Antioxidant Activity and Metabolite Profiling of Xylocarpus granatum Extracts Using Gas Chromatography–Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Instruments
2.2. Extraction
2.3. Antioxidant Activity
2.4. Sample Preparation for GC-MS Analysis
2.5. GC-MS Analysis
2.6. Data Analysis with MS-DIAL
2.7. Data Analysis
3. Results
3.1. X. granatum Antioxidant Activity
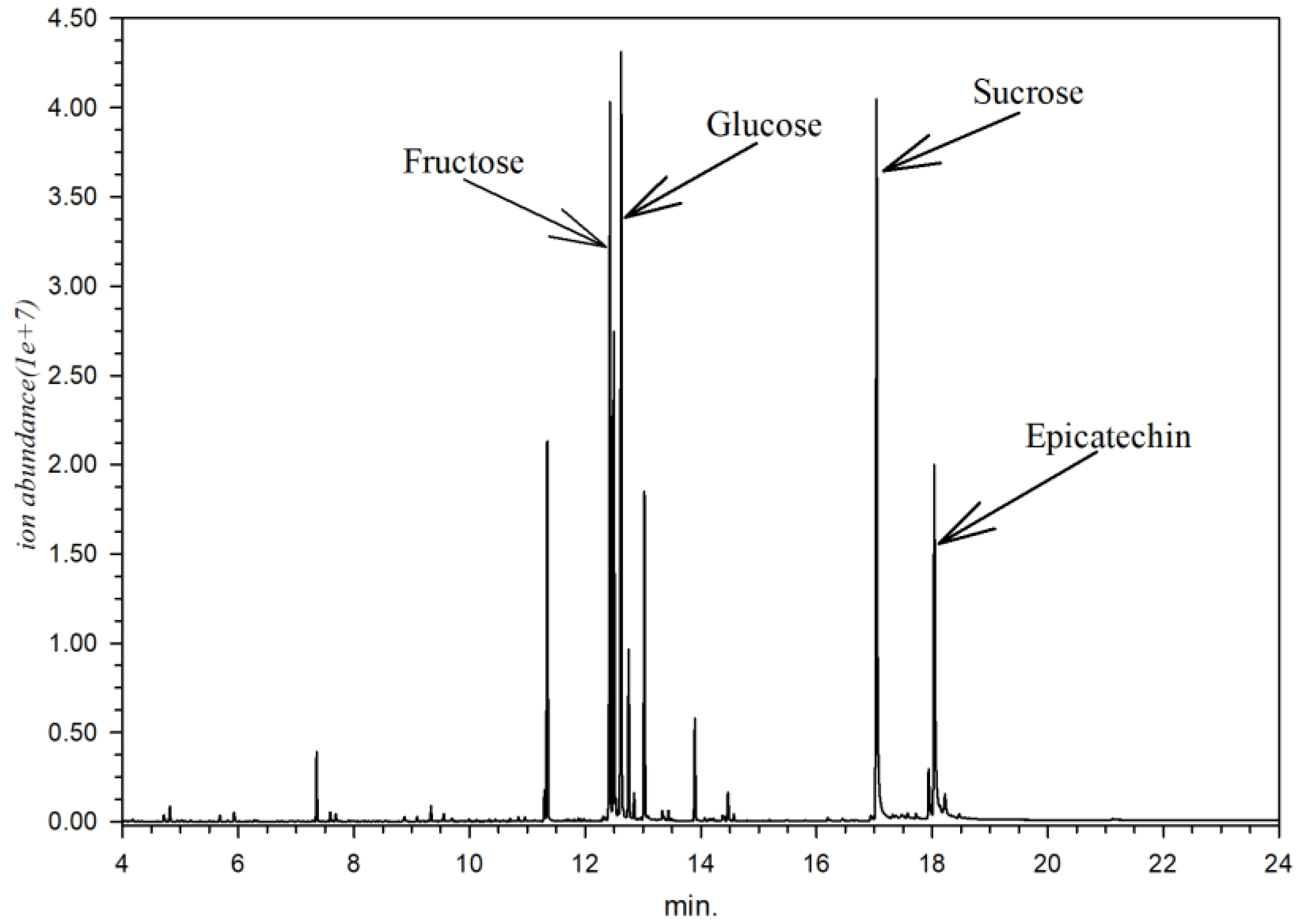
3.2. X. granatum Metabolite Profiling Using GC-MS
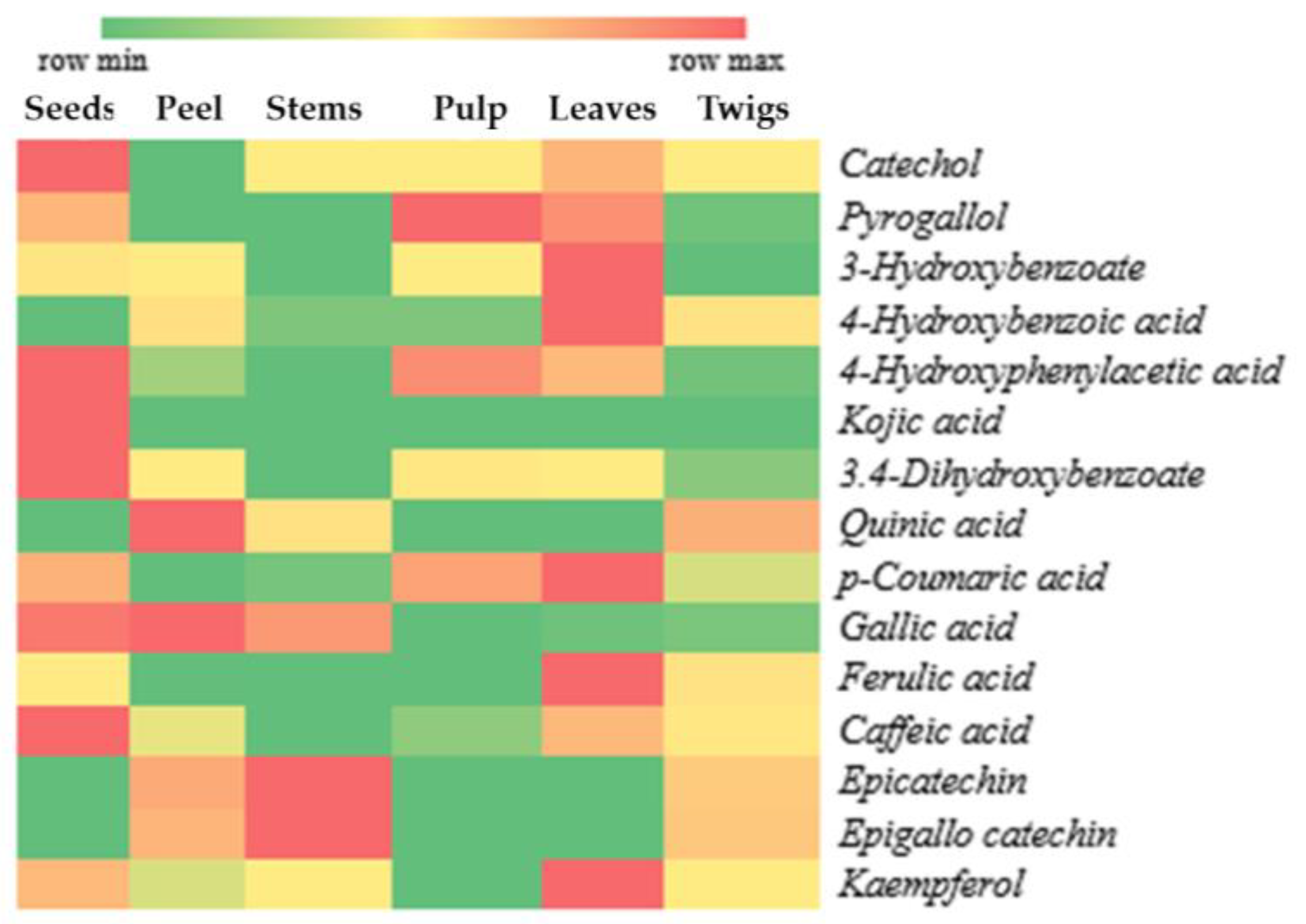
3.3. Prediction of Active Antioxidant Compounds in X. granatum
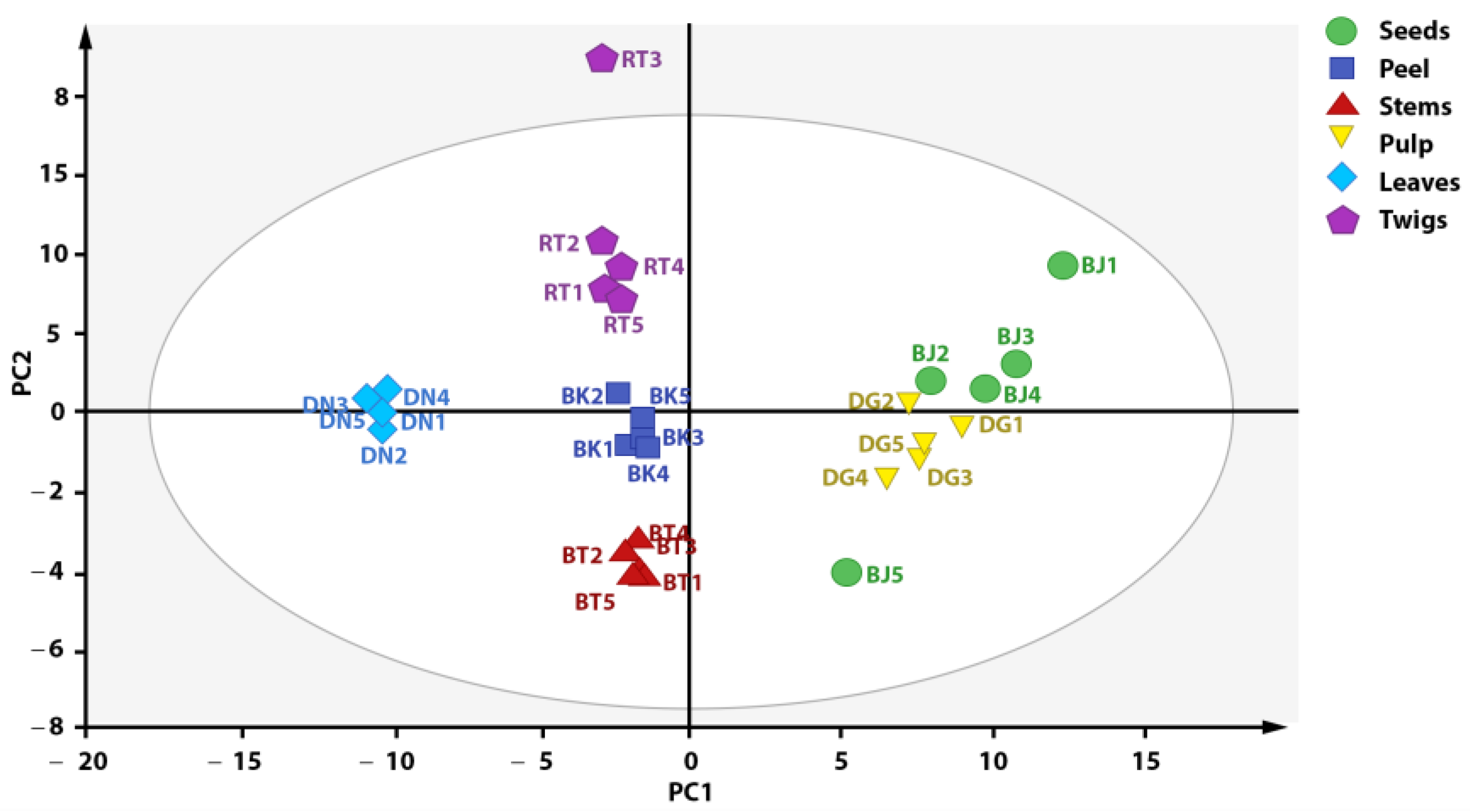
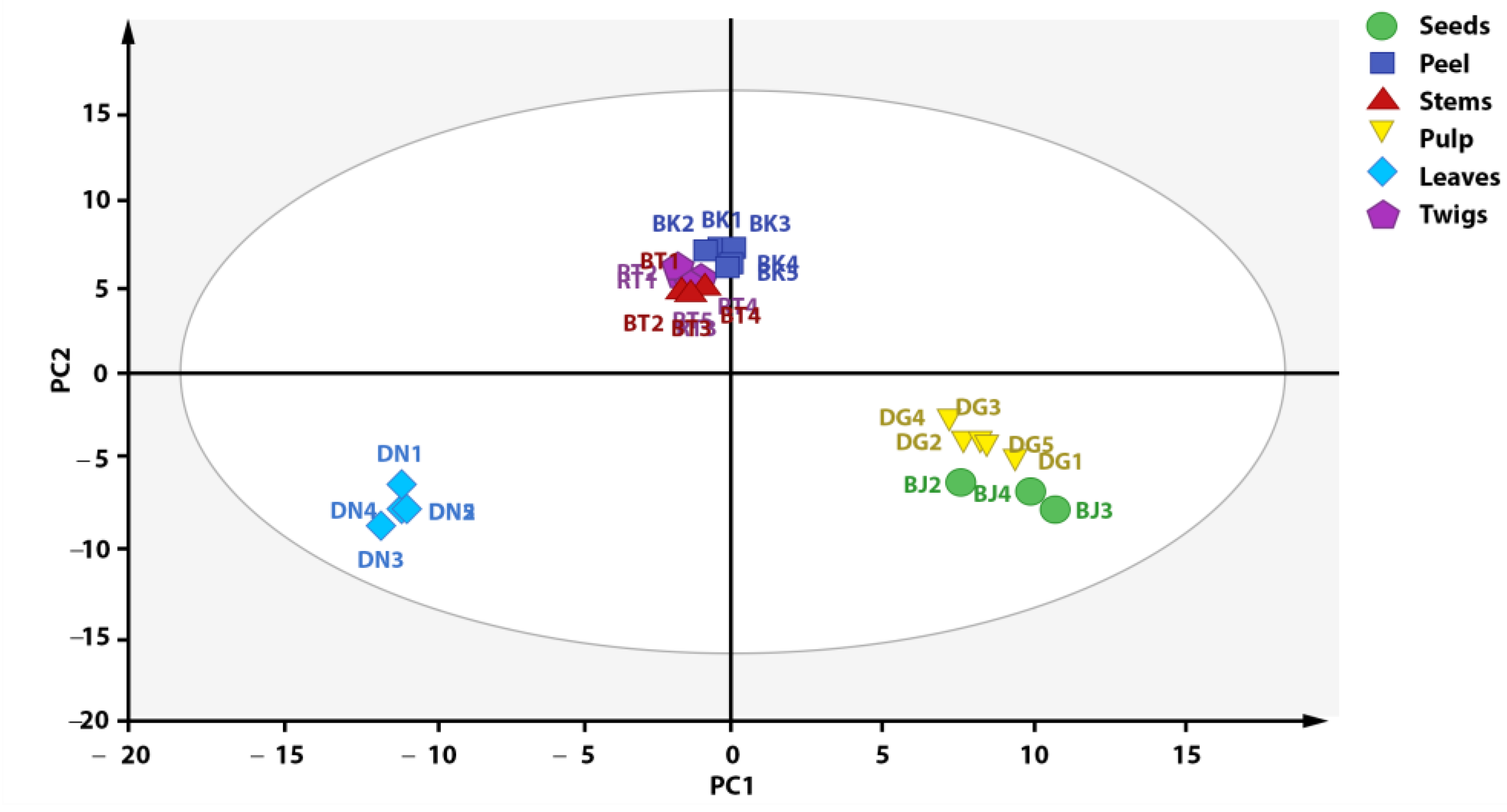
3.4. Discrimination of X. granatum Plant Parts with PCA Based on Metabolite Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Purnobasuki, H. Potensi mangrove sebagai tanaman obat. J. Ilm. Ilmu-Ilmu Hayati 2004, 9, 125–126. [Google Scholar] [CrossRef]
- Zamani, N.P.; Ghazali, M.; Batubara, I. The study of tyrosinase and antioxidant activity of Xylocarpus granatum Koenig seed kernel extract toward evidence based indigenous knowledge from Togean Archipelago, Indonesia. J. Mar. Sci: Res. Dev. 2015, 5, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Das, S.K.; Samantaray, D.; Sahoo, S.K.; Pradhan, S.K.; Samanta, L.; Thatoi, H. Bioactivity guided isolation of antidiabetic and antioxidant compound from Xylocarpus granatum J. Koenig bark. 3 Biotech 2019, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Quispe, C.; Hossain, R.; Jain, D.; Ahmed Khan, R.; Janmeda, P.; Islam, M.T.; Ansar Rasul Suleria, H.; Martorell, M.; Daştan, S.D.; et al. Ethnomedicinal use, phytochemistry, and pharmacology of Xylocarpus granatum J. Koenig. Evid. Based Complement Altern. Med. 2021, 2021, 8922196. [Google Scholar] [CrossRef]
- Gao, Q.; Gao, Y.; Song, H.; Li, J.; Wu, Y.; Shi, X.; Shi, H.; Ma, Y. Cipadesin A, a bioactive ingredient of Xylocarpus granatum, produces antidepressant-like effects in adult mice. Neurosci. Lett. 2016, 633, 33–39. [Google Scholar] [CrossRef]
- Pejin, B.; Glumac, M. New cytotoxic natural products from the mangrove biome: Covering the period 2007-2015. J. Nat. Prod. Res. 2018, 33, 1624–1628. [Google Scholar] [CrossRef]
- Zhou, Z.F.; Kurtán, T.; Mándi, A.; Gu, Y.C.; Yao, L.G.; Xin, G.R.; Li, X.W.; Guo, Y.W. Novel and neuroprotective tetranortriterpenoids from Chinese mangrove Xylocarpus granatum Koenig. Sci. Rep. 2016, 6, 33908. [Google Scholar] [CrossRef]
