Metabolic Features of a Novel Trichoderma asperellum YNQJ1002 with Potent Antagonistic Activity against Fusarium graminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Dual Confrontation Assays
2.3. Antifungal Experiment on Strains’ Culture Filtrates
2.4. Assay of Cell Wall-Degrading Enzyme Activity
2.5. Crude Extraction of Metabolites
2.6. Analytical Procedures for Peptaibols Using Acquity UPLC-QTOF-MS
2.7. DNA Extraction, PCR Amplification, and Sequencing
2.8. Molecular Identification and Phylogenetic Analysis
2.9. Statistical Analysis
3. Results
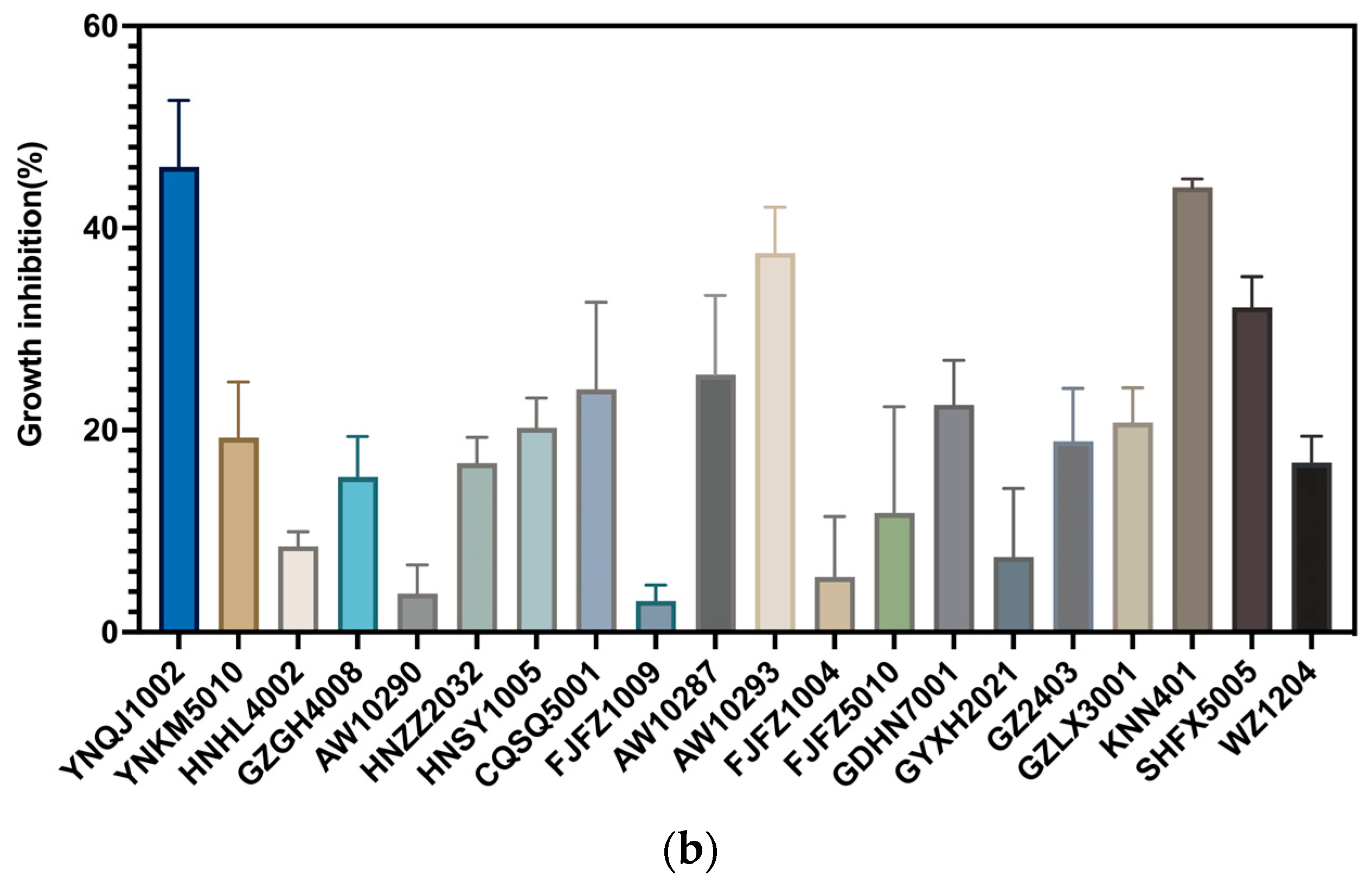
3.1. Screening for Strains Effective against Fungal Pathogens In Vitro
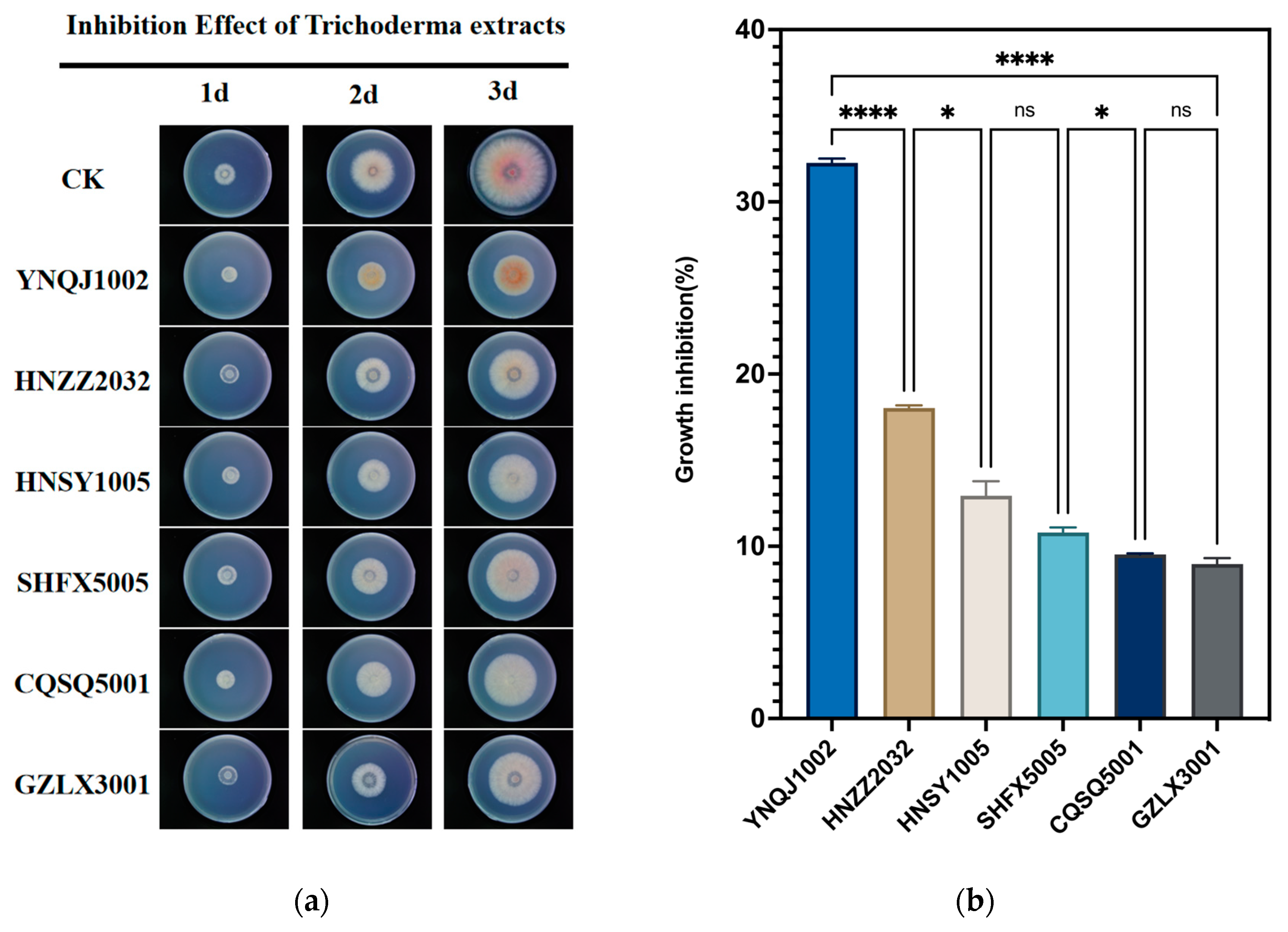
3.2. Effect of Culture Filtrates on F. graminearum
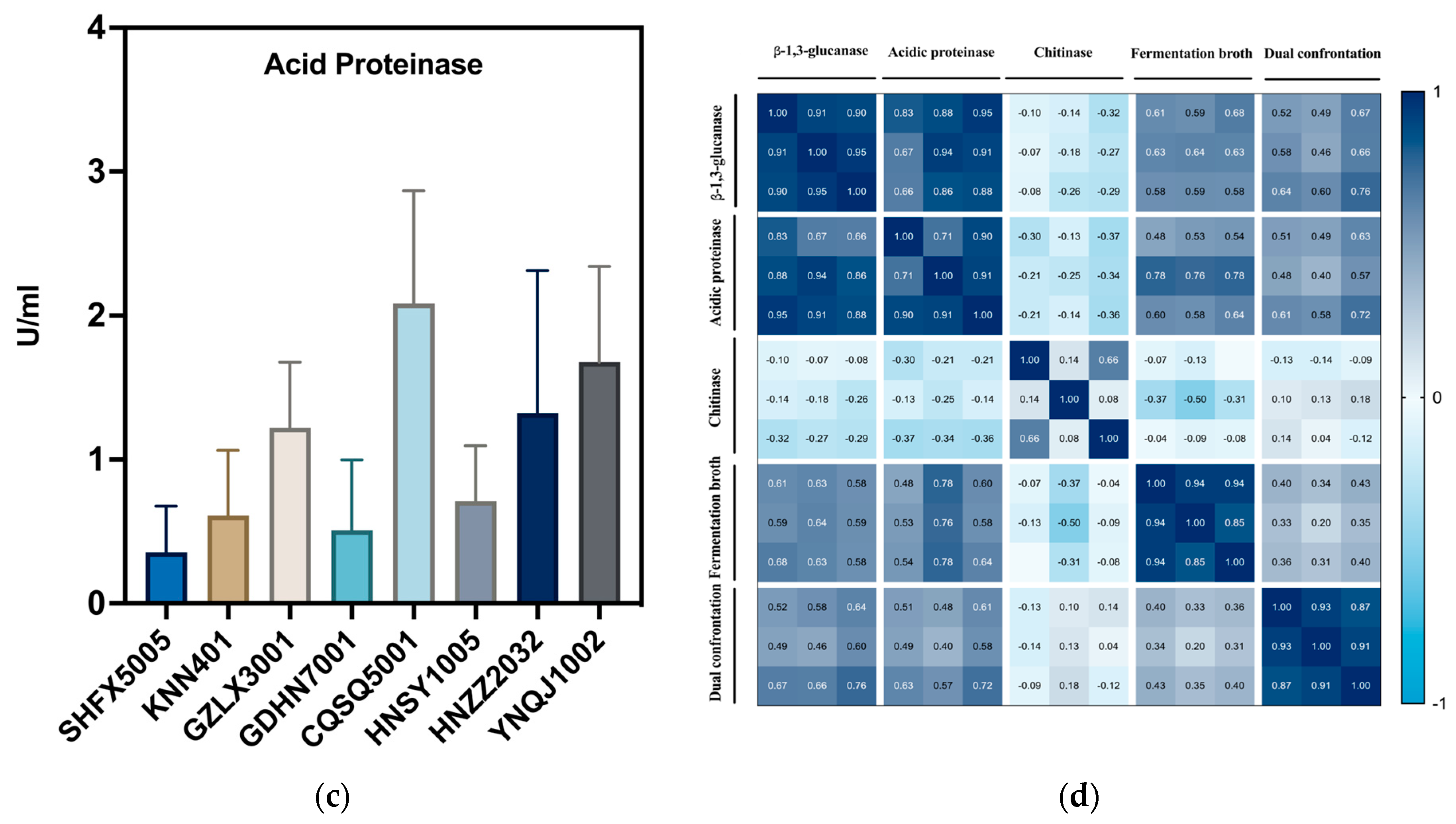
3.3. Cell Wall-Degrading Enzyme Activities
3.4. Effect of Crude Extraction of Metabolites on F. graminearum
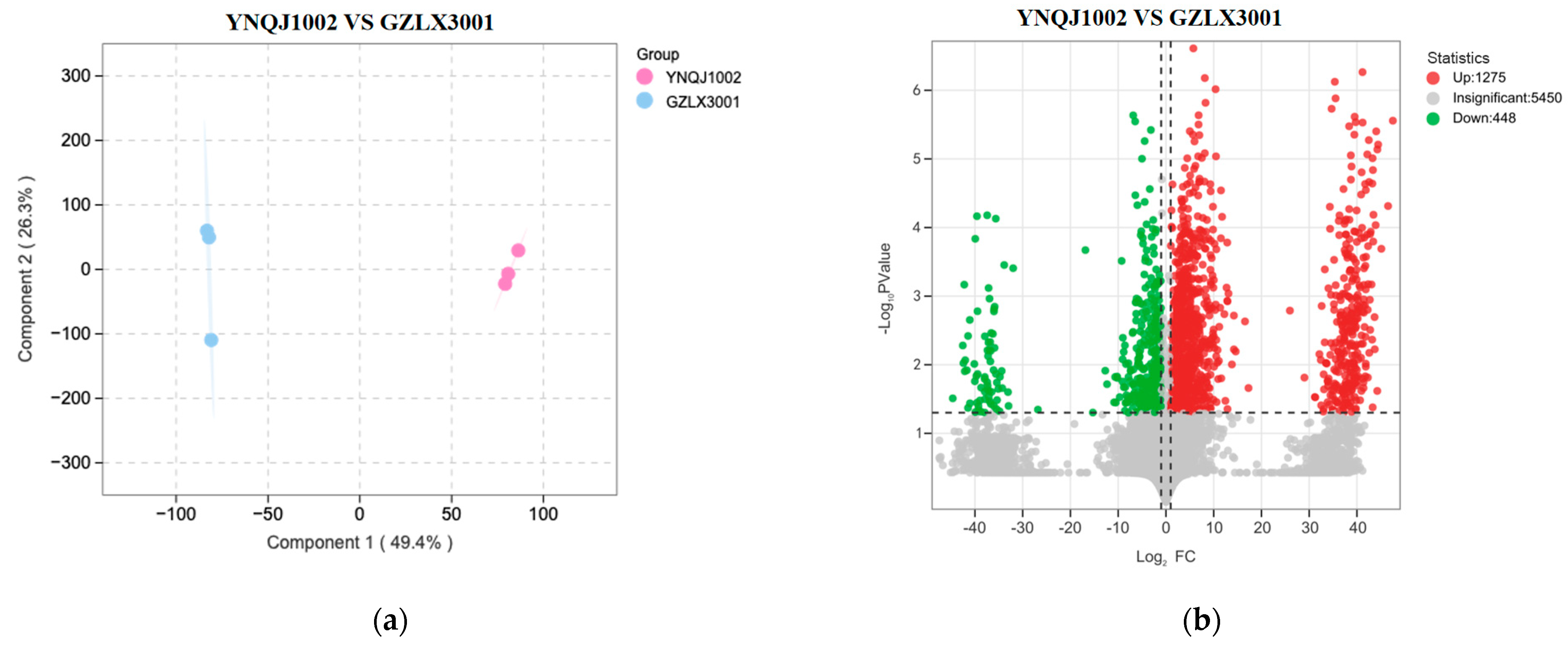
3.5. Analysis of Differential Metabolites between YNQJ1002 and GZLX3001 Using LC–MS/MS
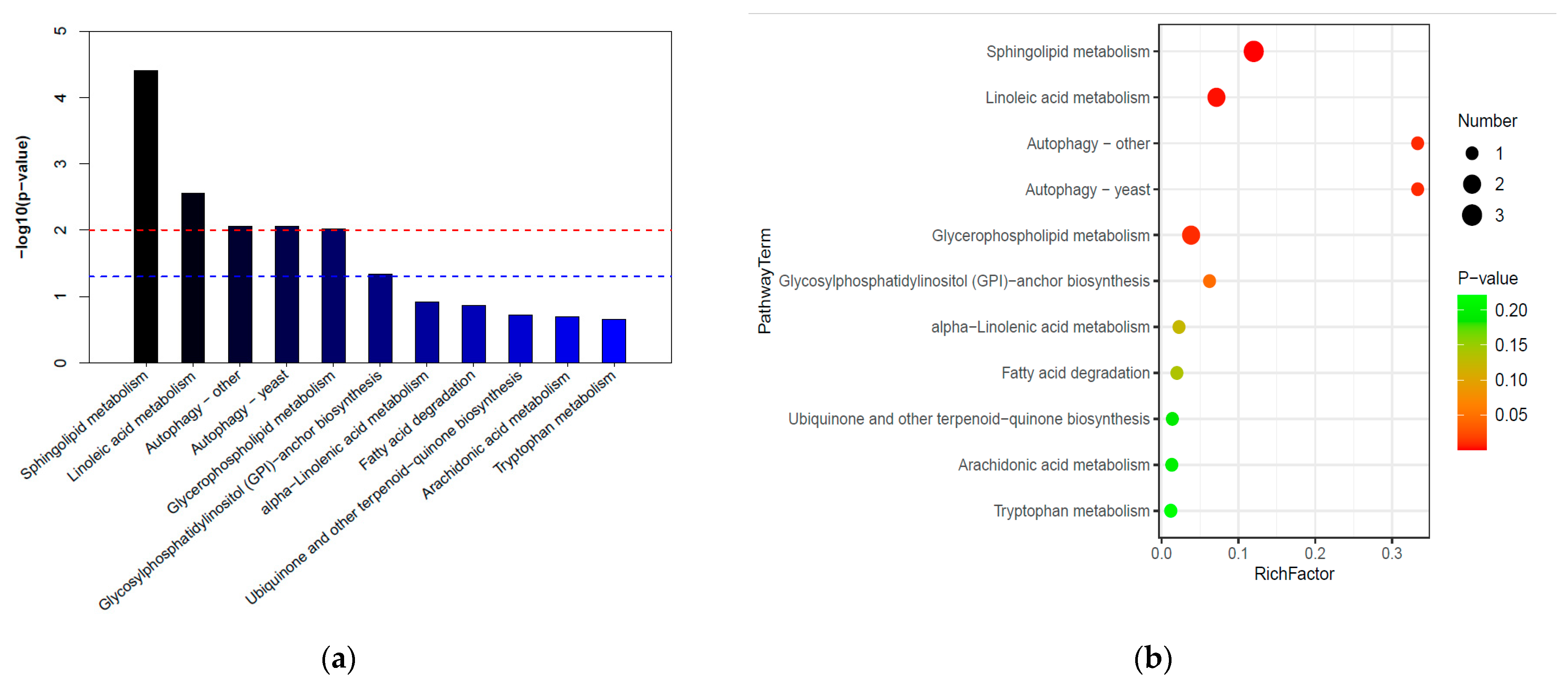
3.6. The KEGG Analysis of Differential Compounds in YNQJ1002 and GZLX3001
3.7. Molecular Identification and Phylogenetic Analysis
4. Discussion
4.1. The Correlation between Cell Wall-Degrading Enzymes and Other Biocontrol Factors
4.2. Abundant and Specific Metabolites Dominate the Trichoderma’s Antagonistic Action
4.3. The KEGG Enrichment of Metabolite Pathways in YNQJ1002
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, T.; Zhou, H.; Zhou, W.; Hu, L.; Chen, J.; Shi, Z. The Fungicidal Activity of Thymol against Fusarium graminearum via Inducing Lipid Peroxidation and Disrupting Ergosterol Biosynthesis. Molecules 2016, 21, 770. [Google Scholar] [CrossRef]
- Haile, J.K.; N’Diaye, A.; Walkowiak, S.; Nilsen, K.T.; Clarke, J.M.; Kutcher, H.R.; Steiner, B.; Buerstmayr, H.; Pozniak, C.J. Fusarium Head Blight in Durum Wheat: Recent Status, Breeding Directions, and Future Research Prospects. Phytopathology 2019, 109, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.D.B.; Nganje, W. Economic costs of Fusarium Head Blight, scab and deoxynivalenol. World Mycotoxin J. 2018, 11, 291–302. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum Trichothecene Mycotoxins: Biosynthesis, Regulation, and Management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Hou, S.; Ma, J.; Cheng, Y.; Wang, H.; Sun, J.; Yan, Y. The toxicity mechanisms of DON to humans and animals and potential biological treatment strategies. Crit. Rev. Food Sci. Nutr. 2023, 63, 790–812. [Google Scholar] [CrossRef]
- Ghimire, B.; Sapkota, S.; Bahri, B.A.; Martinez-Espinoza, A.D.; Buck, J.W.; Mergoum, M. Fusarium Head Blight and Rust Diseases in Soft Red Winter Wheat in the Southeast United States: State of the Art, Challenges and Future Perspective for Breeding. Front. Plant Sci. 2020, 11, 1080. [Google Scholar] [CrossRef]
- Palazzini, J.M.; Alberione, E.; Torres, A.; Donat, C.; Köhl, J.; Chulze, S. Biological control of Fusarium graminearum sensu stricto, causal agent of Fusarium head blight of wheat, using formulated antagonists under field conditions in Argentina. Biol. Control 2016, 94, 56–61. [Google Scholar] [CrossRef]
- Nawrocka, J.; Szczech, M.; Małolepsza, U. Trichoderma atroviride Enhances Phenolic Synthesis and Cucumber Protection against Rhizoctonia solani. Plant Prot. 2018, 54, 17–23. [Google Scholar] [CrossRef]
- Legrand, F.; Picot, A.; Cobo-Díaz, J.F.; Chen, W.; Le Floch, G. Challenges facing the biological control strategies for the management of Fusarium Head Blight of cereals caused by F. graminearum. Biol. Control 2017, 113, 26–38. [Google Scholar] [CrossRef]
- Matarese, F.; Sarrocco, S.; Gruber, S.; Seidl-Seiboth, V.; Vannacci, G. Biocontrol of Fusarium Head Blight: Interactions between Trichoderma and Mycotoxigenic Fusarium. Microbiology 2012, 158, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.L.; Hermosa, R.; Lorito, M.; Monte, E. Trichoderma: A multipurpose, plant-beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. 2023, 21, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Sarrocco, S.; Esteban, P.; Vicente, I.; Bernardi, R.; Plainchamp, T.; Domenichini, S.; Puntoni, G.; Baroncelli, R.; Vannacci, G.; Dufresne, M. Straw Competition and Wheat Root Endophytism of Trichoderma gamsii T6085 as Useful Traits in the Biological Control of Fusarium Head Blight. Phytopathology 2021, 111, 1129–1136. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Li, Y.; Yu, C.; Wang, Q.Q.; Wang, M.; Sun, J.; Gao, J.X.; Chen, J. Effect of Trichoderma harzianum on maize rhizosphere microbiome and biocontrol of Fusarium Stalk rot. Sci. Rep. 2017, 7, 1771. [Google Scholar] [CrossRef]
- Lu, Z.-X.; Tu, G.-P.; Zhang, T.; Li, Y.-Q.; Wang, X.-H.; Zhang, Q.-G.; Song, W.; Chen, J. Screening of antagonistic Trichoderma strains and their application for controlling stalk rot in maize. J. Integr. Agric. 2020, 19, 145–152. [Google Scholar] [CrossRef]
- Li, Y.; Sun, R.; Yu, J.; Saravanakumar, K.; Chen, J. Antagonistic and Biocontrol Potential of Trichoderma asperellum ZJSX5003 Against the Maize Stalk Rot Pathogen Fusarium graminearum. Indian J. Microbiol. 2016, 56, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, K.; Dou, K.; Lu, Z.; Wang, X.; Li, Y.; Chen, J. Enhanced biocontrol activity of cellulase from Trichoderma harzianum against Fusarium graminearum through activation of defense-related genes in maize. Physiol. Mol. Plant Pathol. 2018, 103, 130–136. [Google Scholar] [CrossRef]
- Lombardi, N.; Salzano, A.M.; Troise, A.D.; Scaloni, A.; Vitaglione, P.; Vinale, F.; Marra, R.; Caira, S.; Lorito, M.; d’Errico, G.; et al. Effect of Trichoderma Bioactive Metabolite Treatments on the Production, Quality, and Protein Profile of Strawberry Fruits. J. Agric. Food Chem. 2020, 68, 7246–7258. [Google Scholar] [CrossRef]
- Zhang, J.-L.; Tang, W.-L.; Huang, Q.-R.; Li, Y.-Z.; Wei, M.-L.; Jiang, L.-L.; Liu, C.; Yu, X.; Zhu, H.-W.; Chen, G.-Z.; et al. Trichoderma: A Treasure House of Structurally Diverse Secondary Metabolites With Medicinal Importance. Front. Microbiol. 2021, 12, 723828. [Google Scholar] [CrossRef]
- Zeilinger, S.; Gruber, S.; Bansal, R.; Mukherjee, P.K. Secondary metabolism in Trichoderma—Chemistry meets genomics. Fungal Biol. Rev. 2016, 30, 74–90. [Google Scholar] [CrossRef]
- Keswani, C.; Mishra, S.; Sarma, B.K.; Singh, S.P.; Singh, H.B. Unraveling the efficient applications of secondary metabolites of various Trichoderma spp. Appl. Microbiol. Biotechnol. 2014, 98, 533–544. [Google Scholar] [CrossRef]
