The Metabolomic Characteristics and Dysregulation of Fatty Acid Esters of Hydroxy Fatty Acids in Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Breast Tissue
2.2. Chemicals and Reagents
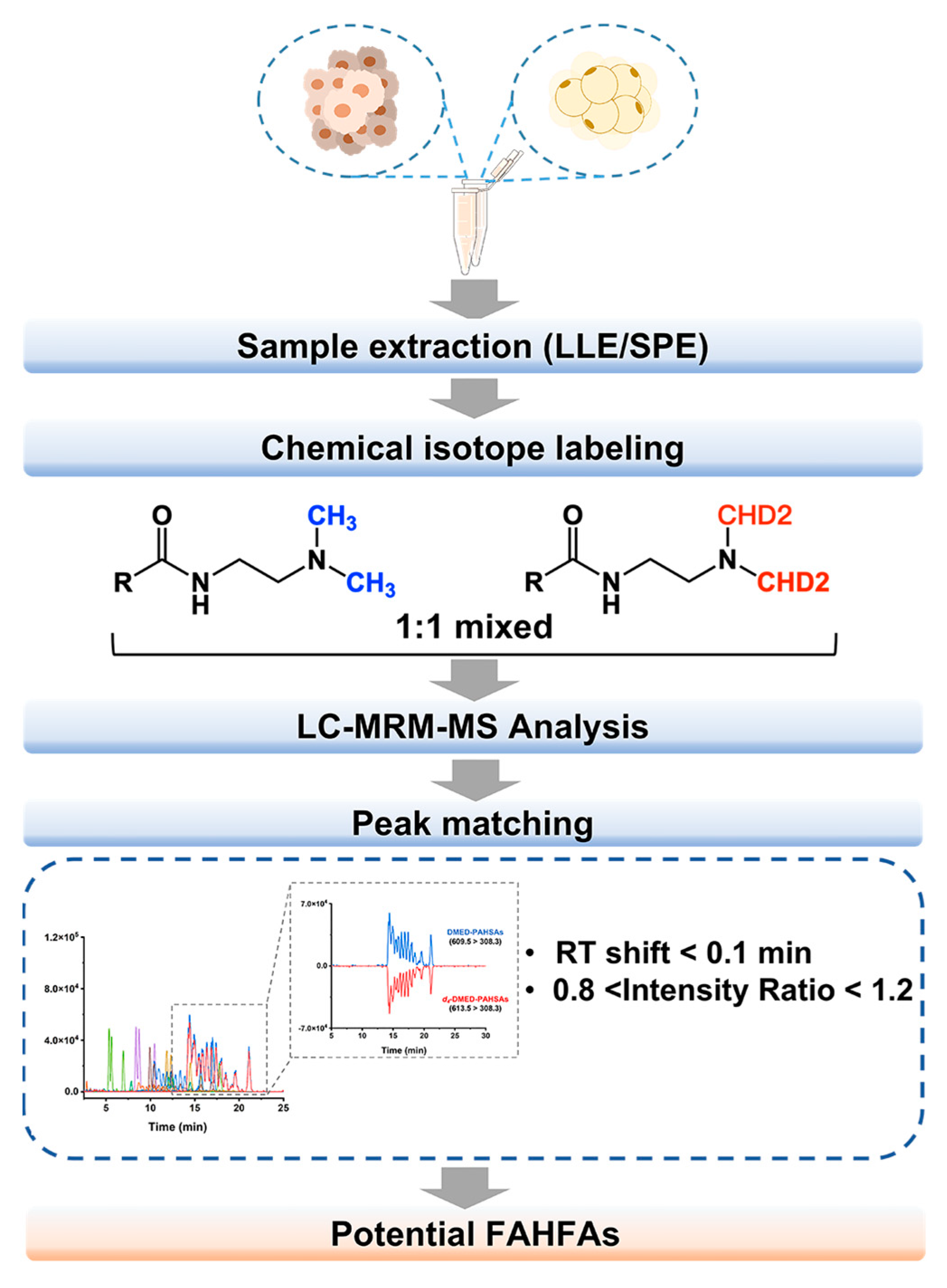
2.3. Sample Pretreatment
2.4. Mass Analysis
2.5. Statistical Analysis
3. Results
3.1. Screening and Annotation of FAHFAs in BC Tissue
3.2. FAHFA Alterations in Breast Cancer Tissue
3.3. Relationship between BC-Associated FAHFAs and HFAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Ginsburg, O.; Bray, F.; Coleman, M.P.; Vanderpuye, V.; Eniu, A.; Kotha, S.R.; Sarker, M.; Huong, T.T.; Allemani, C.; Dvaladze, A.; et al. The Global Burden of Women’s Cancers: A Grand Challenge in Global Health. Lancet 2017, 389, 847–860. [Google Scholar] [CrossRef]
- Guo, R.; Chen, Y.; Borgard, H.; Jijiwa, M.; Nasu, M.; He, M.; Deng, Y. The Function and Mechanism of Lipid Molecules and Their Roles in The Diagnosis and Prognosis of Breast Cancer. Molecules 2020, 25, 4864. [Google Scholar] [CrossRef]
- Yang, K.; Wang, X.; Song, C.; He, Z.; Wang, R.; Xu, Y.; Jiang, G.; Wan, Y.; Mei, J.; Mao, W. The Role of Lipid Metabolic Reprogramming in Tumor Microenvironment. Theranostics 2023, 13, 1774–1808. [Google Scholar] [CrossRef]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a Class of Endogenous Mammalian Lipids with Anti-Diabetic and Anti-Inflammatory Effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef]
- Syed, I.; Lee, J.; Moraes-Vieira, P.M.; Donaldson, C.J.; Sontheimer, A.; Aryal, P.; Wellenstein, K.; Kolar, M.J.; Nelson, A.T.; Siegel, D.; et al. Palmitic Acid Hydroxystearic Acids Activate GPR40, Which Is Involved in Their Beneficial Effects on Glucose Homeostasis. Cell Metab. 2018, 27, 419–427.e4. [Google Scholar] [CrossRef]
- Simone, V.; D’Avenia, M.; Argentiero, A.; Felici, C.; Rizzo, F.M.; De Pergola, G.; Silvestris, F. Obesity and Breast Cancer: Molecular Interconnections and Potential Clinical Applications. Oncologist 2016, 21, 404–417. [Google Scholar] [CrossRef]
- Brown, K.A. Metabolic Pathways in Obesity-Related Breast Cancer. Nat. Rev. Endocrinol. 2021, 17, 350–363. [Google Scholar] [CrossRef]
- Chou, P.; Choi, H.H.; Huang, Y.; Fuentes-Mattei, E.; Velazquez-Torres, G.; Zhang, F.; Phan, L.; Lee, J.; Shi, Y.; Bankson, J.A.; et al. Impact of Diabetes on Promoting the Growth of Breast Cancer. Cancer Commun. 2021, 41, 414–431. [Google Scholar] [CrossRef]