- Das, S.K.; Samanta, L.; Thatoi, H. In vitro antidiabetic and antioxidant potentials of leaf and stem bark extracts of a mangrove plant, Xylocarpus granatum. J. Herbs Spices Med. Plants 2016, 22, 105–117. [Google Scholar] [CrossRef]
- Liao, M.; Pedpradab, P.; Wun, J. Thaixylograins A-H: Eight new limonoids from the thai mangrove, Xylocarpus granatum. Phytochem. Lett. 2017, 19, 126–131. [Google Scholar] [CrossRef]
- Batubara, I.; Darusman, L.K.; Mitsunaga, T.; Rahminiwati, M.; Djauhari, E. Potency of Indonesian medicinal plants as tyrosinase inhibitor and antioxidant agent. J. Bio. Sci. 2010, 10, 138–144. [Google Scholar] [CrossRef]
- Celano, R.; Docimo, T.; Piccinelli, A.L.; Rizzo, S.; Campone, L.; Di Sanzo, R.; Carabetta, S.; Rastrelli, L.; Russo, M. Specialized metabolite profiling of different Glycyrrhiza glabra organs by untargeted UHPLC-HRMS. Ind. Crop. Prod. 2021, 170, 113688. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, W.; Wu, W.; Bai, R.; Kuang, S.; Shi, B.; Li, D. Chemical composition and diversity of the essential oils of Juniperus rigida along the elevations in Helan and Changbai Mountains and correlation with the soil characteristics. Ind. Crop. Prod. 2021, 159, 113032. [Google Scholar] [CrossRef]
- Dadwal, V.; Joshi, R.; Gupta, M. A comparative metabolomic investigation in fruit sections of Citrus medica L. and Citrus maxima L. detecting potential bioactive metabolites using UHPLC-QTOF-IMS. Food Res. Int. 2022, 157, 111486. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Suh, J.H. Metabolomics as a tool to evaluate nut quality and safety. Trends Food Sci. Technol. 2022, 129, 528–543. [Google Scholar] [CrossRef]
- Lebanov, L.; Ghiasvand, A.; Paull, B. Data handling and data analysis in metabolomic studies of essential oils using GC-MS. J. Chromatogr. A 2021, 1640, 461896. [Google Scholar] [CrossRef]
- Putri, S.P.; Fukusaki, E. Mass Spectrometry-Based Metabolomics: A Practical Guide; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Niu, X.; Mi, S.; Jin, Q.; Sang, Y.; Wang, X. Characterization and discrimination of two varieties of eggplants using multi-element and metabolomics profiles coupled with chemometrics analysis. Food Res. Int. 2022, 162, 111976. [Google Scholar] [CrossRef]
- Bro, L.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef] [Green Version]
