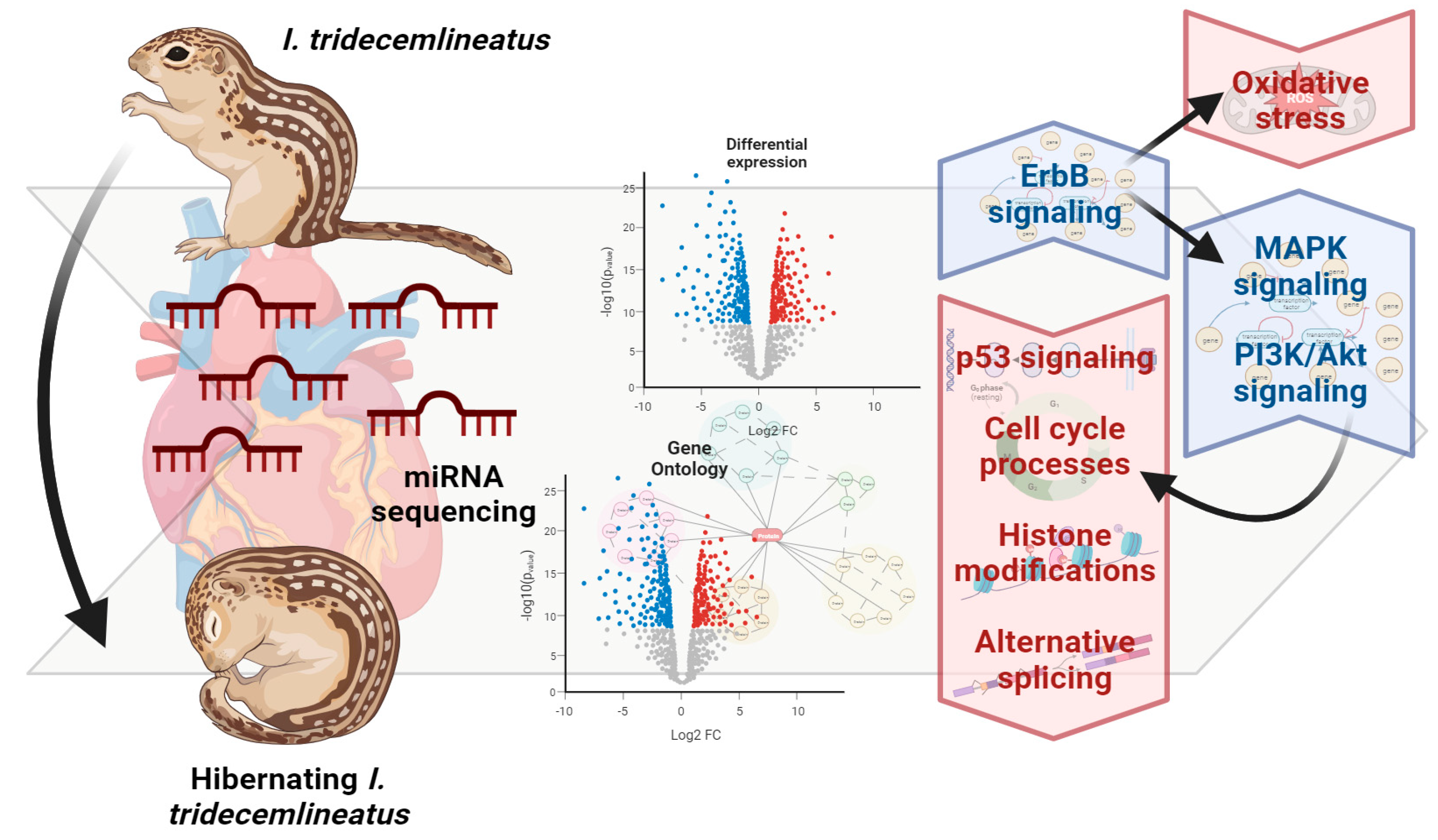
3. Results
3.1. Small RNA Sequencing Summary
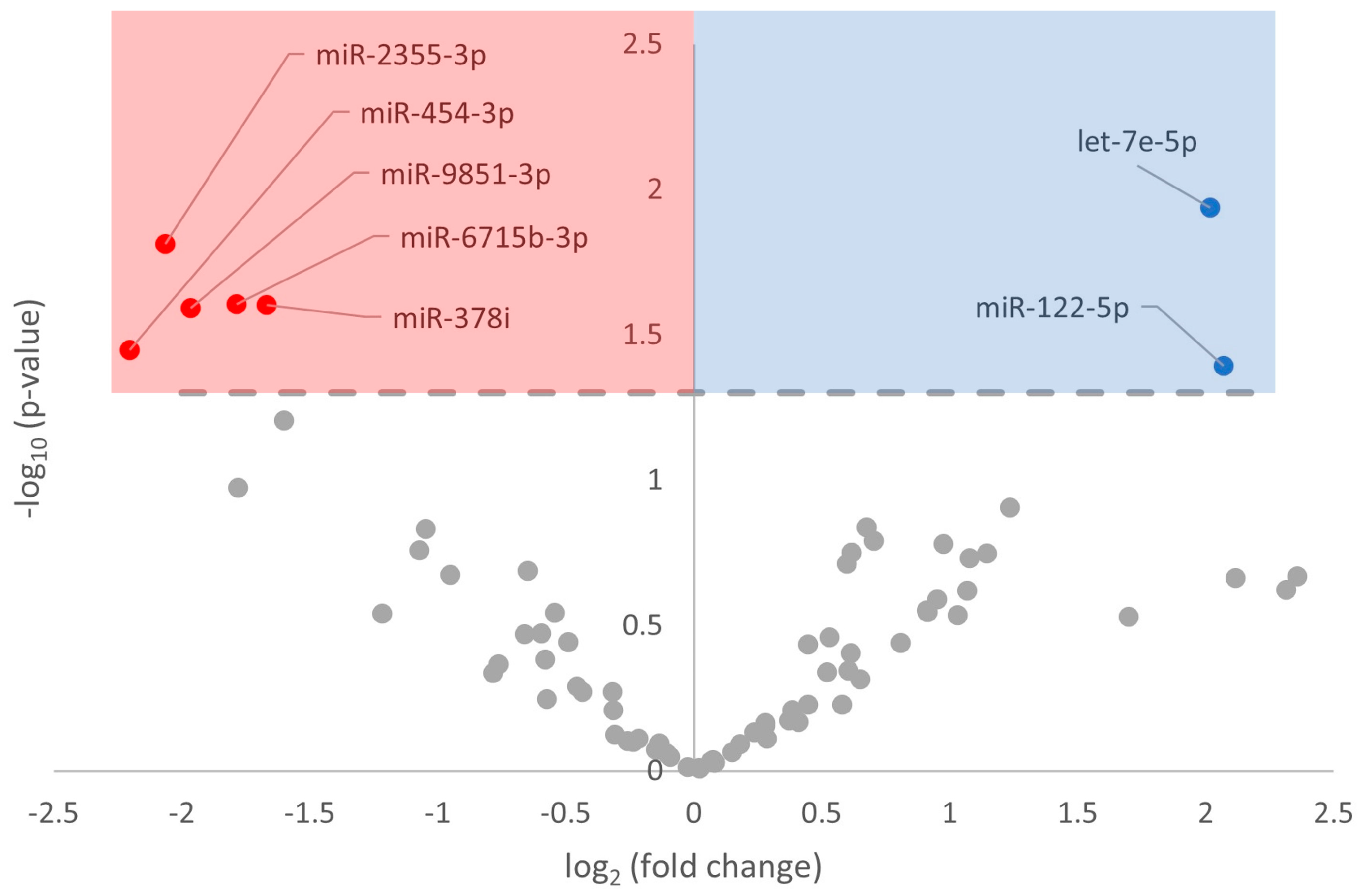
3.2. Differential Expression of miRNA in Response to Hibernation
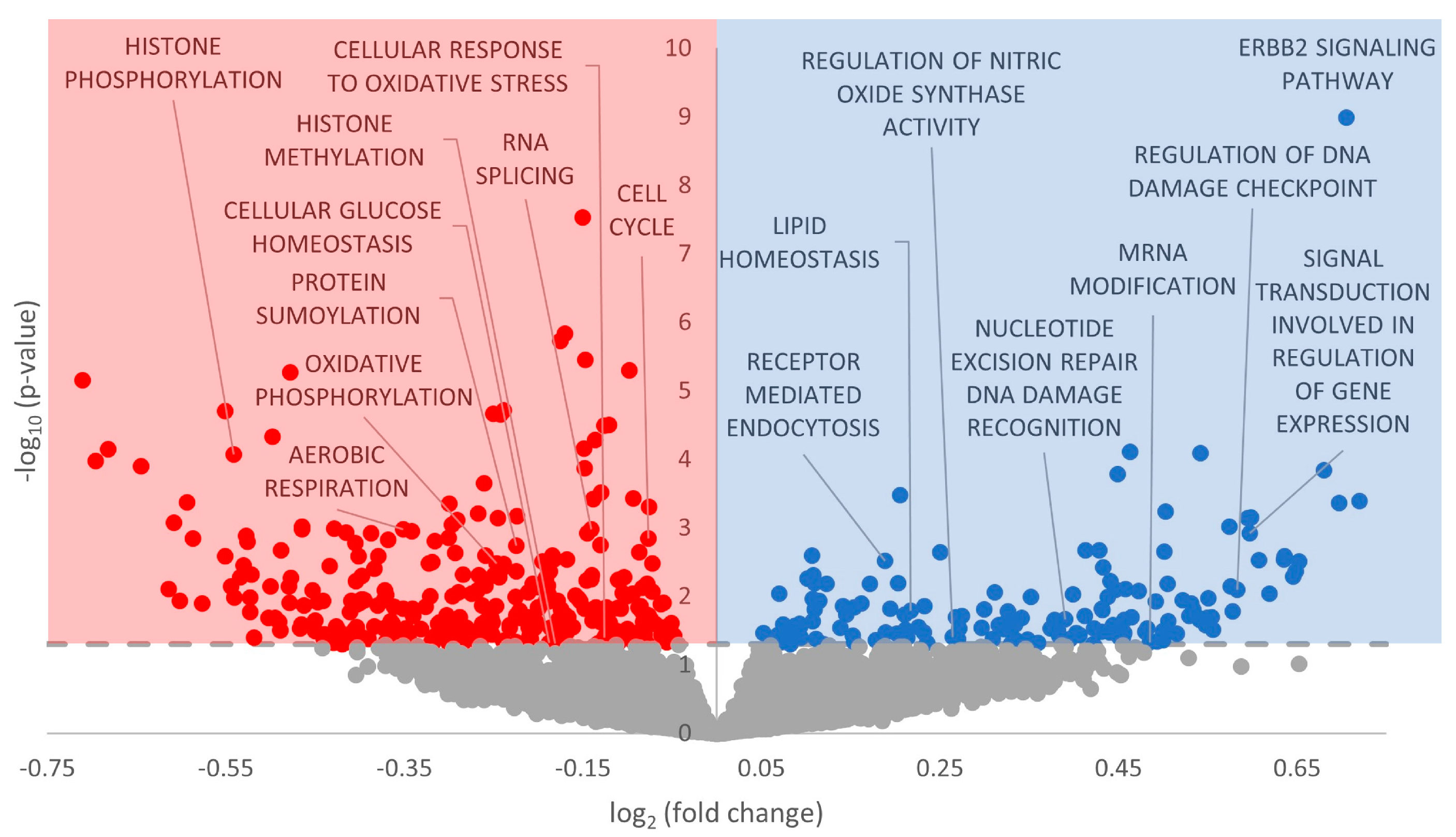
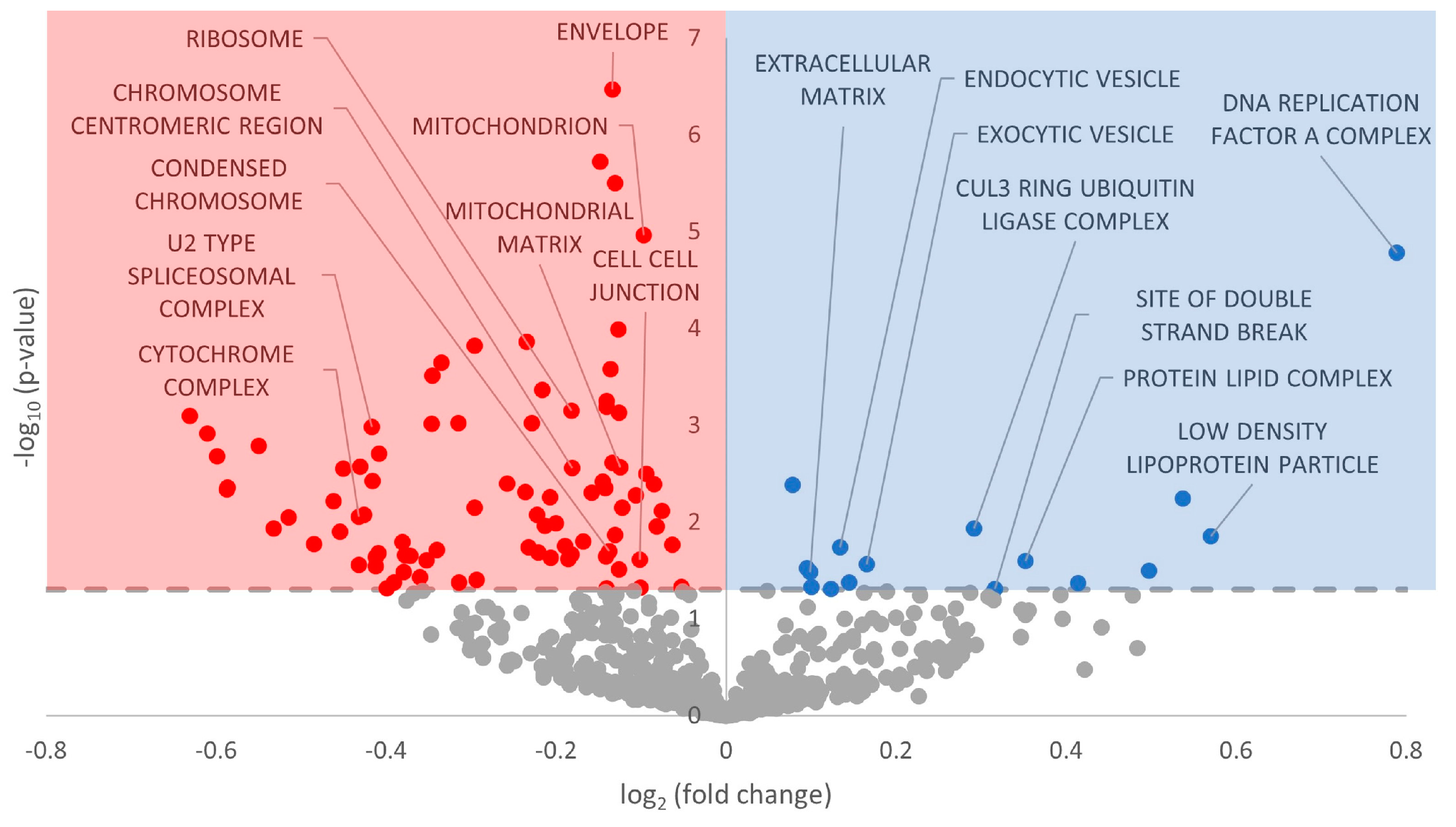
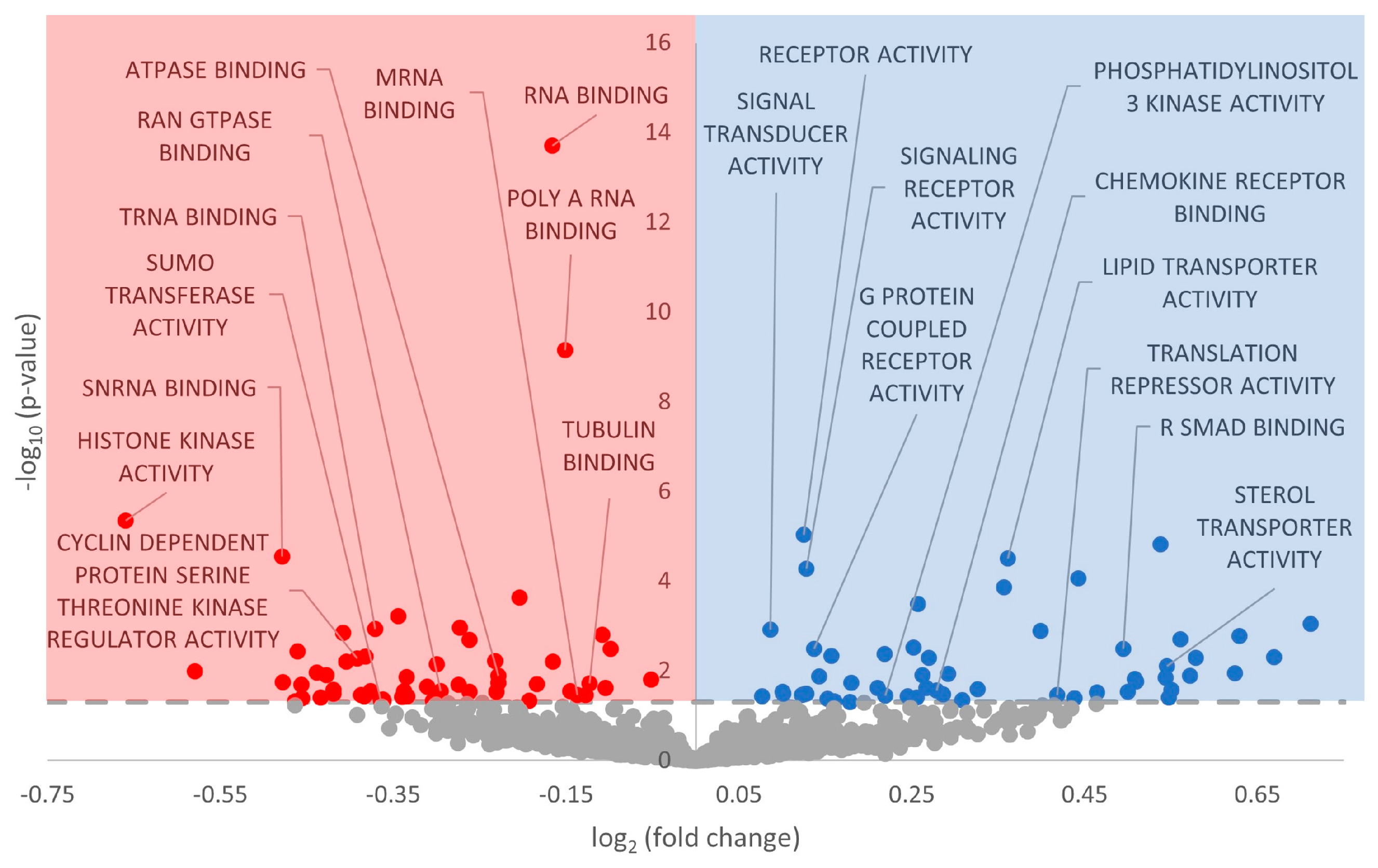
3.3. Gene Ontology Terms Enriched for Differentially Expressed miRNA
3.4. KEGG Pathways with Reduced miRNA Regulation
4. Discussion
4.1. Upregulation of ErbB2 Signaling and Implications in Downstream Signaling
4.2. Cell Cycle Processes Appear Heavily Downregulated by miRNA
4.3. Downregulation of p53 Signaling
4.4. Global MRD and Other Modes of Regulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carey, H.V.; Andrews, M.T.; Martin, S.L. Mammalian Hibernation: Cellular and Molecular Responses to Depressed Metabolism and Low Temperature. Physiol. Rev. 2003, 83, 1153–1181. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.C.H.; Lee, T.F. Torpor and Hibernation in Mammals: Metabolic, Physiological, and Biochemical Adaptations. Compr. Physiol. 1996, 65, 507–532. [Google Scholar] [CrossRef]
- Sonsalla, M.M.; Love, S.L.; Hoh, L.J.; Summers, L.N.; Follett, H.M.; Bojang, A.; Duddleston, K.N.; Kurtz, C.C. Development of Metabolic Inflammation during Pre-Hibernation Fattening in 13-Lined Ground Squirrels (Ictidomys tridecemlineatus). J. Comp. Physiol. B 2021, 191, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Metabolic Rate Depression. The Biochemistry of Mammalian Hibernation. In Advances in Clinical Chemistry; Makowski, G., Ed.; Academic Press: Burlington, MA, USA, 2010; pp. 77–78. [Google Scholar]
- Wang, L.C.H.; Wolowyk, M.W. Torpor in Mammals and Birds. Can. J. Zool. 1988, 66, 133–137. [Google Scholar] [CrossRef]
