Metabolomics Analysis of Different Tissues of Lonicera japonica Thunb. Based on Liquid Chromatography with Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Plant Materials
2.3. Sample Preparation
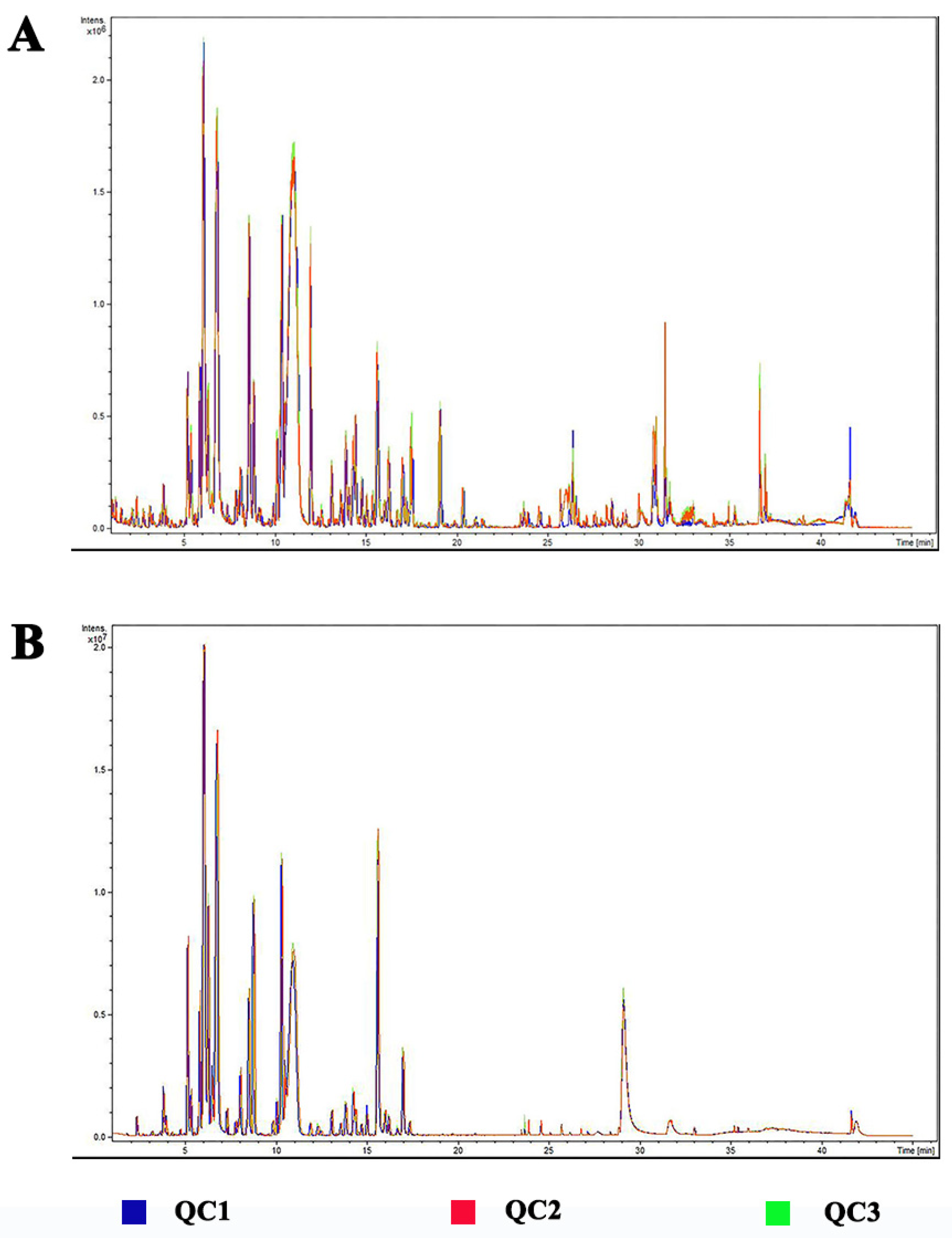
2.4. LC-MS Analysis
2.5. Data Analysis
3. Results and Discussion
3.1. Metabolites Identification in Different Tissues
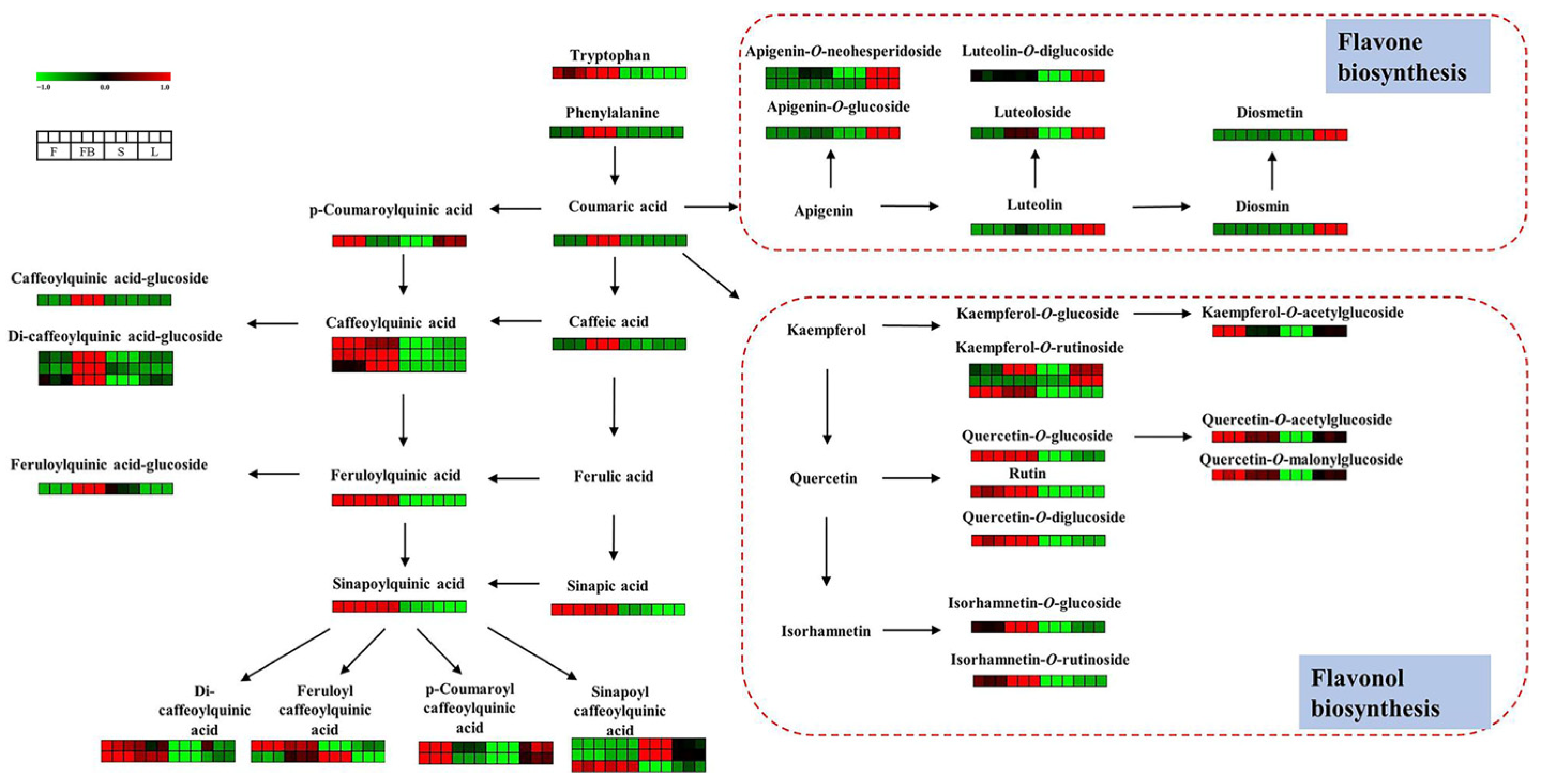
3.1.1. Identification of Flavonoids
3.1.2. Identification of Phenolic Acids
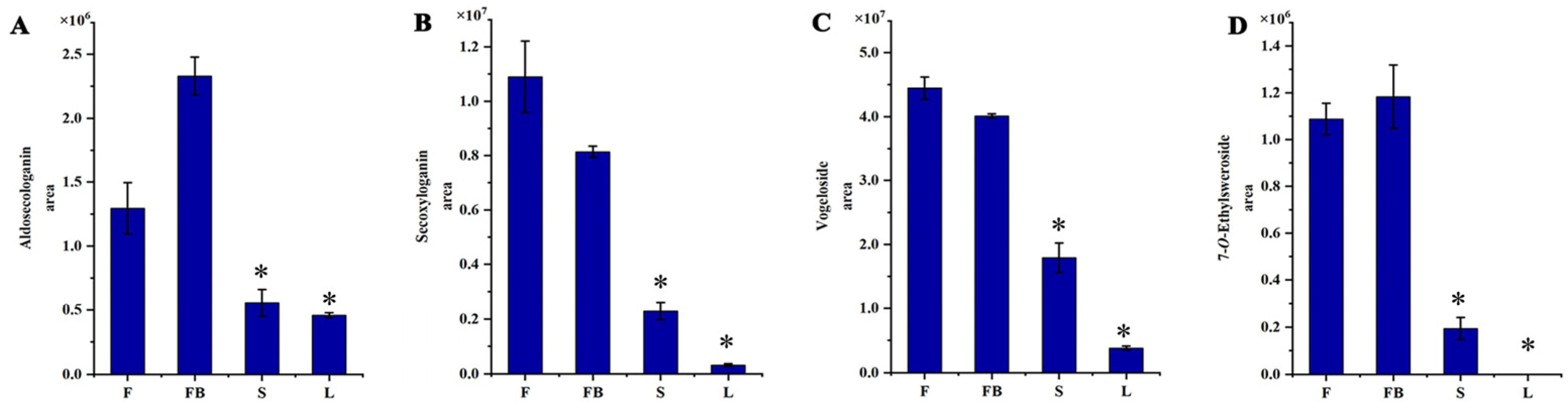
3.1.3. Identification of Iridoids
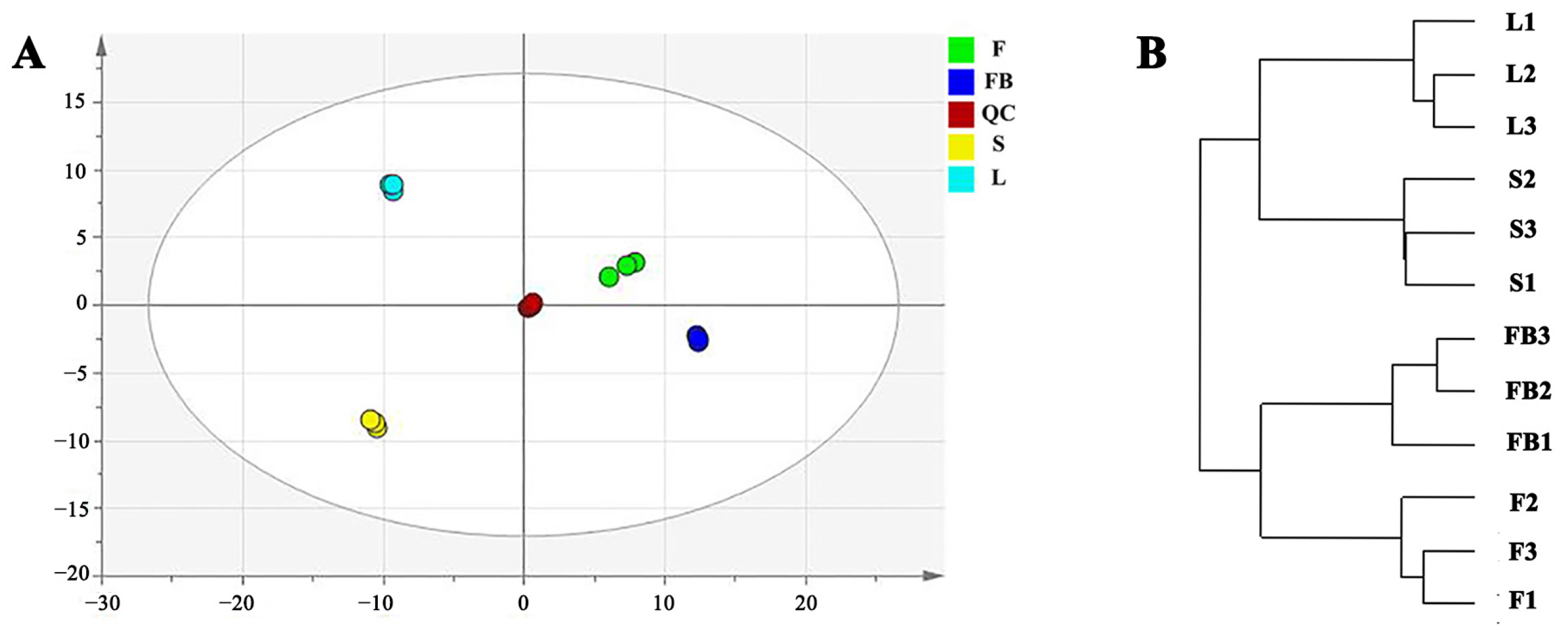
3.2. Metabolomics Difference in Different Tissues of LJT
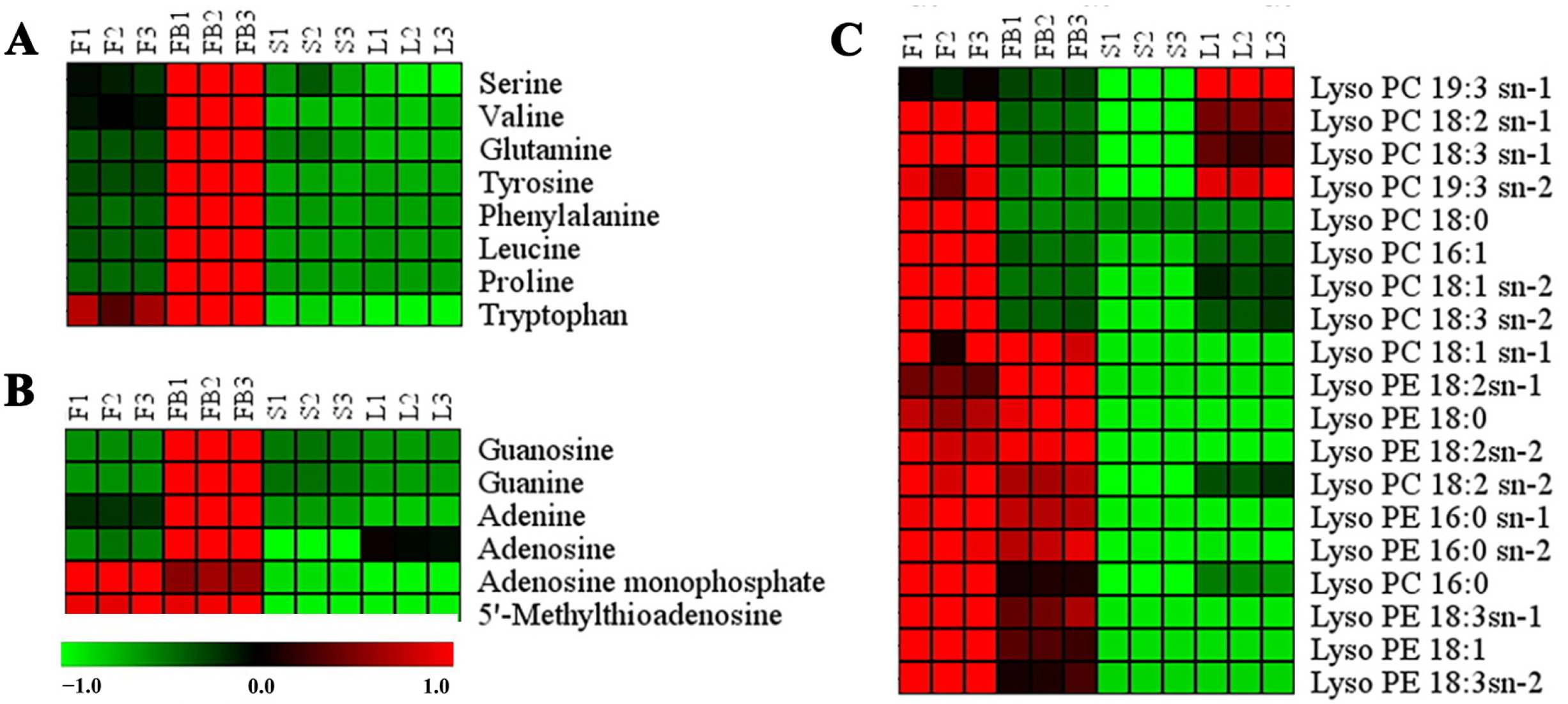
3.2.1. Differences in Primary Metabolites of Different Tissues
3.2.2. Differences in Secondary Metabolites of Different Tissues
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, B.; Zhong, Z.; Wang, T.; Ou, Y.; Tian, J.; Komatsu, S.; Zhang, L. Integrative omics of Lonicera japonica Thunb. Flower development unravels molecular changes regulating secondary metabolites. J. Proteomics 2019, 208, 103470. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Li, C.; Wang, R.; Li, X.; Guo, S.; Zhang, W.; Liu, B. The metabolic profile elucidation of Lonicera japonica flos water extract and the metabolic characteristics evaluation of bioactive compounds in human gastrointestinal tract in vitro. J. Pharm. Biomed. Anal. 2022, 219, 114906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, H.; Bai, X.; Liu, P.; Yang, Y.; Huang, J.; Zhou, L.; Min, X. Fractionation and antioxidant activities of the water-soluble polysaccharides from Lonicera japonica Thunb. Int. J. Biol. Macromol. 2020, 151, 1058–1066. [Google Scholar] [CrossRef]
- Ge, L.; Xie, Q.; Jiang, Y.; Xiao, L.; Wan, H.; Zhou, B.; Wu, S.; Tian, J.; Zeng, X. Genus Lonicera: New drug discovery from traditional usage to modern chemical and pharmacological research. Phytomedicine 2022, 96, 153889. [Google Scholar] [CrossRef]
- Han, M.H.; Lee, W.S.; Nagappan, A.; Hong, S.H.; Jung, J.H.; Park, C.; Kim, H.J.; Kim, G.Y.; Kim, G.; Jung, J.M.; et al. Flavonoids isolated from flowers of Lonicera japonica Thunb. inhibit inflammatory responses in BV2 microglial cells by suppressing TNF-α and IL-β through PI3K/Akt/NF-kb signaling pathways. Phytother. Res. 2016, 30, 1824–1832. [Google Scholar] [CrossRef]