- Feizi, N.; Hashemi-Nasab, F.S.; Golpelichi, F.; Saburouh, N.; Parastar, H. Recent trends in application of chemometric methods for GC-MS and GC×GC-MS-based metabolomic studies. TrAC Trends Anal. Chem. 2021, 138, 116239. [Google Scholar] [CrossRef]
- Zduńska, K.; Dana, A.; Kolodziejczak, A. Antioxidant properties of ferulic acid and its possible application. J. Skin Pharm. Physiol. 2018, 31, 332–336. [Google Scholar] [CrossRef]
- Singh, D.P.; Verma, S.; Prabha, R. Investigation on antioxidant potential of phenolic acid and flavonoid: The common phytochemical ingredients in plant. J. Plant Biochem. Physiol. 2018, 6, 219. [Google Scholar] [CrossRef]
- Rahmawati, S.I.; Izzati, F.N.; Hapsari, Y.; Septiana, E.; Rachman, F.; Bustanussalam; Simanjuntak, P. Endophytic microbes and antioxidant activities of secondary metabolites from mangroves Avicennia marina and Xylocarpus granatum. IOP Conf. Ser. Earth Env. Sci. 2019, 278, 012065. [Google Scholar] [CrossRef]
- Caban, M.; Lewandowska, U. Polyphenols and the potential mechanisms of their therapeutic benefits against inflammatory bowel diseases. J. Funct. Foods 2022, 95, 105181. [Google Scholar] [CrossRef]
- Procházková, D.; Boušová, I.; Wilhelmová, N. Antioxidant and prooxidant properties of flavonoids. Fitoterapia 2011, 82, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Zhong, Y.; Duan, Y.; Chen, Q.; Li, F. Antioxidant mechanism of tea polyphenols and its impact on health benefits. Anim. Nutr. 2020, 6, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lin, L.; Feng, Y.; Zhao, M.; Li, X.; Zhu, Q.; Xiao, Z. Enrichment of antioxidants from soy sauce using macroporous resin and identification of 4-ethylguaiacol, catechol, daidzein, and 4-ethylphenol as key small molecule antioxidants in soy sauce. Food Chem. 2018, 240, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Hang, D.T.N.; Hoa, N.T.; Bich, H.N.; Mechler, A.; Vo, Q.V. The hydroperoxyl radical scavenging activity of natural hydroxybenzoic acids in oil and aqueous environments: Insights into the mechanism and kinetics. Phytochemistry 2022, 201, 113281. [Google Scholar] [CrossRef]
- Saeedi, M.; Eslamifar, M.; Khezri, K. Kojic acid application in cosmetic and pharmaceutical preparations. J. Biomed Pharmacother 2019, 110, 582–593. [Google Scholar] [CrossRef]
- Nicolì, F.; Negro, C.; Vergine, M.; Aprile, A.; Nutricati, E.; Sabella, E.; Miceli, A.; Luvisi, A.; De Bellis, L. Evaluation of phytochemical and antioxidant properties of 15 Italian Olea europaea L. cultivar leaves. Molecules 2019, 24, 1998. [Google Scholar] [CrossRef] [Green Version]