- Barros, R.C.H.; Zimmer, M.E.; Branco, L.G.S.; Milsom, W.K. Hypoxic Metabolic Response of the Golden-Mantled Ground Squirrel. J. Appl. Physiol. 2001, 91, 603–612. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic Rate Depression and Biochemical Adaptation in Anaerobiosis, Hibernation and Estivation. Q. Rev. Biol. 1990, 65, 145–174. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Freeze Tolerance in Animals. Physiol. Rev. 1988, 68, 27–84. [Google Scholar] [CrossRef]
- Guppy, M.; Withers, P. Metabolic Depression in Animals: Physiological Perspectives and Biochemical Generalizations. Biol. Rev. Camb. Philos. Soc. 1999, 74, 1–40. [Google Scholar] [CrossRef]
- Storey, K.B. Mammalian Hibernation: Transcriptional and Translational Controls. In Hypoxia: Through the Lifecycle; Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Storey, K.B.; Storey, J.M. Metabolic Rate Depression in Animals: Transcriptional and Translational Controls. Biol. Rev. Camb. Philos. Soc. 2004, 79, 207–233. [Google Scholar] [CrossRef]
- Buck, L.T.; Hochachka, P.W. Anoxic Suppression of Na(+)-K(+)-ATPase and Constant Membrane Potential in Hepatocytes: Support for Channel Arrest. Am. J. Physiol. 1993, 265, R1020–R1025. [Google Scholar] [CrossRef]
- Hochachka, P.W.; Buck, L.T.; Doll, C.J.; Land, S.C. Unifying Theory of Hypoxia Tolerance: Molecular/Metabolic Defense and Rescue Mechanisms for Surviving Oxygen Lack. Proc. Natl. Acad. Sci. USA 1996, 93, 9493–9498. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.; Storey, K. Perspectives in Cell Cycle Regulation: Lessons from an Anoxic Vertebrate. Curr. Genom. 2009, 10, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Storey, K.B. Pattern of Cellular Quiescence over the Hibernation Cycle in Liver of Thirteen-Lined Ground Squirrels. Cell Cycle 2012, 11, 1714–1726. [Google Scholar] [CrossRef]