- Oboh, G.; Agunloye, O.M.; Akinyemi, A.J.; Ademiluyi, A.O.; Adefegha, S.A. Comparative study on the inhibitory effect of caffeic and chlorogenic acids on key enzymes linked to Alzheimer’s disease and some pro-oxidant induced oxidative stress in rats’ brain-in vitro. Neurochem. Res. 2013, 38, 413–419. [Google Scholar] [CrossRef]
- Kou, Y.; Li, Z.; Yang, T.; Shen, X.; Wang, X.; Li, H.; Zhou, K.; Li, L.; Xia, Z.; Zheng, X.; et al. Therapeutic potential of plant iridoids in depression: A review. Pharm. Biol. 2022, 60, 2167–2181. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Zhan, C.; Yang, C.; Fernie, A.R.; Luo, J. Metabolomics-centered mining of plant metabolic diversity and function: Past decade and future perspectives. Mol. Plant 2023, 16, 43–63. [Google Scholar] [CrossRef]
- Rao, G.; Liu, X.; Zhang, W.; Wu, W.; Zhang, J. Metabolomics reveals variation and correlation among different tissues of olive (Olea europaea L.). Biol. Open 2017, 6, 1317–1323. [Google Scholar]
- Gálvez Ranilla, L. The application of metabolomics for the study of cereal corn (Zea mays L.). Metabolites 2020, 10, 300. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, H.; Huang, Z.; Zhang, C.; Lyu, L.; Li, W.; Wu, W. Metabolite profiling and classification of highbush blueberry leaves under different shade treatments. Metabolites 2022, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Wang, W.T.; Kong, J.Q.; Yue, Y.D.; Dong, Y.Y.; Zhang, J.C.; Liu, L. Application of UHPLC-Q-TOF MS based untargeted metabolomics reveals variation and correlation amongst different tissues of Eucommia ulmoides Oliver. Microchem. J. 2022, 172, 106919. [Google Scholar] [CrossRef]
- Fan, R.; Peng, C.; Zhang, X.; Qiu, D.; Mao, G.; Lu, Y.; Zeng, J. A comparative UPLC-Q-Orbitrap-MS untargeted metabolomics investigation of different parts of Clausena lansium (Lour.) Skeels. Food Sci. Nutr. 2020, 8, 5811–5822. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, Y.; Duan, Y.; Zhao, Y.; Zhang, D.; Zang, L.; Ya, H. Widely targeted metabolomics analysis of different parts of Salsola collina Pall. Molecules 2021, 26, 1126. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Gu, L.; Xu, W.; Yu, X.; Yin, G.; Wang, J.; Jin, Y.; Wang, L.; Wang, B.; Wang, T. Integrating anti-influenza virus activity and chemical pattern recognition to explore the quality evaluation method of Lonicerae japonicae Flos. Molecules 2022, 27, 5789. [Google Scholar] [CrossRef]
- Wang, D.; Du, N.; Wen, L.; Zhu, H.; Liu, F.; Wang, X.; Du, J.; Li, S. An efficient method for the preparative isolation and purification of flavonoid glycosides and caffeoylquinic acid derivatives from leaves of Lonicera japonica Thunb. using high speed counter-current chromatography (HSCCC) and prep-HPLC guided by DPPH-HPLC experiments. Molecules 2017, 22, 229. [Google Scholar]