- Adeyemi, O.S.; Atolani, O.; Banerjee, P.; Arolasafe, G.; Preissner, R.; Etukudoh, P.; Ibraheem, O. Computational and experimental validation of antioxidant properties of synthesized biactive ferulic acid derivatives. Int. J. Food Prop. 2018, 21, 101. [Google Scholar] [CrossRef]
- Prakash, M.; Basavaraj, B.V.; Chidambara Murthy, K.N. Biological functions of epicatechin: Plant cell to human cell health. J. Funct. Foods 2019, 52, 14–24. [Google Scholar] [CrossRef]
- Spagnol, C.M.; de Assis, R.P.; Brunetti, I.L.; Isaac, V.L.B.; Salgado, H.R.N.; Corrêa, M.A. In vitro methods to determine the antioxidant activity of caffeic acid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 219, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Phasha, V.; Senabe, J.; Ndzotoyi, P.; Okole, B.; Fouche, G.; Chuturgoon, A. Review on the use of kojic acid—A skin-lightening ingredient. Cosmetics 2022, 9, 64. [Google Scholar] [CrossRef]
| Plant Part | IC50 (ppm) |
|---|---|
| Seeds | 104.64 e |
| Fruit peel | 9.02 c |
| Stems | 7.73 b |
| Pulp | 44.48 d |
| Leaves | 295.08 f |
| Twigs | 9.86 c |
| Vitamin C | 4.18 a |
| No | Rt (minutes) | RI | Ion (m/z) | Formula | Compound | Area (%) |
|---|---|---|---|---|---|---|
| 1 | 4.33 | 1003 | 117 | C3H8O2 | propylene glycol | 0.028 |
| 2 | 4.60 | 1028 | 174 | C3H9N | n-propylamine | 0.006 |
| 3 | 4.71 | 1038 | 117 | C4H10O2 | butane-2.3-diol | 1.084 |
| 4 | 4.73 | 1039 | 152 | C5H5NO | 2-hydroxypyridine | 0.035 |
| 5 | 4.94 | 1058 | 147 | C3H8O2 | propane-1.3-diol | 0.006 |
| 6 | 4.95 | 1058 | 130 | C3H4O3 | Pyruvic acid | 0.047 |
| 7 | 5.02 | 1064 | 147 | C3H6O3 | Lactic acid | 0.125 |
| 8 | 5.07 | 1069 | 131 | C4H8O3 | 2-Hydroxyisobutyric acid | 0.341 |
| 9 | 5.18 | 1078 | 147 | C2H4O3 | Glycolic acid | 0.356 |
| 10 | 5.39 | 1097 | 174 | C4H11N | n-Butylamine | 0.003 |
| 11 | 5.55 | 1111 | 131 | C4H8O3 | 2-hydroxybutanoic acid | 0.003 |
| 12 | 5.87 | 1141 | 152 | C5H5NO | 4-Hydroxypyridine | 0.007 |
| 13 | 5.93 | 1146 | 147 | C4H8O3 | 3-hydroxybutyric acid | 0.317 |
| 14 | 5.94 | 1147 | 116 | C3H7NO2 | Sarcosine_2TMS | 0.007 |
| 15 | 5.95 | 1148 | 142 | C4H7NO | butyrolactam | 0.036 |
| 16 | 6.68 | 1216 | 131 | C4H8O3 | 3-Hydroxyisovaleric acid | 0.005 |
| 17 | 6.76 | 1224 | 144 | C5H11NO2 | Valine_2TMS | 0.063 |
| 18 | 6.89 | 1237 | 189 | CH4N2O | Urea | 0.005 |
| 19 | 6.91 | 1238 | 116 | C4H8O3 | 4-hydroxybutyric acid | 0.011 |