- Li, W.-Y.; Liu, Y.; Lin, Y.-T.; Liu, Y.-C.; Guo, K.; Li, X.-N.; Luo, S.-H.; Li, S.-H. Antibacterial harziane diterpenoids from a fungal symbiont Trichoderma atroviride isolated from Colquhounia coccinea var. mollis. Phytochemistry 2020, 170, 112198. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, X.; Kong, F.D.; Huang, H.M.; Zhao, Y.N.; Liu, M.; Wang, Z.P.; Han, J. Overexpression of Global Regulator Talae1 Leads to the Discovery of New Antifungal Polyketides From Endophytic Fungus Trichoderma afroharzianum. Front. Microbiol. 2020, 11, 622785. [Google Scholar] [CrossRef] [PubMed]
- Du, F.-Y.; Ju, G.-L.; Xiao, L.; Zhou, Y.-M.; Wu, X. Sesquiterpenes and Cyclodepsipeptides from Marine-Derived Fungus Trichoderma longibrachiatum and Their Antagonistic Activities against Soil-Borne Pathogens. Mar. Drugs 2020, 18, 165. [Google Scholar] [CrossRef]
- Song, X.-Y.; Shen, Q.-T.; Xie, S.-T.; Chen, X.-L.; Sun, C.-Y.; Zhang, Y.-Z. Broad-spectrum antimicrobial activity and high stability of Trichokonins from Trichoderma koningii SMF2 against plant pathogens. FEMS Microbiol. Lett. 2006, 260, 119–125. [Google Scholar]
- Yamazaki, H.; Takahashi, O.; Kirikoshi, R.; Yagi, A.; Ogasawara, T.; Bunya, Y.; Rotinsulu, H.; Uchida, R.; Namikoshi, M. Epipolythiodiketopiperazine and trichothecene derivatives from the NaI-containing fermentation of marine-derived Trichoderma cf. brevicompactum. J Antibiot 2020, 73, 559–567. [Google Scholar] [CrossRef]
- Chen, L.; Hao, D.; Dou, K.; Lang, B.; Wang, X.; Li, Y.; Chen, J. Preparation of High Water-Soluble Trichoderma Co-Culture Metabolite Powder and Its Effects on Seedling Emergence Rate and Growth of Crops. J. Fungi 2023, 9, 767. [Google Scholar] [CrossRef]
- Marik, T.; Tyagi, C.; Racic, G.; Rakk, D.; Szekeres, A.; Vagvolgyi, C.; Kredics, L. New 19-Residue Peptaibols from Trichoderma Clade Viride. Microorganisms 2018, 6, 85. [Google Scholar] [CrossRef]