- Storey, K.B. Hypometabolism and the Cell Cycle. Cell Cycle 2012, 11, 1665. [Google Scholar] [CrossRef][Green Version]
- Chazarin, B.; Storey, K.B.; Ziemianin, A.; Chanon, S.; Plumel, M.; Chery, I.; Durand, C.; Evans, A.L.; Arnemo, J.M.; Zedrosser, A.; et al. Metabolic Reprogramming Involving Glycolysis in the Hibernating Brown Bear Skeletal Muscle. Front. Zool. 2019, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Churchill, T.A.; Cheetham, K.M.; Simpkin, S.; Green, C.J.; Wang, L.C.H.; Fuller, B.J. Liver Metabolism in Cold Hypoxia: A Comparison of Energy Metabolism and Glycolysis in Cold-Sensitive and Cold-Resistant Mammals. J. Comp. Physiol. B 1994, 164, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hadj-Moussa, H.; Storey, K.B. Marine Periwinkle Stress-Responsive MicroRNAs: A Potential Factor to Reflect Anoxia and Freezing Survival Adaptations. Genomics 2020, 112, 4385–4398. [Google Scholar] [CrossRef]
- English, S.G.; Hadj-Moussa, H.; Storey, K.B. MicroRNAs Regulate Survival in Oxygen-Deprived Environments. J. Exp. Biol. 2018, 221, jeb190579. [Google Scholar] [CrossRef]
- Lyons, P.J.; Poitras, J.J.; Courteau, L.A.; Storey, K.B.; Morin, P.J. Identification of Differentially Regulated MicroRNAs in Cold-Hardy Insects. Cryo-Letters 2013, 34, 83–89. [Google Scholar]
- Biggar, K.K.; Storey, K.B. The Emerging Roles of MicroRNAs in the Molecular Responses of Metabolic Rate Depression. J. Mol. Cell Biol. 2011, 3, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Luu, B.E.; Lefai, E.; Giroud, S.; Swenson, J.E.; Chazarin, B.; Gauquelin-Koch, G.; Arnemo, J.M.; Evans, A.L.; Bertile, F.; Storey, K.B. MicroRNAs Facilitate Skeletal Muscle Maintenance and Metabolic Suppression in Hibernating Brown Bears. J. Cell Physiol. 2020, 235, 3984–3993. [Google Scholar] [CrossRef] [PubMed]