| 20 | 6.95 | 1242 | 174 | C2H7NO | Ethanolamine | 0.008 |
| 21 | 7.01 | 1249 | 175 | C6H10O4 | ethylsuccinate | 0.007 |
| 22 | 7.04 | 1252 | 179 | C7H6O2 | Benzoic acid | 0.017 |
| 23 | 7.07 | 1255 | 147 | C3H6O3 | Dihydroxyacetone | 0.013 |
| 24 | 7.17 | 1264 | 116 | C3H7NO3 | Serine_2TMS | 0.003 |
| 25 | 7.29 | 1276 | 174 | C2H7NO | 2-Aminoethanol | 0.070 |
| 26 | 7.33 | 1280 | 158 | C6H13NO2 | Leucine_2TMS | 0.033 |
| 27 | 7.35 | 1282 | 147 | C3H8O3 | Glycerol | 0.880 |
| 28 | 7.50 | 1297 | 180 | C6H5NO2 | Nicotinic acid | 0.008 |
| 29 | 7.53 | 1300 | 158 | C6H13NO2 | Isoleucine_2TMS | 0.033 |
| 30 | 7.69 | 1317 | 147 | C4H6O4 | Succinic acid | 0.068 |
| 31 | 7.80 | 1328 | 254 | C6H6O2 | Catechol | 0.007 |
| 32 | 7.91 | 1340 | 147 | C3H6O4 | Glyceric acid | 0.075 |
| 33 | 7.97 | 1347 | 241 | C4H4N2O2 | Uracil | 0.010 |
| 34 | 7.99 | 1349 | 254 | C4H4O4 | Fumaric acid | 0.027 |
| 35 | 8.14 | 1365 | 240 | C5H5NO2 | pyrrole-2-carboxylic acid | 0.013 |
| 36 | 8.21 | 1372 | 188 | C3H7NO2 | Alanine_3TMS | 0.020 |
| 37 | 8.25 | 1376 | 156 | C6H11NO2 | DL-Pipecolic acid | 0.158 |
| 38 | 8.32 | 1384 | 147 | C4H6O4 | erythronic acid lactone | 0.032 |
| 39 | 8.46 | 1398 | 218 | C4H9NO3 | Threonine_3TMS | 0.007 |
| 40 | 8.55 | 1408 | 239 | C6H6O2 | hydroquinone | 0.021 |
| 41 | 8.55 | 1409 | 147 | C5H8O4 | Glutaric acid | 0.069 |
| 42 | 8.61 | 1415 | 255 | C5H6N2O2 | Thymine | 0.001 |
| 43 | 8.66 | 1422 | 103 | C4H9NO3 | homoserine | 0.016 |
| 44 | 8.83 | 1440 | 233 | C4H8O4 | 2-deoxytetronic acid | 0.004 |
| 45 | 8.86 | 1443 | 174 | C4H9NO2 | 3-aminoisobutyric acid | 0.080 |
| 46 | 8.96 | 1456 | 117 | C10H20O2 | Decanoic acid | 0.004 |
| 47 | 9.17 | 1480 | 158 | C5H9NO3 | trans-4-hydroxy-L-proline | 0.012 |
| 48 | 9.23 | 1487 | 247 | C5H8O5 | Citramalic acid | 0.003 |
| 49 | 9.33 | 1498 | 147 | C4H6O5 | Malic acid | 0.751 |
| 50 | 9.34 | 1499 | 223 | C7H6O2 | p-Hydroxybenzal dehyde | 0.011 |
| 51 | 9.43 | 1509 | 244 | C4H8N2O3 | asparagine dehydrated | 0.004 |
| 52 | 9.45 | 1512 | 117 | C4H8O5 | isothreonic acid | 0.022 |
| 53 | 9.48 | 1516 | 147 | C4H10O4 | Threitol | 0.031 |
| 54 | 9.53 | 1521 | 174 | C4H12N2 | putrescine 3tms | 0.018 |
| 55 | 9.54 | 1523 | 267 | C9H8O4 | Acetylsalicylic acid | 0.014 |
| 56 | 9.55 | 1524 | 217 | C4H10O4 | Meso erythritol | 0.109 |
| 57 | 9.59 | 1528 | 232 | C4H7NO4 | Aspartic acid_3TMS | 0.010 |
| 58 | 9.63 | 1534 | 156 | C5H7NO3 | Pyroglutamic acid_2TMS | 0.168 |
