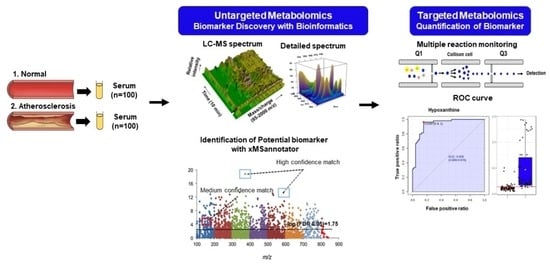
Identification of Novel Biomarkers for Early Diagnosis of Atherosclerosis Using High-Resolution Metabolomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Sample Collection
2.3. Sample Preparation
2.4. LC-MS/MS Analysis
2.5. Untargeted Metabolite Profiling
2.6. Untargeted Pathway Analysis
2.7. Targeted Metabolite Profiling of the Significantly Altered in Untargeted Metabolomics
3. Results
3.1. Characteristics of the Study Population
3.2. Determination of Metabolomic Signature in Two Groups
3.3. Identification of Potential Metabolites between Normal and AS Groups
3.4. Validation of the Levels of Metabolites with Targeted Metabolomics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhattacharya, P.; Kanagasooriyan, R.; Subramanian, M. Tackling inflammation in atherosclerosis: Are we there yet and what lies beyond? Curr. Opin. Pharmacol. 2022, 66, 102283. [Google Scholar] [CrossRef] [PubMed]
- Močnik, M.; Varda, N.M. Lipid Biomarkers and Atherosclerosis—Old and New in Cardiovascular Risk in Childhood. Int. J. Mol. Sci. 2023, 24, 2237. [Google Scholar] [CrossRef] [PubMed]
- Na, J.; Hwang, H.-J.; Shin, M.-S.; Kang, M.; Lee, J.; Bang, G.; Kim, Y.J.; Hwang, Y.-J.; Hwang, K.-A.; Park, Y.H. Extract of radish (R. Sativus Linn) promotes anti-atherosclerotic effect using urine metabolomics in ApoE−/− mice. J. Funct. Foods 2021, 78, 104368. [Google Scholar] [CrossRef]
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgözoğlu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, 56. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, X.; Qiu, J.; Zhou, A.; Xu, P.; Liu, Y.; Wu, H. A UHPLC-Q-TOF-MS-based serum and urine metabolomics approach reveals the mechanism of Gualou-Xiebai herb pair intervention against atherosclerosis process in ApoE−/− mice. J. Chromatogr. B 2023, 1215, 123567. [Google Scholar] [CrossRef]
- Farthing, D.E.; Farthing, C.A.; Xi, L. Inosine and hypoxanthine as novel biomarkers for cardiac ischemia: From bench to point-of-care. Exp. Biol. Med. 2015, 240, 821–831. [Google Scholar] [CrossRef]
- Uemura, S. Invasive imaging of vulnerable atherosclerotic plaques in coronary artery disease. Circ. J. 2013, 77, 869–875. [Google Scholar] [CrossRef]
- Gao, X.; Ke, C.; Liu, H.; Liu, W.; Li, K.; Yu, B.; Sun, M. Large-scale Metabolomic Analysis Reveals Potential Biomarkers for Early Stage Coronary Atherosclerosis. Sci. Rep. 2017, 7, 11817. [Google Scholar] [CrossRef]
- Iida, M.; Harada, S.; Takebayashi, T. Application of metabolomics to epidemiological studies of atherosclerosis and cardiovascular disease. J. Atheroscler. Thromb. 2019, 26, 747–757. [Google Scholar] [CrossRef]
- Khan, A.; Choi, S.A.; Na, J.; Pamungkas, A.D.; Jung, K.J.; Jee, S.H.; Park, Y.H. Noninvasive Serum Metabolomic Profiling Reveals Elevated Kynurenine Pathway’s Metabolites in Humans with Prostate Cancer. J. Proteome Res. 2019, 18, 1532–1541. [Google Scholar] [CrossRef]