- Hadj-Moussa, H.; Zhang, J.; Pifferi, F.; Perret, M.; Storey, K.B. Profiling Torpor-Responsive MicroRNAs in Muscles of the Hibernating Primate Microcebus Murinus. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194473. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Biggar, K.K.; Storey, K.B. Expression Profiling and Structural Characterization of MicroRNAs in Adipose Tissues of Hibernating Ground Squirrels. Genom. Proteom. Bioinform. 2014, 12, 284–291. [Google Scholar] [CrossRef]
- Kornfeld, S.F.; Biggar, K.K.; Storey, K.B. Differential Expression of Mature MicroRNAs Involved in Muscle Maintenance of Hibernating Little Brown Bats, Myotis Lucifugus: A Model of Muscle Atrophy Resistance. Genom. Proteom. Bioinform. 2012, 10, 295–301. [Google Scholar] [CrossRef]
- Logan, S.M.; Storey, K.B. MicroRNA Expression Patterns in the Brown Fat of Hibernating 13-Lined Ground Squirrels. Genomics 2021, 113, 769–781. [Google Scholar] [CrossRef]
- Morin, P., Jr.; Dubuc, A.; Storey, K.B. Differential Expression of MicroRNA Species in Organs of Hibernating Ground Squirrels: A Role in Translational Suppression during Torpor. Biochim. Biophys. Acta Gene Regul. Mech. 2008, 1779, 628–633. [Google Scholar] [CrossRef]
- Wu, C.W.; Biggar, K.K.; Luu, B.E.; Szereszewski, K.E.; Storey, K.B. Analysis of MicroRNA Expression during the Torpor-Arousal Cycle of a Mammalian Hibernator, the 13-Lined Ground Squirrel. Physiol. Genom. 2016, 48, 388–396. [Google Scholar] [CrossRef]
- Ingelson-Filpula, W.A.; Storey, K.B. Muscles in Winter: The Epigenetics of Metabolic Arrest. Epigenomes 2021, 5, 28. [Google Scholar] [CrossRef]