- Quail, D.F.; Dannenberg, A.J. The Obese Adipose Tissue Microenvironment in Cancer Development and Progression. Nat. Rev. Endocrinol. 2019, 15, 139–154. [Google Scholar] [CrossRef]
- Kang, C.; LeRoith, D.; Gallagher, E.J. Diabetes, Obesity, and Breast Cancer. Endocrinology 2018, 159, 3801–3812. [Google Scholar] [CrossRef] [PubMed]
- Brezinova, M.; Kuda, O.; Hansikova, J.; Rombaldova, M.; Balas, L.; Bardova, K.; Durand, T.; Rossmeisl, M.; Cerna, M.; Stranak, Z.; et al. Levels of Palmitic Acid Ester of Hydroxystearic Acid (PAHSA) Are Reduced in the Breast Milk of Obese Mothers. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2018, 1863, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Holm, J.B.; Rosendahl, A.H.; Borgquist, S. Local Biomarkers Involved in the Interplay between Obesity and Breast Cancer. Cancers 2021, 13, 6286. [Google Scholar] [CrossRef] [PubMed]
- Benlebna, M.; Balas, L.; Gaillet, S.; Durand, T.; Coudray, C.; Casas, F.; Feillet-Coudray, C. Potential Physio-Pathological Effects of Branched Fatty Acid Esters of Hydroxy Fatty Acids. Biochimie 2021, 182, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Tan, Z.; Yu, J.; Peng, F.; Guo, J.; Meng, W.; Chen, Y.; Rao, T.; Liu, Z.; Peng, J. A Conjunctive Lipidomic Approach Reveals Plasma Ethanolamine Plasmalogens and Fatty Acids as Early Diagnostic Biomarkers for Colorectal Cancer Patients. Expert Rev. Proteom. 2020, 17, 233–242. [Google Scholar] [CrossRef]
- Lee, J.; Moraes-Vieira, P.M.; Castoldi, A.; Aryal, P.; Yee, E.U.; Vickers, C.; Parnas, O.; Donaldson, C.J.; Saghatelian, A.; Kahn, B.B. Branched Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs) Protect against Colitis by Regulating Gut Innate and Adaptive Immune Responses. J. Biol. Chem. 2016, 291, 22207–22217. [Google Scholar] [CrossRef]
- Rodríguez, J.P.; Guijas, C.; Astudillo, A.M.; Rubio, J.M.; Balboa, M.A.; Balsinde, J. Sequestration of 9-Hydroxystearic Acid in FAHFA (Fatty Acid Esters of Hydroxy Fatty Acids) as a Protective Mechanism for Colon Carcinoma Cells to Avoid Apoptotic Cell Death. Cancers 2019, 11, 524. [Google Scholar] [CrossRef]
- Cheng, H.; Wang, M.; Su, J.; Li, Y.; Long, J.; Chu, J.; Wan, X.; Cao, Y.; Li, Q. Lipid Metabolism and Cancer. Life 2022, 12, 784. [Google Scholar] [CrossRef]