- Lee, E.-J.; Kim, D.J.; Kang, D.-W.; Yang, W.; Jeong, H.-Y.; Kim, J.-M.; Ko, S.-B.; Lee, S.-H.; Yoon, B.-W.; Cho, J.-Y.; et al. Targeted Metabolomic Biomarkers for Stroke Subtyping. Transl. Stroke Res. 2023. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Khan, A.; Cho, S.; Na, J.; Lee, Y.; Bang, G.; Yu, W.-J.; Jeong, J.-S.; Jee, S.H.; Park, Y.H. Effect of developmental exposure to bisphenol A on steroid hormone and vitamin D3 metabolism. Chemosphere 2019, 237, 124469. [Google Scholar] [CrossRef] [PubMed]
- Munjal, Y.; Tonk, R.K.; Sharma, R. Analytical Techniques Used in Metabolomics: A Review. Syst. Rev. Pharm. 2022, 13, 550–556. [Google Scholar]
- Yang, Q.; Zhang, A.-H.; Miao, J.-H.; Sun, H.; Han, Y.; Yan, G.-L.; Wu, F.-F.; Wang, X.-J. Metabolomics biotechnology, applications, and future trends: A systematic review. RSC Adv. 2019, 9, 37245–37257. [Google Scholar] [CrossRef]
- Sardar, S.W.; Choi, Y.; Park, N.; Jeon, J. Occurrence and Concentration of Chemical Additives in Consumer Products in Korea. Int. J. Environ. Res. Public Health 2019, 16, 5075. [Google Scholar] [CrossRef]
- Park, M.; Lee, Y.; Khan, A.; Aleta, P.; Cho, Y.; Park, H.; Park, Y.H.; Kim, S. Metabolite tracking to elucidate the effects of environmental pollutants. J. Hazard. Mater. 2019, 376, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Park, Y.; Johnson, J.M.; Jones, D.P. apLCMS—Adaptive processing of high-resolution LC/MS data. Bioinformatics 2009, 25, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Uppal, K.; Soltow, Q.A.; Strobel, F.H.; Pittard, W.S.; Gernert, K.M.; Yu, T.; Jones, D.P. xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinform. 2013, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Bang, G.; Park, J.H.; Park, C.; Kim, K.-J.; Kim, J.K.; Lee, S.Y.; Kim, J.Y.; Park, Y.H. High-resolution metabolomics-based biomarker discovery using exhaled breath condensate from patients with lung cancer. J. Anal. Sci. Technol. 2022, 13, 37. [Google Scholar] [CrossRef]
- Scheubert, K.; Hufsky, F.; Petras, D.; Wang, M.; Nothias, L.-F.; Dührkop, K.; Bandeira, N.; Dorrestein, P.C.; Böcker, S. Significance estimation for large scale metabolomics annotations by spectral matching. Nat. Commun. 2017, 8, 1494. [Google Scholar] [CrossRef]
- Uppal, K.; Walker, D.I.; Jones, D.P. xMSannotator: An R package for network-based annotation of high-resolution metabolomics data. Anal. Chem. 2017, 89, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Fujibuchi, W.; Kanehisa, M. Computation with the KEGG pathway database. Biosystems 1998, 47, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, S.; Yang, J.-S.; Lee, J.-H.; Joo, C.; Jung, H.-I. Real-time measurement of human salivary cortisol for the assessment of psychological stress using a smartphone. Sens. Bio-Sens. Res. 2014, 2, 8–11. [Google Scholar] [CrossRef]
- Danese, E.; Negrini, D.; Pucci, M.; De Nitto, S.; Ambrogi, D.; Donzelli, S.; Lievens, P.M.-J.; Salvagno, G.L.; Lippi, G. Bile acids quantification by liquid chromatography–tandem mass spectrometry: Method validation, reference range, and interference study. Diagnostics 2020, 10, 462. [Google Scholar] [CrossRef]
- Kennedy, A.; Bivens, A. Methods for the Analysis of Underivatized Amino Acids by LC/MS. Agil Trust Answers. Published online. 2017. Available online: https://www.agilent.com/cs/library/applications/5991-8582EN_HILIC_Underivatized_Amino_Acid_application.pdf (accessed on 16 September 2023).