- Luu, B.E.; Biggar, K.K.; Wu, C.W.; Storey, K.B. Torpor-Responsive Expression of Novel MicroRNA Regulating Metabolism and Other Cellular Pathways in the Thirteen-Lined Ground Squirrel, Ictidomys tridecemlineatus. FEBS Lett. 2016, 590, 3574–3582. [Google Scholar] [CrossRef]
- Lang-Ouellette, D.; Morin, P.J. Differential Expression of MiRNAs with Metabolic Implications in Hibernating Thirteen-Lined Ground Squirrels, Ictidomys tridecemlineatus. Mol. Cell Biochem. 2014, 394, 291–298. [Google Scholar] [CrossRef]
- Rourke, B.C.; Cotton, C.J.; Harlow, H.J.; Caiozzo, V.J. Maintenance of Slow Type I Myosin Protein and MRNA Expression in Overwintering Prairie Dogs (Cynomys leucurus and ludovicianus) and Black Bears (Ursus americanus). J. Comp. Physiol. B 2006, 176, 709–720. [Google Scholar] [CrossRef]
- Shavlakadze, T.; Grounds, M. Of Bears, Frogs, Meat, Mice and Men: Complexity of Factors Affecting Skeletal Muscle Mass and Fat. Bioessays 2006, 28, 994–1009. [Google Scholar] [CrossRef]
- Merriman, D.K.; Lahvis, G.; Jooss, M.; Gesicki, J.A.; Schill, K. Current Practices in a Captive Breeding Colony of 13-Lined Ground Squirrels (Ictidomys tridecemlineatus). Lab Anim. 2012, 41, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Hadj-Moussa, H.; Pamenter, M.E.; Storey, K.B. Hypoxic Naked Mole-Rat Brains Use MicroRNA to Coordinate Hypometabolic Fuels and Neuroprotective Defenses. J. Cell Physiol. 2021, 236, 5080–5097. [Google Scholar] [CrossRef]
- Ingelson-Filpula, W.A.; Cheng, H.; Eaton, L.; Pamenter, M.E.; Storey, K.B. Small RNA Sequencing in Hypoxic Naked Mole-Rat Hearts Suggests MicroRNA Regulation of RNA- and Translation-Related Processes. FEBS Lett. 2022, 596, 2821–2833. [Google Scholar] [CrossRef]