| 59 | 9.68 | 1539 | 230 | C5H9NO3 | Hydroxyproline | 0.187 |
| 60 | 9.70 | 1542 | 174 | C4H9NO2 | 4-Aminobutyric acid | 0.455 |
| 61 | 9.81 | 1556 | 263 | C6H6O4 | 5-hydroxymethyl-2-furoic acid | 0.002 |
| 62 | 9.84 | 1560 | 239 | C6H6O3 | Pyrogallol | 0.004 |
| 63 | 9.96 | 1574 | 267 | C7H6O3 | 3-Hydroxybenzoate | 0.007 |
| 64 | 9.99 | 1577 | 147 | C4H8O5 | Threonic acid | 0.097 |
| 65 | 10.01 | 1580 | 179 | C8H10O2 | 4-Hydroxyphenethyl alcohol | 0.012 |
| 66 | 10.04 | 1584 | 129 | C5H8O5 | 2-hydroxyglutaric acid | 0.015 |
| 67 | 10.12 | 1594 | 147 | C7H12O5 | 2-Isopropylmalic acid | 0.124 |
| 68 | 10.28 | 1614 | 147 | C6H10O5 | 3-Hydroxy-3-methylglutarate | 0.032 |
| 69 | 10.31 | 1617 | 217 | C5H8O5 | xylonolactone | 0.009 |
| 70 | 10.35 | 1622 | 117 | C4H6O6 | L-(+)-Tartaric acid | 0.034 |
| 71 | 10.41 | 1630 | 246 | C5H9NO4 | Glutamic acid_3TMS | 0.007 |
| 72 | 10.46 | 1636 | 267 | C7H6O3 | 4-Hydroxybenzoic acid | 0.094 |
| 73 | 10.50 | 1642 | 200 | C6H11NO2 | Pipecolic acid | 0.037 |
| 74 | 10.55 | 1648 | 179 | C8H8O3 | 4-Hydroxyphenylacetic acid | 0.004 |
| 75 | 10.74 | 1672 | 103 | C5H10O5 | Xylose | 0.013 |
| 76 | 10.82 | 1683 | 103 | C5H10O5 | Lyxose | 0.273 |
| 77 | 10.84 | 1686 | 271 | C6H6O4 | Kojic acid | 0.285 |
| 78 | 10.98 | 1703 | 103 | C5H10O5 | Ribose | 0.095 |
| 79 | 11.14 | 1726 | 204 | C6H10O5 | 1.6-Anhydroglucose | 0.087 |
| 80 | 11.20 | 1734 | 217 | C5H12O5 | Xylitol | 0.016 |
| 81 | 11.22 | 1736 | 219 | C6H14O5 | diglycerol | 0.005 |
| 82 | 11.29 | 1746 | 117 | C6H12O5 | Rhamnose | 0.252 |
| 83 | 11.30 | 1746 | 217 | C5H12O5 | Arabitol | 1.275 |
| 84 | 11.34 | 1752 | 117 | C6H14O5 | 6-deoxyglucitol | 0.971 |
| 85 | 11.48 | 1771 | 147 | C6H12O5 | 2-Deoxy-D-glucose | 0.065 |
| 86 | 11.51 | 1775 | 297 | C8H8O4 | vanillic acid | 0.020 |
| 87 | 11.52 | 1777 | 231 | C6H10O5 | 3.6-anhydro-D-galactose | 0.021 |
| 88 | 11.60 | 1787 | 156 | C5H10N2O3 | Glutamine_3TMS | 0.007 |
| 89 | 11.62 | 1790 | 129 | C6H14O5 | 3-deoxyhexitol | 0.077 |
| 90 | 11.65 | 1794 | 147 | C5H10O6 | Xylonic acid | 0.056 |
| 91 | 11.84 | 1821 | 204 | C7H10O5 | Shikimic acid | 0.434 |
| 92 | 11.93 | 1834 | 193 | C7H6O4 | 3.4-Dihydroxybenzoate | 0.267 |
| 93 | 11.98 | 1841 | 204 | C6H12O5 | 1.5-Anhydro-D-glucitol | 0.067 |
| 94 | 12.26 | 1881 | 159 | C6H13NO5 | D-(+)-Galactosamine | 0.268 |
| 95 | 12.29 | 1886 | 103 | C6H12O6 | Psicose | 0.118 |
| 96 | 12.30 | 1887 | 103 | C6H12O6 | Tagatose | 0.084 |