- Su, X.; Zhu, Z.H.; Zhang, L.; Wang, Q.; Xu, M.M.; Lu, C.; Zhu, Y.; Zeng, J.; Duan, J.A.; Zhao, M. Anti-inflammatory property and functional substances of Lonicerae japonicae Caulis. J. Ethnopharmacol. 2021, 267, 113502. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Liu, X.; Fan, X.; Wang, X.; Liu, R.; Meng, C.; Wang, C. Structural characterization and screening of chemical markers of flavonoids in Lysimachiae Herba and Desmodii Styracifolii Herba by ultra high-performance liquid chromatography quadrupole time-of-flight tandem mass spectrometry based metabolomics approach. J. Pharm. Biomed. Anal. 2019, 171, 52–64. [Google Scholar] [CrossRef]
- Ali, A.; Cottrell, J.J.; Dunshea, F.R. LC-MS/MS Characterization of phenolic metabolites and their antioxidant activities from australian native plants. Metabolites 2022, 12, 1016. [Google Scholar] [CrossRef]
- Wu, S.; Shen, D.; Wang, R.; Li, Q.; Mo, R.; Zheng, Y.; Zhou, Y.; Liu, Y. Phenolic profiles and antioxidant activities of free, esterified and bound phenolic compounds in walnut kernel. Food Chem. 2021, 350, 129217. [Google Scholar] [CrossRef]
- Zhang, F.X.; Li, Z.T.; Li, M.; Yuan, Y.L.; Cui, S.S.; Chen, J.X.; Li, R.M. Dissection of the potential anti-influenza materials and mechanism of Lonicerae japonicae flos based on in vivo substances profiling and network pharmacology. J. Pharm. Biomed. Anal. 2021, 193, 113721. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tegeder, M. Transporters for amino acids in plant cells: Some functions and many unknowns. Curr. Opin. Plant Biol. 2012, 15, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.B.; El-Azaz, J.; de la Torre, F.N.; Cañas, R.A.; Avila, C.; Cánovas, F.M. Biosynthesis and metabolic fate of phenylalanine in conifers. Front. Plant Sci. 2016, 7, 1030. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kang, Y.; Ma, C.; Miao, R.; Wu, C.; Long, Y.; Ge, T.; Wu, Z.; Hou, X.; Zhang, J.; et al. CNGC2 is a Ca2+ influx channel that prevents accumulation of apoplastic Ca2+ in the leaf. Plant Physiol. 2017, 173, 1342–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty acid- and lipid-mediated signaling in plant defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ruan, Z.; Fei, Z.; Yan, J.; Tang, G. Integrated transcriptome and metabolome analysis revealed that flavonoid biosynthesis may dominate the resistance of Zanthoxylum bungeanum against stem canker. J. Agric. Food Chem. 2021, 69, 6360–6378. [Google Scholar] [CrossRef]