- Zhang, J.; Hadj-Moussa, H.; Storey, K.B. Current Progress of High-Throughput MicroRNA Differential Expression Analysis and Random Forest Gene Selection for Model and Non-Model Systems: An R Implementation. J. Integr. Bioinform. 2016, 13, 35–46. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FASTQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2015. [Google Scholar]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a Genome-Centric Resource for Non-Coding RNA Families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Sai Lakshmi, S.; Agrawal, S. PiRNABank: A Web Resource on Classified and Clustered Piwi-Interacting RNAs. Nucleic Acids Res. 2008, 36 (Suppl. S1), D173–D177. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Zhang, J.; Storey, K.B. RBiomirGS: An All-in-One MiRNA Gene Set Analysis Solution Featuring Target MRNA Mapping and Expression Profile Integration. PeerJ 2018, 2018, e4262. [Google Scholar] [CrossRef]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New Approach for Understanding Genome Variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Garcia-Garcia, F.; Panadero, J.; Dopazo, J.; Montaner, D. Integrated Gene Set Analysis for MicroRNA Studies. Bioinformatics 2016, 32, 2809–2816. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.; Botvinnik, O.; Hobson, P.; Warmenhoven, J.; Cole, J.B.; Halchenko, Y.; Vanderplas, J.; Hoyer, S.; Villalba, S.; Quintero, E.; et al. Seaborn: Statistical Data Visualization. 2014. Available online: https://seaborn.pydata.org/ (accessed on 5 October 2023).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Liaw, W.H.A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M.; et al. Gplots: Various R Programming Tools for Plotting Data. R Package Version 3.0.1.1. 2019. Available online: http://CRAN.R-project.org/package=gplots (accessed on 16 May 2023).