- Go, A.; Shim, G.; Park, J.; Hwang, J.; Nam, M.; Jeong, H.; Chung, H. Analysis of hypoxanthine and lactic acid levels in vitreous humor for the estimation of post-mortem interval (PMI) using LC–MS/MS. Forensic Sci. Int. 2019, 299, 135–141. [Google Scholar] [CrossRef]
- Liu, X.; Gao, J.; Chen, J.; Wang, Z.; Shi, Q.; Man, H.; Guo, S.; Wang, Y.; Li, Z.; Wang, W. Identification of metabolic biomarkers in patients with type 2 diabetic coronary heart diseases based on metabolomic approach. Sci. Rep. 2016, 6, 30785. [Google Scholar] [CrossRef]
- Yin, J.; Xie, J.; Guo, X.; Ju, L.; Li, Y.; Zhang, Y. Plasma metabolic profiling analysis of cyclophosphamide-induced cardiotoxicity using metabolomics coupled with UPLC/Q–TOF–MS and ROC curve. J. Chromatogr. B 2016, 1033–1034, 428–435. [Google Scholar] [CrossRef]
- Randrianarisoa, E.; Lehn-Stefan, A.; Wang, X.; Hoene, M.; Peter, A.; Heinzmann, S.S.; Zhao, X.; Königsrainer, I.; Königsrainer, A.; Balletshofer, B.; et al. Relationship of serum trimethylamine N-oxide (TMAO) levels with early atherosclerosis in humans. Sci. Rep. 2016, 6, 26745. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, X.; Ma, Y.; Liu, Y.; Zhang, L.; He, Y.; Chen, Y. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem. Biophys. Res. Commun. 2016, 481, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chiang, J.Y.L. Regulation of bile acid and cholesterol metabolism by PPARs. PPAR Res. 2009, 2009, 501739. [Google Scholar] [CrossRef]
- Staels, B.; Fonseca, V.A. Bile acids and metabolic regulation: Mechanisms and clinical responses to bile acid sequestration. Diabetes Care 2009, 32, 2. [Google Scholar] [CrossRef]
- Pathak, P.; Liu, H.; Boehme, S.; Xie, C.; Krausz, K.W.; Gonzalez, F.; Chiang, J.Y.L. Farnesoid X receptor induces Takeda G-protein receptor 5 cross-talk to regulate bile acid synthesis and hepatic metabolism. J. Biol. Chem. 2017, 292, 11055–11069. [Google Scholar] [CrossRef]
- Poznyak, A.; Grechko, A.V.; Poggio, P.; Myasoedova, V.A.; Alfieri, V.; Orekhov, A.N. The diabetes mellitus–atherosclerosis connection: The role of lipid and glucose metabolism and chronic inflammation. Int. J. Mol. Sci. 2020, 21, 1835. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M.; Wu, Y.; Boehme, S. Bile Acid and Cholesterol Metabolism in Atherosclerotic Cardiovascular Disease and Therapy. Cardiol. Plus 2020, 5, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Claudel, T.; Staels, B.; Kuipers, F. The Farnesoid X receptor: A molecular link between bile acid and lipid and glucose metabolism. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 2020–2031. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Wu, W.; Zhu, Y.; Liu, X. The Role of Bile Acids in Cardiovascular Diseases: From Mechanisms to Clinical Implications. Aging Dis. 2023, 14, 261–282. [Google Scholar] [CrossRef]
- Uchida, K.; Chikai, T.; Takase, H.; Nomura, Y.; Seo, S.; Nakao, H.; Takeuchi, N. Age–related changes of bile acid metabolism in rats. Arch. Gerontol. Geriatr. 1990, 10, 37–48. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, H.; Tang, X.; Zhao, J.; Zhang, H.; Chen, Y.Q.; Chen, W. The relationship between amino acid and lipid metabolism in oleaginous eukaryotic microorganism. Appl. Microbiol. Biotechnol. 2022, 106, 3405–3417. [Google Scholar] [CrossRef]
- Zaric, B.L.; Radovanovic, J.N.; Gluvic, Z.; Stewart, A.J.; Essack, M.; Motwalli, O.; Gojobori, T.; Isenovic, E.R. Atherosclerosis Linked to Aberrant Amino Acid Metabolism and Immunosuppressive Amino Acid Catabolizing Enzymes. Front. Immunol. 2020, 11, 551758. [Google Scholar] [CrossRef]
- Grajeda-Iglesias, C.; Rom, O.; Aviram, M. Branched-chain amino acids and atherosclerosis: Friends or foes? Curr. Opin. Lipidol. 2018, 29, 166–169. [Google Scholar] [CrossRef]
- Tobias, D.K.; Lawler, P.R.; Harada, P.H.; Demler, O.V.; Ridker, P.M.; Manson, J.E.; Cheng, S.; Mora, S. Circulating Branched-Chain Amino Acids and Incident Cardiovascular Disease in a Prospective Cohort of US Women. Circ. Genom. Precis. Med. 2018, 11, e002157. [Google Scholar] [CrossRef]