- Aziz, N.; Kim, M.Y.; Cho, J.Y. Anti-inflammatory effects of luteolin: A review of in vitro, in vivo, and in silico studies. J. Ethnopharmacol. 2018, 225, 342–358. [Google Scholar] [CrossRef]
- Wang, W.; Yue, R.F.; Jin, Z.; He, L.M.; Shen, R.; Du, D.; Tang, Y.Z. Efficiency comparison of apigenin-7-O-glucoside and trolox in antioxidative stress and anti-inflammatory properties. J. Pharm. Pharmacol. 2020, 72, 1645–1656. [Google Scholar] [CrossRef]
- Devi, V.G.; Rooban, B.N.; Sasikala, V.; Sahasranamam, V.; Abraham, A. Isorhamnetin-3-glucoside alleviates oxidative stress and opacification in selenite cataract in vitro. Toxicol. In Vitro 2010, 24, 1662–1669. [Google Scholar] [CrossRef]
- Lee, J.; Lee, J.; Kim, S.J.; Kim, J.H. Quercetin-3-O-glucoside suppresses pancreatic cancer cell migration induced by tumor-deteriorated growth factors in vitro. Oncol. Rep. 2016, 35, 2473–2479. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Lüscher, J.; Brillante, L.; Kurtural, S.K. Flavonol profile is a reliable indicator to assess canopy architecture and the exposure of red wine grapes to solar radiation. Front. Plant Sci. 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, X.; Zhang, M.; Li, X.; Lai, F.; Zhao, G. Biocatalytic synthesis of acylated derivatives of troxerutin: Their bioavailability and antioxidant properties in vitro. Microb. Cell Fact. 2018, 17, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Kersh, D.M.; Abou El-Ezz, R.F.; Fouad, M.; Farag, M.A. Unveiling natural and semisynthetic acylated flavonoids: Chemistry and biological actions in the context of molecular docking. Molecules 2022, 27, 5501. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Bamunuarachchi, N.I.; Tabassum, N.; Kim, Y.M. Caffeic acid and its derivatives: Antimicrobial drugs toward microbial pathogens. J. Agric. Food Chem. 2021, 69, 2979–3004. [Google Scholar] [CrossRef]
- Naveed, M.; Hejazi, V.; Abbas, M.; Kamboh, A.A.; Khan, G.J.; Shumzaid, M.; Ahmad, F.; Babazadeh, D.; FangFang, X.; Modarresi-Ghazani, F.; et al. Chlorogenic acid (CGA): A pharmacological review and call for further research. Biomed. Pharmacother. 2018, 97, 67–74. [Google Scholar] [CrossRef]
- Gao, C.Y.; Tian, C.R.; Zhou, R.; Zhang, R.G.; Lu, Y.H. Phenolic composition, DNA damage protective activity and hepatoprotective effect of free phenolic extract from Sphallerocarpus gracilis seeds. Int. Immunopharmacol. 2014, 20, 238–247. [Google Scholar] [CrossRef]