- Szekeres, A.; Leitgeb, B.; Kredics, L.; Manczinger, L.; Vagvolgyi, C. A novel, image analysis-based method for the evaluation of in vitro antagonism. J. Microbiol. Methods 2006, 65, 619–622. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Yu, C.; Dou, K.; Wang, M.; Li, Y.; Chen, J. Synergistic effect of Trichoderma-derived antifungal metabolites and cell wall degrading enzymes on enhanced biocontrol of Fusarium oxysporum f. sp. cucumerinum. Biol. Control 2016, 94, 37–46. [Google Scholar] [CrossRef]
- Dou, K.; Lu, Z.; Wu, Q.; Ni, M.; Yu, C.; Wang, M.; Li, Y.; Wang, X.; Xie, H.; Chen, J.; et al. MIST: A Multilocus Identification System for Trichoderma. Appl. Environ. Microbiol. 2020, 86, e01532-20. [Google Scholar] [CrossRef]
- Varjas, T.; Nowrasteh, G.; Budan, F.; Horvath, G.; Cseh, J.; Gyongyi, Z.; Makai, S.; Ember, I. The effect of fenugreek on the gene expression of arachidonic acid metabolizing enzymes. Phytother. Res. 2011, 25, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Guo, R.; Yu, F.; Chen, X.; Zhao, H.; Li, H.; Wu, J. Indole-3-Acetic Acid Biosynthesis Pathways in the Plant-Beneficial Bacterium Arthrobacter pascens ZZ21. Int. J. Mol. Sci. 2018, 19, 443. [Google Scholar] [CrossRef]
- Hasskarl, J. Everolimus. Recent Results Cancer Res. 2018, 211, 101–123. [Google Scholar]
- Pinches, S.E.; Apps, P. Production in food of 1,3-pentadiene and styrene by Trichoderma species. Int. J. Food Microbiol. 2007, 116, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, A.J.; Wang, J.; Zhang, Z.; Gold, A.; Jung, D.; Zeng, D.; Pollard, A.; Coronell, O. Grafting of bioactive 2-aminoimidazole into active layer makes commercial RO/NF membranes anti-biofouling. J. Membr. Sci. 2018, 556, 85–97. [Google Scholar] [CrossRef]
- Abhishek, R.U.; Thippeswamy, S.; Manjunath, K.; Mohana, D.C. Antifungal and antimycotoxigenic potency of Solanum torvum Swartz. leaf extract: Isolation and identification of compound active against mycotoxigenic strains of Aspergillus flavus and Fusarium verticillioides. J. Appl. Microbiol. 2015, 119, 1624–1636. [Google Scholar] [CrossRef]
- Negi, J.S.; Bisht, V.K.; Bhandari, A.K.; Bhatt, V.P.; Singh, P.; Singh, N. Paris polyphylla: Chemical and Biological Prospectives. ACAMC 2014, 14, 833–839. [Google Scholar] [CrossRef]
- Nayak, N.; Ramprasad, J.; Dalimba, U. Synthesis and antitubercular and antibacterial activity of some active fluorine containing quinoline–pyrazole hybrid derivatives. J. Fluor. Chem. 2016, 183, 59–68. [Google Scholar] [CrossRef]