- Graus-Porta, D.; Beerli, R.R.; Daly, J.M.; Hynes, N.E. ErbB-2, the Preferred Heterodimerization Partner of All ErbB Receptors, Is a Mediator of Lateral Signaling. EMBO J. 1997, 16, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Birchmeier, C. Multiple Essential Functions of Neuregulin in Development. Nature 1995, 378, 386–390. [Google Scholar] [CrossRef]
- D’Uva, G.; Aharonov, A.; Lauriola, M.; Kain, D.; Yahalom-Ronen, Y.; Carvalho, S.; Weisinger, K.; Bassat, E.; Rajchman, D.; Yifa, O.; et al. ERBB2 Triggers Mammalian Heart Regeneration by Promoting Cardiomyocyte Dedifferentiation and Proliferation. Nat. Cell Biol. 2015, 17, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Okoshi, K.; Nakayama, M.; Yan, X.; Okoshi, M.P.; Schuldt, A.J.T.; Marchionni, M.A.; Lorell, B.H. Neuregulins Regulate Cardiac Parasympathetic Activity: Muscarinic Modulation of Beta-Adrenergic Activity in Myocytes from Mice with Neuregulin-1 Gene Deletion. Circulation 2004, 110, 713–717. [Google Scholar] [CrossRef]
- Kuramochi, Y.; Cote, G.M.; Guo, X.; Lebrasseur, N.K.; Cui, L.; Liao, R.; Sawyer, D.B. Cardiac Endothelial Cells Regulate Reactive Oxygen Species-Induced Cardiomyocyte Apoptosis through Neuregulin-1beta/ErbB4 Signaling. J. Biol. Chem. 2004, 279, 51141–51147. [Google Scholar] [CrossRef]
- Baliga, R.R.; Pimental, D.R.; Zhao, Y.Y.; Simmons, W.W.; Marchionni, M.A.; Sawyer, D.B.; Kelly, R.A. NRG-1-Induced Cardiomyocyte Hypertrophy. Role of PI-3-Kinase, P70(S6K), and MEK-MAPK-RSK. Am. J. Physiol. 1999, 277, H2026–H2037. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Sawyer, D.R.; Baliga, R.R.; Opel, D.J.; Han, X.; Marchionni, M.A.; Kelly, R.A. Neuregulins Promote Survival and Growth of Cardiac Myocytes. Persistence of ErbB2 and ErbB4 Expression in Neonatal and Adult Ventricular Myocytes. J. Biol. Chem. 1998, 273, 10261–10269. [Google Scholar] [CrossRef]
- Lemmens, K.; Segers, V.F.M.; Demolder, M.; De Keulenaer, G.W. Role of Neuregulin-1/ErbB2 Signaling in Endothelium-Cardiomyocyte Cross-Talk. J. Biol. Chem. 2006, 281, 19469–19477. [Google Scholar] [CrossRef]
- Klichkhanov, N.K.; Nikitina, E.R.; Shihamirova, Z.M.; Astaeva, M.D.; Chalabov, S.I.; Krivchenko, A.I. Erythrocytes of Little Ground Squirrels Undergo Reversible Oxidative Stress During Arousal from Hibernation. Front. Physiol. 2021, 12, 730657. [Google Scholar] [CrossRef]
- Orr, A.L.; Lohse, L.A.; Drew, K.L.; Hermes-Lima, M. Physiological Oxidative Stress after Arousal from Hibernation in Arctic Ground Squirrel. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2009, 153, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, J.; Xu, S.; Peng, X.; Yan, X.; Li, X.; Wang, H.; Chang, H.; Gao, Y. Controllable Oxidative Stress and Tissue Specificity in Major Tissues during the Torpor-Arousal Cycle in Hibernating Daurian Ground Squirrels. Open Biol. 2018, 8, 180068. [Google Scholar] [CrossRef] [PubMed]
- Carey, H.V.; Frank, C.L.; Seifert, J.P. Hibernation Induces Oxidative Stress and Activation of NK-KappaB in Ground Squirrel Intestine. J. Comp. Physiol. B 2000, 170, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Hermes-Lima, M.; Zenteno-Savín, T. Animal Response to Drastic Changes in Oxygen Availability and Physiological Oxidative Stress. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2002, 133, 537–556. [Google Scholar] [CrossRef]
- Duffy, B.M.; Staples, J.F. Arousal from Torpor Increases Oxidative Damage in the Hibernating Thirteen-Lined Ground Squirrel (Ictidomys tridecemlineatus). Physiol. Biochem. Zool. 2022, 95, 229–238. [Google Scholar] [CrossRef]
- Childers, C.L.; Tessier, S.N.; Storey, K.B. The Heart of a Hibernator: EGFR and MAPK Signaling in Cardiac Muscle during the Hibernation of Thirteen-Lined Ground Squirrels, Ictidomys tridecemlineatus. PeerJ 2019, 7, e7587. [Google Scholar] [CrossRef]
- Yue, T.L.; Wang, C.; Gu, J.L.; Ma, X.L.; Kumar, S.; Lee, J.C.; Feuerstein, G.Z.; Thomas, H.; Maleeff, B.; Ohlstein, E.H. Inhibition of Extracellular Signal-Regulated Kinase Enhances Ischemia/Reoxygenation-Induced Apoptosis in Cultured Cardiac Myocytes and Exaggerates Reperfusion Injury in Isolated Perfused Heart. Circ. Res. 2000, 86, 692–699. [Google Scholar] [CrossRef]
- Huang, S.; Lv, Z.; Guo, Y.; Li, L.; Zhang, Y.; Zhou, L.; Yang, B.; Wu, S.; Zhang, Y.; Xie, C.; et al. Identification of Blood Let-7e-5p as a Biomarker for Ischemic Stroke. PLoS ONE 2016, 11, e0163951. [Google Scholar] [CrossRef]
- Abnous, K.; Dieni, C.A.; Storey, K.B. Regulation of Akt during Hibernation in Richardson’s Ground Squirrels. Biochim. Biophys. Acta BBA Gen. Subj. 2008, 1780, 185–193. [Google Scholar] [CrossRef]