- Wang, C.; Gong, X.; Bo, A.; Zhang, L.; Zhang, M.; Zang, E.; Zhang, C.; Li, M. Iridoids: Research advances in their phytochemistry, biological activities, and pharmacokinetics. Molecules 2020, 25, 287. [Google Scholar] [CrossRef] [Green Version]
- Meragelman, T.L.; Renteria, B.S.; Silva, G.L.; Sotomayor, C.; Gil, R.R. Modified secoiridoid from Acicarpha tribuloides and inhibition of nitric oxide production in LPS-activated macrophages. Phytochemistry 2006, 67, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
| No. | Name | tR/min | m/z | Formula | Classification | Mode | MS Fragments | Tissues |
|---|---|---|---|---|---|---|---|---|
| 1 | Apigenin-5-O-neohesperidoside | 15.59 | 577.1558 | C27H30O14 | Flavonoids | [M−H]− | 431, 269, 251, 223 | F/FB/L |
| 2 | Apigenin-7-O-neohesperidoside | 16.30 | 577.1558 | C27H30O14 | Flavonoids | [M−H]− | 431, 269, 251, 223 | F/FB/S/L |
| 3 | Apigenin-7-glucoside | 16.23 | 431.0976 | C21H20O10 | Flavonoids | [M−H]− | 269, 251, 223 | F/FB/L |
| 4 | Isorhamnetin-3-O-glucoside | 16.25 | 477.1030 | C22H22O12 | Flavonoids | [M−H]− | 315, 300, 271, 243, 179, 151 | F/FB/L |
| 5 | Luteoloside | 13.93 | 447.0924 | C21H20O11 | Flavonoids | [M−H]− | 285, 241, 151, 133 | F/FB/S/L |
| 6 | Quercetin-3-O-glucoside | 13.65 | 463.0874 | C21H20O12 | Flavonoids | [M−H]− | 301, 300, 271, 255, 243, 151 | F/FB/S/L |
| 7 | Isorhamnetin-O-rutinoside Ⅰ | 15.66 | 623.1608 | C28H32O16 | Flavonoids | [M−H]− | 315, 299, 271, 243, 178, 151 | F/FB/L |
| 8 | Isorhamnetin-O-rutinoside Ⅱ | 16.30 | 623.1608 | C28H32O16 | Flavonoids | [M−H]− | 315, 299, 271, 243, 178, 151 | F/FB/L |
| 9 | Luteolin-O-diglucoside | 10.42 | 609.1452 | C27H30O16 | Flavonoids | [M−H]− | 285, 199, 151, 133 | F/FB/L |
| 10 | Kaempferol-O-rutinoside Ⅰ | 13.58 | 593.1502 | C27H30O15 | Flavonoids | [M−H]− | 285, 255 | F/FB/S/L |
| 11 | Kaempferol-O-rutinoside Ⅱ | 14.35 | 593.1502 | C27H30O15 | Flavonoids | [M−H]− | 285, 255 | F/FB/S/L |
| 12 | kaempferol-O-rutinoside Ⅲ | 15.12 | 593.1502 | C27H30O15 | Flavonoids | [M−H]− | 285, 255 | F/FB/L |
| 13 | Quercetin-O-diglucoside | 7.51 | 625.1396 | C27H30O17 | Flavonoids | [M−H]− | 301, 271, 213, 193, 150, 117 | F/FB/S/L |
| 14 | Quercetin-7-O-rutinosie | 8.10 | 609.1447 | C27H30O16 | Flavonoids | [M−H]− | 301, 271, 255, 178 | F/FB/S/L |
| 15 | Rutin | 13.16 | 609.1447 | C27H30O16 | Flavonoids | [M−H]− | 300, 271, 255, 178, 151 | F/FB/S/L |