- Hao, Y.H.; Zhang, Z.; Wang, L.; Liu, C.; Lei, A.W.; Yuan, B.F.; Feng, Y.Q. Stable Isotope Labeling Assisted Liquid Chromatography–Electrospray Tandem Mass Spectrometry for Quantitative Analysis of Endogenous Gibberellins. Talanta 2015, 144, 341–348. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Zhu, Q.-F.; Yan, J.-W.; Ni, J.; Feng, Y.-Q. FAHFA Footprint in the Visceral Fat of Mice across Their Lifespan. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2020, 1865, 158639. [Google Scholar] [CrossRef]
- Zheng, S.-J.; Liu, S.-J.; Zhu, Q.-F.; Guo, N.; Wang, Y.-L.; Yuan, B.-F.; Feng, Y.-Q. Establishment of Liquid Chromatography Retention Index Based on Chemical Labeling for Metabolomic Analysis. Anal. Chem. 2018, 90, 8412–8420. [Google Scholar] [CrossRef]
- Zhu, Q.-F.; Yan, J.-W.; Gao, Y.; Zhang, J.-W.; Yuan, B.-F.; Feng, Y.-Q. Highly Sensitive Determination of Fatty Acid Esters of Hydroxyl Fatty Acids by Liquid Chromatography-Mass Spectrometry. J. Chromatogr. B 2017, 1061–1062, 34–40. [Google Scholar] [CrossRef]
- Zhu, Q.-F.; Yan, J.-W.; Zhang, T.-Y.; Xiao, H.-M.; Feng, Y.-Q. Comprehensive Screening and Identification of Fatty Acid Esters of Hydroxy Fatty Acids in Plant Tissues by Chemical Isotope Labeling-Assisted Liquid Chromatography–Mass Spectrometry. Anal. Chem. 2018, 90, 10056–10063. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, M.; Yamada, A.; Katsuta, E.; Aoyagi, T.; Huang, W.-C.; Terracina, K.P.; Hait, N.C.; Allegood, J.C.; Tsuchida, J.; Yuza, K.; et al. Targeting the SphK1/S1P/S1PR1 Axis That Links Obesity, Chronic Inflammation, and Breast Cancer Metastasis. Cancer Res. 2018, 78, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Kothari, C.; Diorio, C.; Durocher, F. The Importance of Breast Adipose Tissue in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 5760. [Google Scholar] [CrossRef]
- Syed, I.; Rubin de Celis, M.F.; Mohan, J.F.; Moraes-Vieira, P.M.; Vijayakumar, A.; Nelson, A.T.; Siegel, D.; Saghatelian, A.; Mathis, D.; Kahn, B.B. PAHSAs Attenuate Immune Responses and Promote β Cell Survival in Autoimmune Diabetic Mice. J. Clin. Investig. 2019, 129, 3717–3731. [Google Scholar] [CrossRef]