- Appuhamy, J.A.D.R.N.; Knoebel, N.A.; Nayananjalie, W.A.D.; Escobar, J.; Hanigan, M.D. Isoleucine and leucine independently regulate mTOR signaling and protein synthesis in MAC-T cells and bovine mammary tissue slices. J. Nutr. 2012, 142, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.L.; Liu, J.; Wang, C.X.; Ni, J.; Zhang, Y.; Wu, Y.; Lv, L.L.; Ruan, X.Z.; Liu, B.C. Increased mTORC1 activity contributes to atherosclerosis in apolipoprotein E knockout mice and in vascular smooth muscle cells. Int. J. Cardiol. 2013, 168, 5450–5453. [Google Scholar] [CrossRef] [PubMed]
- Nemkov, T.; Sun, K.; Reisz, J.A.; Song, A.; Yoshida, T.; Dunham, A.; Wither, M.J.; Francis, R.O.; Roach, C.R.; Dzieciatkowska, M.; et al. Hypoxia modulates the purine salvage pathway and decreases red blood cell and supernatant levels of hypoxanthine during refrigerated storage. Haematologica 2018, 103, 361–372. [Google Scholar] [CrossRef]
- Ryu, H.; Kim, Y.; Oh, E.; Oh, S.; Choi, J.; Cho, J.; Kim, C.; Park, S.; Kim, Y. Hypoxanthine induces cholesterol accumulation and incites atherosclerosis in apolipoprotein E-deficient mice and cells. J. Cell. Mol. Med. 2016, 20, 2160–2172. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Ma, Z.; Li, F.; Duan, C.; Yuan, Y.; Zhu, C.; Wang, L.; Zhu, X.; Wang, S.; Gao, P.; et al. Hypoxanthine Induces Muscular ATP Depletion and Fatigue via UCP2. Front. Physiol. 2021, 12, 647743. [Google Scholar] [CrossRef]
- Dekker, M.J.H.J.; Koper, J.W.; van Aken, M.O.; Pols, H.A.P.; Hofman, A.; de Jong, F.H.; Kirschbaum, C.; Witteman, J.C.M.; Lamberts, S.W.J.; Tiemeier, H. Salivary cortisol is related to atherosclerosis of carotid arteries. J. Clin. Endocrinol. Metab. 2008, 93, 3741–3747. [Google Scholar] [CrossRef]
- Hajat, A.; Diez-Roux, A.V.; Sánchez, B.N.; Holvoet, P.; Lima, J.A.; Merkin, S.S.; Polak, J.F.; Seeman, T.E.; Wu, M. Examining the association between salivary cortisol levels and subclinical measures of atherosclerosis: The Multi-Ethnic Study of Atherosclerosis. Psychoneuroendocrinology 2013, 38, 1036–1046. [Google Scholar] [CrossRef]
- Lazzarino, A.I.; Hamer, M.; Gaze, D.; Collinson, P.; Steptoe, A. The association between cortisol response to mental stress and high-sensitivity cardiac troponin T plasma concentration in healthy adults. J. Am. Coll. Cardiol. 2013, 62, 1694–1701. [Google Scholar] [CrossRef] [PubMed]








| Normal | AS | p-Value | |
|---|---|---|---|
| Number of patients | 100 | 100 | |
| Gender | Male (100%) | Male (79%), Female (21%) | |
| Age (year) | 19–44 | 41–101 | |
| Body mass index (kg/m2) | 25.61 ± 1.799 | 22.67 ± 0.4179 | 0.1141 |
| Systolic blood pressure (mmHg) | 120.3 ± 0.8022 | 133.6 ± 2.395 | <0.0001 |
| Diastolic blood pressure (mmHg) | 76.62 ± 0.6925 | 73.68 ± 1.185 | 0.0337 |
| Fasting blood glucose (mg/dL) | 92.51 ± 0.6717 | 128.8 ± 4.71 | <0.0001 |
| Total triglyceride (mg/dL) | 101.9 ± 2.72 | 116.7 ± 6.8 | 0.0449 |
| Total cholesterol (mg/dL) | 182.4 ± 2.561 | 137 ± 4.271 | <0.0001 |
| Low-density lipoprotein cholesterol (mg/dL) | 107.6 ± 2 | 79.27 ± 3.292 | <0.0001 |
| High-density lipoprotein cholesterol (mg/dL) | 52.73 ± 0.973 | 73.43 ± 4.911 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sardar, S.W.; Nam, J.; Kim, T.E.; Kim, H.; Park, Y.H. Identification of Novel Biomarkers for Early Diagnosis of Atherosclerosis Using High-Resolution Metabolomics. Metabolites 2023, 13, 1160. https://doi.org/10.3390/metabo13111160
Sardar SW, Nam J, Kim TE, Kim H, Park YH. Identification of Novel Biomarkers for Early Diagnosis of Atherosclerosis Using High-Resolution Metabolomics. Metabolites. 2023; 13(11):1160. https://doi.org/10.3390/metabo13111160
Chicago/Turabian StyleSardar, Syed Wasim, Jeonghun Nam, Tae Eun Kim, Hyunil Kim, and Youngja H. Park. 2023. "Identification of Novel Biomarkers for Early Diagnosis of Atherosclerosis Using High-Resolution Metabolomics" Metabolites 13, no. 11: 1160. https://doi.org/10.3390/metabo13111160
APA StyleSardar, S. W., Nam, J., Kim, T. E., Kim, H., & Park, Y. H. (2023). Identification of Novel Biomarkers for Early Diagnosis of Atherosclerosis Using High-Resolution Metabolomics. Metabolites, 13(11), 1160. https://doi.org/10.3390/metabo13111160