| 97 | 12.33 | 1891 | 345 | C7H12O6 | Quinic acid | 1.910 |
| 98 | 12.46 | 1910 | 104 | C6H10O6 | L-Gulcono-1.4-lactone | 3.503 |
| 99 | 12.50 | 1916 | 103 | C6H12O6 | Fructose | 6.983 |
| 100 | 12.53 | 1921 | 319 | C6H12O6 | Mannose | 0.179 |
| 101 | 12.56 | 1925 | 204 | C7H14O6 | 1-methylgalactose | 0.121 |
| 102 | 12.57 | 1927 | 319 | C6H12O6 | Galactose | 0.328 |
| 103 | 12.58 | 1928 | 217 | C6H10O6 | Glucono-1.5-lactone | 0.998 |
| 104 | 12.62 | 1935 | 319 | C6H12O6 | Glucose | 7.042 |
| 105 | 12.71 | 1947 | 203 | C6H13NO5 | Glucosamine | 0.023 |
| 106 | 12.74 | 1953 | 293 | C9H8O3 | p-Coumaric acid | 0.035 |
| 107 | 12.78 | 1959 | 218 | C9H11NO3 | Tyrosine | 0.086 |
| 108 | 12.80 | 1962 | 275 | C6H12O7 | galactonic acid | 0.014 |
| 109 | 12.82 | 1963 | 204 | C6H12O6 | hexose | 1.079 |
| 110 | 12.84 | 1968 | 319 | C6H14O6 | Mannitol | 0.181 |
| 111 | 12.89 | 1975 | 281 | C7H6O5 | Gallic acid | 0.034 |
| 112 | 12.90 | 1977 | 217 | C6H14O6 | Galactitol | 0.025 |
| 113 | 13.02 | 1994 | 318 | C6H10O5 | conduritol-beta-expoxide | 2.131 |
| 114 | 13.10 | 2006 | 147 | C6H14O6 | hexitol | 0.099 |
| 115 | 13.31 | 2040 | 353 | C5H4N4O2 | Xanthine | 0.006 |
| 116 | 13.33 | 2043 | 147 | C6H12O7 | Gluconic acid | 0.154 |
| 117 | 13.35 | 2046 | 117 | C16H32O2 | palmitic acid | 0.103 |
| 118 | 13.44 | 2059 | 204 | C8H15NO6 | N-Acetyl-D-glucosamine | 0.363 |
| 119 | 13.48 | 2067 | 147 | C6H12O6 | myo-inositol | 0.019 |
| 120 | 13.53 | 2073 | 129 | C8H15NO6 | N-acetyl-D-mannosamine | 0.007 |
| 121 | 13.73 | 2105 | 338 | C10H10O4 | Ferulic acid | 0.016 |
| 122 | 13.84 | 2124 | 204 | C8H15NO6 | n-acetyl-d-hexosamine | 0.114 |
| 123 | 13.89 | 2132 | 217 | C6H12O6 | Inositol | 1.004 |
| 124 | 14.00 | 2149 | 219 | C9H8O4 | Caffeic acid | 0.011 |
| 125 | 14.07 | 2160 | 319 | C8H15NO6 | N-Acetyl galactosamine | 0.035 |
| 126 | 14.30 | 2198 | 204 | C9H16O9 | beta-mannosylglycerate | 0.027 |
| 127 | 14.57 | 2245 | 117 | C18H36O2 | Stearic acid | 0.196 |
| 128 | 14.61 | 2251 | 202 | C11H12N2O2 | Tryptophan_3TMS | 0.011 |
| 129 | 14.69 | 2266 | 204 | C9H18O8 | glycerol-3-galactoside | 0.021 |
| 130 | 14.79 | 2282 | 361 | C10H17NO9S2 | Sinigrin | 0.004 |
| 131 | 15.12 | 2341 | 217 | C11H15N5O4 | 1-methyladenosine | 0.013 |
| 132 | 15.18 | 2352 | 204 | C6H13O9P | galactose-6-phosphate | 0.056 |
| 134 | 15.85 | 2474 | 217 | C9H12N2O6 | Uridine_4TMS | 0.010 |
| 135 | 16.19 | 2538 | 204 | C12H22O11 | Leucrose | 0.130 |