- Paluchova, V.; Vik, A.; Cajka, T.; Brezinova, M.; Brejchova, K.; Bugajev, V.; Draberova, L.; Draber, P.; Buresova, J.; Kroupova, P.; et al. Triacylglycerol-Rich Oils of Marine Origin Are Optimal Nutrients for Induction of Polyunsaturated Docosahexaenoic Acid Ester of Hydroxy Linoleic Acid (13-DHAHLA) with Anti-Inflammatory Properties in Mice. Mol. Nutr. Food Res. 2020, 64, 1901238. [Google Scholar] [CrossRef] [PubMed]
- Kolar, M.J.; Konduri, S.; Chang, T.; Wang, H.; McNerlin, C.; Ohlsson, L.; Härröd, M.; Siegel, D.; Saghatelian, A. Linoleic Acid Esters of Hydroxy Linoleic Acids Are Anti-Inflammatory Lipids Found in Plants and Mammals. J. Biol. Chem. 2019, 294, 10698–10707. [Google Scholar] [CrossRef]
- Aryal, P.; Syed, I.; Lee, J.; Patel, R.; Nelson, A.T.; Siegel, D.; Saghatelian, A.; Kahn, B.B. Distinct Biological Activities of Isomers from Several Families of Branched Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs). J. Lipid Res. 2021, 62, 100108. [Google Scholar] [CrossRef]
- Kuda, O.; Brezinova, M.; Silhavy, J.; Landa, V.; Zidek, V.; Dodia, C.; Kreuchwig, F.; Vrbacky, M.; Balas, L.; Durand, T.; et al. Nrf2-Mediated Antioxidant Defense and Peroxiredoxin 6 Are Linked to Biosynthesis of Palmitic Acid Ester of 9-Hydroxystearic Acid. Diabetes 2018, 67, 1190–1199. [Google Scholar] [CrossRef]
- Patel, R.; Santoro, A.; Hofer, P.; Tan, D.; Oberer, M.; Nelson, A.T.; Konduri, S.; Siegel, D.; Zechner, R.; Saghatelian, A.; et al. ATGL Is a Biosynthetic Enzyme for Fatty Acid Esters of Hydroxy Fatty Acids. Nature 2022, 606, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Sammarco, A.; Eyme, K.; Jha, R.; Silva, W.D.N.; Neustadt, R.; Ford, I.; Bensinger, S.J.; Zappulli, V.; Breakefield, X.; Badr, C.E. TMET-02. Therapeutic Efficacy of a Novel Brain Penetrant SCD Inhibitor on Breast Cancer Brain Metastases. Neuro-Oncology 2022, 24, vii261. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of Fatty Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Kuda, O.; Brezinova, M.; Rombaldova, M.; Slavikova, B.; Posta, M.; Beier, P.; Janovska, P.; Veleba, J.; Kopecky, J.; Kudova, E.; et al. Docosahexaenoic Acid–Derived Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs) With Anti-Inflammatory Properties. Diabetes 2016, 65, 2580–2590. [Google Scholar] [CrossRef]
- Kellerer, T.; Kleigrewe, K.; Brandl, B.; Hofmann, T.; Hauner, H.; Skurk, T. Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs) Are Associated With Diet, BMI, and Age. Front. Nutr. 2021, 8, 691401. [Google Scholar] [CrossRef]