- Martinez-Correa, H.A.; Paula, J.T.; Kayano, A.C.A.V.; Queiroga, C.L.; Magalhães, P.M.; Costa, F.T.M.; Cabral, F.A. Composition and antimalarial activity of extracts of Curcuma longa L. obtained by a combination of extraction processes using supercritical CO2, ethanol and water as solvents. J. Supercrit. Fluids 2017, 119, 122–129. [Google Scholar] [CrossRef]
- Emile, A.; Waikedre, J.; Herrenknecht, C.; Fourneau, C.; Gantier, J.C.; Hnawia, E.; Cabalion, P.; Hocquemiller, R.; Fournet, A. Bioassay-guided isolation of antifungal alkaloids from Melochia odorata. Phytother. Res. 2007, 21, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Yun, T.; Jing, T.; Zang, X.; Zhou, D.; Li, K.; Zhao, Y.; Wang, W.; Xie, J. Antimicrobial mechanisms and secondary metabolite profiles of Streptomyces hygroscopicus subsp. hygroscopicus 5-4 against banana fusarium wilt disease using metabolomics. Front. Microbiol. 2023, 14, 1159534. [Google Scholar] [CrossRef]
- Bakun, P.; Czarczynska-Goslinska, B.; Goslinski, T.; Lijewski, S. In vitro and in vivo biological activities of azulene derivatives with potential applications in medicine. Med. Chem. Res. 2021, 30, 834–846. [Google Scholar] [CrossRef]
- Jiang, Y.-Q.; Lin, J.-P. Recent progress in strategies for steroid production in yeasts. World J. Microbiol. Biotechnol. 2022, 38, 93. [Google Scholar] [CrossRef]
- Kebede, B.H.; Forsido, S.F.; Tola, Y.B.; Astatkie, T. Free radical scavenging capacity, antibacterial activity and essential oil composition of turmeric (Curcuma domestica) varieties grown in Ethiopia. Heliyon 2021, 7, e06239. [Google Scholar] [CrossRef]
- Takano, M.; Simbol, A.B.; Yasin, M.; Shibasaki, I. Bactericidal Effect of Freezing with Chemical Agents. J. Food Sci. 1979, 44, 112–115. [Google Scholar] [CrossRef]
- Pan, J.; Hu, C.; Yu, J.-H. Lipid Biosynthesis as an Antifungal Target. J. Fungi 2018, 4, 50. [Google Scholar] [CrossRef]