- Hadj-Moussa, H.; Moggridge, J.A.; Luu, B.E.; Quintero-Galvis, J.F.; Gaitán-Espitia, J.D.; Nespolo, R.F.; Storey, K.B. The Hibernating South American Marsupial, Dromiciops gliroides, Displays Torpor-Sensitive MicroRNA Expression Patterns. Sci. Rep. 2016, 6, 24627. [Google Scholar] [CrossRef]
- Tessier, S.N.; Zhang, J.; Biggar, K.K.; Wu, C.W.; Pifferi, F.; Perret, M.; Storey, K.B. Regulation of the PI3K/AKT Pathway and Fuel Utilization During Primate Torpor in the Gray Mouse Lemur, Microcebus murinus. Genom. Proteom. Bioinform. 2015, 13, 91–102. [Google Scholar] [CrossRef]
- Fleck, C.C.; Carey, H.V. Modulation of Apoptotic Pathways in Intestinal Mucosa during Hibernation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 289, R586–R595. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Storey, K.B. Evidence for Cell Cycle Suppression and MicroRNA Regulation of Cyclin D1 during Anoxia Exposure in Turtles. Cell Cycle 2012, 11, 1705–1713. [Google Scholar] [CrossRef]
- Zhang, J.; Storey, K.B. Cell Cycle Regulation in the Freeze-Tolerant Wood Frog, Rana sylvatica. Cell Cycle 2012, 11, 1727–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, M.; Yin, Y.; Storey, K.B. MiR-200-3p Is Potentially Involved in Cell Cycle Arrest by Regulating Cyclin A during Aestivation in Apostichopus japonicus. Cells 2019, 8, 843. [Google Scholar] [CrossRef]
- Zhang, J.; Biggar, K.K.; Storey, K.B. Regulation of P53 by Reversible Post-Transcriptional and Post-Translational Mechanisms in Liver and Skeletal Muscle of an Anoxia Tolerant Turtle, Trachemys Scripta Elegans. Gene 2013, 513, 147–155. [Google Scholar] [CrossRef]
- Hefler, J.; Wu, C.W.; Storey, K.B. Transcriptional Activation of P53 during Cold Induced Torpor in the 13-Lined Ground Squirrel Ictidomys tridecemlineatus. Biochem. Res. Int. 2015, 2015, 731595. [Google Scholar] [CrossRef]
- Pan, P.; Treat, M.D.; Van Breukelen, F. A Systems-Level Approach to Understanding Transcriptional Regulation by P53 during Mammalian Hibernation. J. Exp. Biol. 2014, 217, 2489–2498. [Google Scholar] [CrossRef]
- Tessier, S.N.; Luu, B.E.; Smith, J.C.; Storey, K.B. The Role of Global Histone Post-Translational Modifications during Mammalian Hibernation. Cryobiology 2017, 75, 28–36. [Google Scholar] [CrossRef]
- Chuang, J.C.; Jones, P.A. Epigenetics and MicroRNAs. Pediatr. Res. 2007, 61, 24–29. [Google Scholar] [CrossRef]
- Storey, K.B. Regulation of Hypometabolism: Insights into Epigenetic Controls. J. Exp. Biol. 2015, 218, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, O.; Convertini, P.; Zhang, Z.; Wen, Y.; Shen, M.; Falaleeva, M.; Stamm, S. Function of Alternative Splicing. Gene 2013, 514, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Hallenbeck, J.M. SUMO and Ischemic Tolerance. Neuromol. Med. 2013, 15, 771–781. [Google Scholar] [CrossRef]
- Lee, Y.j.; Johnson, K.R.; Hallenbeck, J.M. Global Protein Conjugation by Ubiquitin-like-Modifiers during Ischemic Stress Is Regulated by MicroRNAs and Confers Robust Tolerance to Ischemia. PLoS ONE 2012, 7, e47787. [Google Scholar] [CrossRef]
- Rouble, A.N.; Hawkins, L.J.; Storey, K.B. Roles for Lysine Acetyltransferases during Mammalian Hibernation. J. Therm. Biol. 2018, 74, 71–76. [Google Scholar] [CrossRef]
- Watts, A.J.; Storey, K.B. Hibernation Impacts Lysine Methylation Dynamics in the 13-Lined Ground Squirrel, Ictidomys tridecemlineatus. J. Exp. Zool. A Ecol. Integr. Physiol. 2019, 331, 234–244. [Google Scholar] [CrossRef]
- Biggar, Y.; Storey, K.B. Global DNA Modifications Suppress Transcription in Brown Adipose Tissue during Hibernation. Cryobiology 2014, 69, 333–338. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ingelson-Filpula, W.A.; Storey, K.B. Hibernation-Induced microRNA Expression Promotes Signaling Pathways and Cell Cycle Dysregulation in Ictidomys tridecemlineatus Cardiac Tissue. Metabolites 2023, 13, 1096. https://doi.org/10.3390/metabo13101096
Ingelson-Filpula WA, Storey KB. Hibernation-Induced microRNA Expression Promotes Signaling Pathways and Cell Cycle Dysregulation in Ictidomys tridecemlineatus Cardiac Tissue. Metabolites. 2023; 13(10):1096. https://doi.org/10.3390/metabo13101096
Chicago/Turabian StyleIngelson-Filpula, W. Aline, and Kenneth B. Storey. 2023. "Hibernation-Induced microRNA Expression Promotes Signaling Pathways and Cell Cycle Dysregulation in Ictidomys tridecemlineatus Cardiac Tissue" Metabolites 13, no. 10: 1096. https://doi.org/10.3390/metabo13101096
APA StyleIngelson-Filpula, W. A., & Storey, K. B. (2023). Hibernation-Induced microRNA Expression Promotes Signaling Pathways and Cell Cycle Dysregulation in Ictidomys tridecemlineatus Cardiac Tissue. Metabolites, 13(10), 1096. https://doi.org/10.3390/metabo13101096