- Tan, D.; Ertunc, M.E.; Konduri, S.; Zhang, J.; Pinto, A.M.; Chu, Q.; Kahn, B.B.; Siegel, D.; Saghatelian, A. Discovery of FAHFA-Containing Triacylglycerols and Their Metabolic Regulation. J. Am. Chem. Soc. 2019, 141, 8798–8806. [Google Scholar] [CrossRef]




| No. | Analytes | VIP 1 | FC 2 | log2(FC) | p-Value 3 | FDR 4 | log10 (p-Value) | Regulation |
|---|---|---|---|---|---|---|---|---|
| 1 | 9-OAHSA | 1.5 | 3.2 | 1.7 | 3.2 × 10−8 | 2.0 × 10−5 | 7.5 | up |
| 2 | 11-OAHSA | 1.5 | 3.2 | 1.7 | 1.7 × 10−6 | 1.8 × 10−4 | 5.8 | up |
| 3 | 5-SAHMA | 1.3 | 2.6 | 1.4 | 1.1 × 10−4 | 1.4 × 10−3 | 4.0 | up |
| 4 | 13-OAHSA | 1.3 | 2.1 | 1.1 | 1.2 × 10−4 | 8.3 × 10−4 | 3.9 | up |
| 5 | 9-POHSA | 1.1 | 2.0 | 1 | 5.6 × 10−4 | 1.7 × 10−3 | 3.3 | up |
| 6 | 6-SAHSA | 1.3 | 2.6 | 1.4 | 8.8 × 10−3 | 1.6 × 10−2 | 2.1 | up |
| 7 | 12-PDAHPA | 1.6 | 0.5 | −1 | 1.2 × 10−7 | 2.6 × 10−6 | 6.9 | down |
| 8 | 7-PDAHPA | 1.5 | 0.6 | −0.8 | 1.1 × 10−6 | 2.1 × 10−5 | 5.9 | down |
| 9 | 8-PDAHPA | 1.6 | 0.5 | −0.9 | 3.7 × 10−6 | 2.7 × 10−5 | 5.4 | down |
| 10 | 13-PDAHPA | 1.4 | 0.6 | −0.8 | 5.4 × 10−6 | 2.0 × 10−5 | 5.3 | down |
| 11 | 6-PDAHPA | 1.3 | 0.6 | −0.7 | 3.0 × 10−4 | 8.3 × 10−4 | 3.5 | down |
| 12 | 11-PDAHPA | 1.4 | 0.6 | −0.7 | 6.6 × 10−4 | 2.7 × 10−3 | 3.2 | down |
| 13 | 3-PAHPA | 1.0 | 0.7 | −0.6 | 3.2 × 10−3 | 1.7 × 10−2 | 2.5 | down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; An, N.; Yuan, B.; Zhu, Q.; Feng, Y. The Metabolomic Characteristics and Dysregulation of Fatty Acid Esters of Hydroxy Fatty Acids in Breast Cancer. Metabolites 2023, 13, 1108. https://doi.org/10.3390/metabo13111108
Qin L, An N, Yuan B, Zhu Q, Feng Y. The Metabolomic Characteristics and Dysregulation of Fatty Acid Esters of Hydroxy Fatty Acids in Breast Cancer. Metabolites. 2023; 13(11):1108. https://doi.org/10.3390/metabo13111108
Chicago/Turabian StyleQin, Linlin, Na An, Bifeng Yuan, Quanfei Zhu, and Yuqi Feng. 2023. "The Metabolomic Characteristics and Dysregulation of Fatty Acid Esters of Hydroxy Fatty Acids in Breast Cancer" Metabolites 13, no. 11: 1108. https://doi.org/10.3390/metabo13111108
APA StyleQin, L., An, N., Yuan, B., Zhu, Q., & Feng, Y. (2023). The Metabolomic Characteristics and Dysregulation of Fatty Acid Esters of Hydroxy Fatty Acids in Breast Cancer. Metabolites, 13(11), 1108. https://doi.org/10.3390/metabo13111108