| 16 | Diosmin | 17.02 | 607.1658 | C28H32O15 | Flavonoids | [M−H]− | 300, 299, 284 | F/FB/L |
| 17 | Luteolin | 21.30 | 285.0395 | C15H10O6 | Flavonoids | [M−H]− | 151, 133, 121, 107 | F/FB/S/L |
| 18 | Kaempferol-O-acetylglucoside | 17.30 | 489.1030 | C23H22O12 | Flavonoids | [M−H]− | 285, 255, 227, 153 | F/FB/S/L |
| 19 | Quercetin-O-acetylglucoside | 14.89 | 505.0975 | C23H22O13 | Flavonoids | [M−H]− | 300, 271, 255, 234, 179, 151 | F/FB/S/L |
| 20 | Quercetin-O-malonylglucoside | 14.89 | 549.0873 | C24H22O15 | Flavonoids | [M−H]− | 300, 271, 255, 234, 179, 151 | F/FB/S/L |
| 21 | Diosmetin | 24.12 | 299.0548 | C16H12O6 | Flavonoids | [M−H]− | 269, 151 | F/FB/L |
| 22 | Apigenin | 29.81 | 269.0447 | C15H10O5 | Flavonoids | [M−H]− | 251, 241, 223 | F/FB/S/L |
| 23 | Diosmetin-O-glucoside Ⅰ | 17.10 | 463.1236 | C22H22O11 | Flavonoids | [M+H]+ | 301, 286, 258, 153 | F/FB/S/L |
| 24 | Diosmetin-O-glucoside Ⅱ | 17.70 | 463.1236 | C22H22O11 | Flavonoids | [M+H]+ | 301, 286, 258, 153 | F/FB/S/L |
| 25 | 5-Hydroxyl-3’,4’,7-trimethoxy flavone | 28.60 | 329.1018 | C18H16O6 | Flavonoids | [M+H]+ | 314, 313 | F/FB/S |
| 26 | Corymbosin | 29.00 | 359.1123 | C19H18O7 | Flavonoids | [M+H]+ | 329 | F/FB |
| 27 | Flavoyadorinin B | 23.30 | 477.1390 | C23H24O11 | Flavonoids | [M+H]+ | 315, 300 | F/FB |
| 28 | Eriodictryol-7-O-glucoside | 13.93 | 449.1080 | C21H22O11 | Flavonoids | [M−H]− | 415, 315, 299, 298, 163 | F/FB/S/L |
| 29 | Neochlorogenic acid | 3.94 | 353.0869 | C16H18O9 | Phenolic acids | [M−H]− | 191, 179, 163, 145, 135 | F/FB/S/L |
| 30 | Chlorogenic acid | 6.16 | 353.0869 | C16H18O9 | Phenolic acids | [M−H]− | 191, 179, 163, 145, 135 | F/FB/S/L |
| 31 | Cryptochlorogenic acid | 8.15 | 353.0869 | C16H18O9 | Phenolic acids | [M−H]− | 191, 179, 173, 163, 145 | F/FB/S/L |
| 32 | 3-O-p-Coumaroylquinic acid | 8.52 | 337.0915 | C16H18O8 | Phenolic acids | [M−H]− | 191, 173, 163, 93 | F/FB/S/L |
| 33 | 4-O-p-Coumaroylquinic acid | 10.44 | 337.0915 | C16H18O8 | Phenolic acids | [M−H]− | 191, 173, 163, 93 | F/FB/S/L |
| 34 | 3-O-Feruloylquinic acid | 10.81 | 367.1025 | C17H20O9 | Phenolic acids | [M−H]− | 193, 191, 173, 93 | F/FB/S/L |
| 35 | 4-O-Feruloylquinic acid | 11.54 | 367.1025 | C17H20O9 | Phenolic acids | [M−H]− | 193, 191, 173, 93 | F/FB/S/L |
| 36 | Sinapoylquinic acid | 10.42 | 397.1130 | C18H22O10 | Phenolic acids | [M−H]− | 223, 191, 173 | F/FB/S/L |
| 37 | Isochlorogenic acid B | 15.10 | 515.1186 | C25H24O12 | Phenolic acids | [M−H]− | 353, 191, 179, 173 | F/FB/S/L |