| 136 | 16.89 | 2674 | 361 | C12H22O11 | beta-Lactose | 0.015 |
| 137 | 17.03 | 2702 | 361 | C12H22O11 | Sucrose | 5.839 |
| 138 | 17.05 | 2706 | 204 | C12H22O11 | D-(+)-Maltose | 0.236 |
| 139 | 17.16 | 2729 | 217 | C12H22O11 | D-(+)-Turanose | 0.361 |
| 140 | 17.36 | 2770 | 217 | C12H24O11 | lactitol | 0.105 |
| 141 | 17.53 | 2806 | 217 | C12H22O12 | lactobionic acid | 0.047 |
| 142 | 17.56 | 2814 | 361 | C12H22O11 | Trehalose | 0.137 |
| 143 | 17.76 | 2855 | 355 | C21H22O10 | 4.5-dihydroxy-7-glucosyloxyflavanone | 0.012 |
| 144 | 17.92 | 2889 | 361 | C12H24O11 | Palatinitol | 0.009 |
| 145 | 17.94 | 2893 | 368 | C15H14O6 | (−)-Epicatechin | 0.422 |
| 146 | 18.08 | 2925 | 361 | C12H22O11 | Gentiobiose | 0.033 |
| 147 | 18.14 | 2937 | 204 | C12H22O11 | Melibiose | 0.051 |
| 148 | 18.31 | 2975 | 456 | C15H14O7 | (−)-Epigallo catechin | 0.032 |
| 149 | 18.74 | 3072 | 204 | C12H22O11 | Galactinol | 0.021 |
| 150 | 18.87 | 3100 | 219 | C16H18O9 | Chlorogenic acid | 0.008 |
| 151 | 18.93 | 3109 | 487 | C15H10O6 | Kaempferol | 0.005 |
| 152 | 20.61 | 3349 | 204 | C18H32O16 | Maltotriose | 0.003 |
| 153 | 20.93 | 3396 | 361 | C18H32O16 | Kestose | 0.010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heryanto, R.; Putra, C.A.; Khalil, M.; Rafi, M.; Putri, S.P.; Karomah, A.H.; Batubara, I. Antioxidant Activity and Metabolite Profiling of Xylocarpus granatum Extracts Using Gas Chromatography–Mass Spectrometry. Metabolites 2023, 13, 156. https://doi.org/10.3390/metabo13020156
Heryanto R, Putra CA, Khalil M, Rafi M, Putri SP, Karomah AH, Batubara I. Antioxidant Activity and Metabolite Profiling of Xylocarpus granatum Extracts Using Gas Chromatography–Mass Spectrometry. Metabolites. 2023; 13(2):156. https://doi.org/10.3390/metabo13020156
Chicago/Turabian StyleHeryanto, Rudi, Cecep Abdurohman Putra, Munawar Khalil, Mohamad Rafi, Sastia Prama Putri, Alfi Hudatul Karomah, and Irmanida Batubara. 2023. "Antioxidant Activity and Metabolite Profiling of Xylocarpus granatum Extracts Using Gas Chromatography–Mass Spectrometry" Metabolites 13, no. 2: 156. https://doi.org/10.3390/metabo13020156
APA StyleHeryanto, R., Putra, C. A., Khalil, M., Rafi, M., Putri, S. P., Karomah, A. H., & Batubara, I. (2023). Antioxidant Activity and Metabolite Profiling of Xylocarpus granatum Extracts Using Gas Chromatography–Mass Spectrometry. Metabolites, 13(2), 156. https://doi.org/10.3390/metabo13020156