- McEvoy, K.; Normile, T.G.; Del Poeta, M. Antifungal Drug Development: Targeting the Fungal Sphingolipid Pathway. J. Fungi 2020, 6, 142. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, Y.; Zhang, H.; Wang, X.; Chen, J.; Li, Y. Integrated Transcriptome and Untargeted Metabolomic Analyses Revealed the Role of Methyltransferase Lae1 in the Regulation of Phospholipid Metabolism in Trichoderma atroviride. J. Fungi 2023, 9, 120. [Google Scholar] [CrossRef]
- Choi, G.-J.; Jang, K.-S.; Choi, Y.-H.; Yu, J.-H.; Kim, J.-C. Antifungal Activity of Lower Alkyl Fatty Acid Esters against Powdery Mildews. Plant Pathol. J. 2010, 26, 360–366. [Google Scholar] [CrossRef]
- Liu, S.; Ruan, W.; Li, J.; Xu, H.; Wang, J.; Gao, Y.; Wang, J. Biological Control of Phytopathogenic Fungi by Fatty Acids. Mycopathologia 2008, 166, 93–102. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, H.; Ding, J.-L.; Feng, M.-G.; Ying, S.-H. Transcriptomic investigation reveals a physiological mechanism for Beauveria bassiana to survive under linoleic acid stress. iScience 2023, 26, 106551. [Google Scholar] [CrossRef] [PubMed]
- Boguś, M.I.; Czygier, M.; Gołębiowski, M.; Kędra, E.; Kucińska, J.; Mazgajska, J.; Samborski, J.; Wieloch, W.; Włóka, E. Effects of insect cuticular fatty acids on in vitro growth and pathogenicity of the entomopathogenic fungus Conidiobolus coronatus. Exp. Parasitol. 2010, 125, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, C.; Sun, Q.; Xu, J.; Chai, Y.; Berg, G.; Cernava, T.; Ma, Z.; Chen, Y. Post-translational regulation of autophagy is involved in intra-microbiome suppression of fungal pathogens. Microbiome 2021, 9, 131. [Google Scholar] [CrossRef] [PubMed]




| Metabolites | VIP | p-Value | log2(FC) | Super Class | Biological Activities |
|---|---|---|---|---|---|
| YNQJ1002 | |||||
| Trigoneoside Xb | 1.4662 | 3.96 × 106 | 44.0422 | Fatty acyl glycosides | Antifungal activity [31] |
| 5-Hydroxyindoleacetaldehyde | 1.431 | 1.58 × 103 | 42.256 | Hydroxyindoles | Promote plant growth [32] |
| Everolimus | 1.4621 | 1.40 × 104 | 41.554 | Unclassified | An mTOR inhibitor [33] |
| trans, trans-hepta-2, 4, 6-trienoic acid | 1.4675 | 8.87 × 106 | 38.7295 | Fatty acids and conjugates | Antifungal activity [34] |
| 2-AI | 1.4683 | 7.51 × 107 | 35.3728 | Unclassified | Antimicroalgal activity [35] |