| 38 | Isochlorogenic acid A | 15.68 | 515.1186 | C25H24O12 | Phenolic acids | [M−H]− | 353, 191, 179, 173 | F/FB/S/L |
| 39 | Isochlorogenic acid C | 17.06 | 515.1186 | C25H24O12 | Phenolic acids | [M−H]− | 353, 191, 179, 173 | F/FB/S/L |
| 40 | Feruloylcaffeoylquinic acid Ⅰ | 19.23 | 529.1338 | C26H26O12 | Phenolic acids | [M−H]− | 367, 353, 173 | F/FB/S/L |
| 41 | Feruloylcaffeoylquinic acid Ⅱ | 20.01 | 529.1338 | C26H26O12 | Phenolic acids | [M−H]− | 367, 353, 173 | F/FB/S/L |
| 42 | Feruloylcaffeoylquinic acid Ⅲ | 20.38 | 529.1338 | C26H26O12 | Phenolic acids | [M−H]− | 367, 353, 173 | F/FB/S/L |
| 43 | p-Coumaroyl caffeoylquinic acid Ⅰ | 18.22 | 499.1230 | C25H24O11 | Phenolic acids | [M−H]− | 353, 337, 335 | F/FB/S/L |
| 44 | p-Coumaroyl caffeoylquinic acid Ⅱ | 18.65 | 499.1230 | C25H24O11 | Phenolic acids | [M−H]− | 353, 337, 335 | F/FB/S/L |
| 45 | p-Coumaroyl caffeoylquinic acid Ⅲ | 19.77 | 499.1230 | C25H24O11 | Phenolic acids | [M−H]− | 353, 337, 335 | F/FB/S/L |
| 46 | Sinapoyl caffeoylquinic acid Ⅰ | 16.88 | 559.1448 | C27H28O13 | Phenolic acids | [M−H]− | 397, 353, 223, 173 | F/FB/S/L |
| 47 | Sinapoyl caffeoylquinic acid Ⅱ | 17.44 | 559.1448 | C27H28O13 | Phenolic acids | [M−H]− | 397, 353, 223, 173 | F/FB/S/L |
| 48 | Sinapoyl caffeoylquinic acid Ⅲ | 19.68 | 559.1448 | C27H28O13 | Phenolic acids | [M−H]− | 397, 353, 223, 173 | F/FB/S/L |
| 49 | Caffeoylquinic acid-glucoside | 3.78 | 515.1397 | C22H28O14 | Phenolic acids | [M−H]− | 353, 191, 179 | F/FB/S/L |
| 50 | Feruloylquinic acid-glucoside | 4.98 | 529.1555 | C23H30O14 | Phenolic acids | [M−H]− | 367, 193, 191 | F/FB/S/L |
| 51 | Sinapoylquinic acid-glucoside | 10.69 | 559.1659 | C24H32O15 | Phenolic acids | [M−H]− | 397, 223, 191 | F/FB/S/L |
| 52 | 3,4-O-caffeoylquinic acid-glucoside | 11.64 | 677.1920 | C28H38O19 | Phenolic acids | [M−H]− | 515, 353, 191 | F/FB/S/L |
| 53 | 3,5-O-caffeoylquinic acid-glucoside | 12.74 | 677.1920 | C28H38O19 | Phenolic acids | [M−H]− | 515, 353, 191 | F/FB/S/L |
| 54 | 4,5-O-caffeoylquinic acid-glucoside | 13.48 | 677.1920 | C28H38O19 | Phenolic acids | [M−H]− | 515, 353, 191 | F/FB/S/L |
| 55 | p-Coumaroyl-glucoside | 6.23 | 325.0919 | C15H18O8 | Phenolic acids | [M−H]− | 163 | F/FB/S/L |
| 56 | Protocatechuic acid-4-glucoside | 2.54 | 315.0712 | C13H16O9 | Phenolic acids | [M−H]− | 153 | F/FB/S/L |
| 57 | Isoeugenol | 20.52 | 163.0756 | C10H12O2 | Phenolic acids | [M−H]− | 148 | F/FB/S/L |