| Torvoside A | 1.4699 | 1.52 × 106 | 8.294 | Terpene lactones | Antifungal and anti-mycotoxigenic potency [36] |
| Trigofoenoside B | 1.4489 | 5.56 × 104 | 6.671 | Terpene glycosides | Antimicrobial [37] |
| 6-Methoxyquinoline | 1.4549 | 1.23 × 104 | 6.0625 | Unclassified | Antibacterial activity [38] |
| (R)-ar-Turmerone | 1.4233 | 1.85 × 103 | 5.4807 | Sesquiterpenoids | Induced biochemical defenses [39] |
| Frangulanine | 1.4705 | 1.74 × 105 | 5.0688 | Amino acids, peptides, and analogues | Antifungal activity [40] |
| Torvoside G | 1.4443 | 1.11 × 103 | 3.2429 | Triacyclglycerols | Antimicrobial [41] |
| Chamazulene | 1.3990 | 5.12 × 103 | 1.9201 | Sesquiterpenoids | Antioxidant and radical scavenging activities [42] |
| GZLX3001 | |||||
| Phytosphingosine | 1.4413 | 5.99 × 104 | −1.0791 | Amines | Cell proliferation, recognition, adhesion, and signal transduction [43] |
| Curlone | 1.4428 | 3.07 × 104 | −9.2727 | Sesquiterpenoids | Antibacterial activity [44] |
| Sorbitan laurate | 1.4364 | 1.59 × 103 | −3.5284 | Fatty acid esters | Antibacterial activity [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, H.; Yu, R.; Liu, H.; Zhang, H.; Wang, X.; Chen, J.; Li, Y. Metabolic Features of a Novel Trichoderma asperellum YNQJ1002 with Potent Antagonistic Activity against Fusarium graminearum. Metabolites 2023, 13, 1144. https://doi.org/10.3390/metabo13111144
Ji H, Yu R, Liu H, Zhang H, Wang X, Chen J, Li Y. Metabolic Features of a Novel Trichoderma asperellum YNQJ1002 with Potent Antagonistic Activity against Fusarium graminearum. Metabolites. 2023; 13(11):1144. https://doi.org/10.3390/metabo13111144
Chicago/Turabian StyleJi, Huimin, Ruohan Yu, Hongyi Liu, Hui Zhang, Xinhua Wang, Jie Chen, and Yaqian Li. 2023. "Metabolic Features of a Novel Trichoderma asperellum YNQJ1002 with Potent Antagonistic Activity against Fusarium graminearum" Metabolites 13, no. 11: 1144. https://doi.org/10.3390/metabo13111144
APA StyleJi, H., Yu, R., Liu, H., Zhang, H., Wang, X., Chen, J., & Li, Y. (2023). Metabolic Features of a Novel Trichoderma asperellum YNQJ1002 with Potent Antagonistic Activity against Fusarium graminearum. Metabolites, 13(11), 1144. https://doi.org/10.3390/metabo13111144