| 58 | Cinnamic acid | 28.9 | 149.0598 | C9H8O2 | Phenolic acids | [M+H]+ | 131, 103 | F/FB/S/L |
| 59 | Caffeic acid | 5.80 | 181.0496 | C9H8O4 | Phenolic acids | [M+H]+ | 163, 145, 89 | F/FB/S/L |
| 60 | Sinapic acid | 10.50 | 225.0759 | C11H12O5 | Phenolic acids | [M+H]+ | 207, 175 | F/FB/S/L |
| 61 | Ferulic acid | 6.90 | 195.0654 | C10H10O4 | Phenolic acids | [M+H]+ | 177, 145 | F/FB/S/L |
| 62 | Coumaric acid | 6.20 | 165.0548 | C9H8O3 | Phenolic acids | [M+H]+ | 147, 119 | F/FB/S/L |
| 63 | 3-Hydroxybenzoic acid | 5.14 | 137.0234 | C7H6O3 | Phenolic acids | [M−H]− | 137, 93 | F/FB/S/L |
| 64 | Lonijapospiroside B Ⅰ | 16.72 | 560.1764 | C27H31NO12 | Iridoids | [M−H]− | 398, 380, 328, 296, 284 | F/FB/S |
| 65 | Lonijapospiroside B Ⅱ | 19.42 | 560.1764 | C27H31NO12 | Iridoids | [M−H]− | 398, 380, 328, 296, 284 | F/FB/S |
| 66 | Lonijapospiroside B Ⅲ | 20.92 | 560.1764 | C27H31NO12 | Iridoids | [M−H]− | 398, 380, 328, 296, 284 | F/FB/S |
| 67 | Morroniside Ⅰ | 8.62 | 405.1393 | C17H26O11 | Iridoids | [M−H]− | 373, 225, 179 | F/FB/S/L |
| 68 | Morroniside Ⅱ | 10.42 | 405.1393 | C17H26O11 | Iridoids | [M−H]− | 373, 225, 179 | F/FB/S/L |
| 69 | Secologanoside Ⅰ | 2.48 | 389.1081 | C16H22O11 | Iridoids | [M−H]− | 209, 165, 121 | F/FB/S/L |
| 70 | Secologanoside Ⅱ | 3.33 | 389.1081 | C16H22O11 | Iridoids | [M−H]− | 209, 165, 121 | F/FB/S/L |
| 71 | Secologanoside Ⅲ | 6.40 | 389.1081 | C16H22O11 | Iridoids | [M−H]− | 209, 165, 121 | F/FB/S/L |
| 72 | Dimethylsecologanoside Ⅰ | 11.57 | 417.1394 | C18H26O11 | Iridoids | [M−H]− | 255, 237, 185, 163, 155 | F/FB/S/L |
| 73 | Dimethylsecologanoside Ⅱ | 13.52 | 417.1394 | C18H26O11 | Iridoids | [M−H]− | 255, 237, 185, 163, 155 | F/FB/S/L |
| 74 | (E)-Aldosecologanin | 16.77 | 757.2550 | C34H46O19 | Iridoids | [M−H]− | 595, 525, 493 | F/FB/S/L |
| 75 | (Z)-Aldosecologanin | 17.83 | 757.2550 | C34H46O19 | Iridoids | [M−H]− | 595, 525, 493 | F/FB/S/L |
| 76 | Swertiamarin | 6.86 | 373.1131 | C16H22O10 | Iridoids | [M−H]− | 211, 193, 167, 149, 123 | F/FB/S/L |
| 77 | Genameside A Ⅰ | 3.58 | 421.1342 | C17H26O12 | Iridoids | [M−H]− | 241, 197 | F/FB/S/L |
| 78 | Genameside A Ⅱ | 4.09 | 421.1342 | C17H26O12 | Iridoids | [M−H]− | 241, 197 | F/FB/S/L |
| 79 | Genameside A Ⅲ | 5.27 | 421.1342 | C17H26O12 | Iridoids | [M−H]− | 241, 197 | F/FB/S/L |
| 80 | 8-Epi-loganic acid | 4.41 | 375.1288 | C16H24O10 | Iridoids | [M−H]− | 213, 169, 151, 125 | F/FB/S/L |
| 81 | 7-Epi-loganic acid | 5.27 | 375.1288 | C16H24O10 | Iridoids | [M−H]− | 213, 169, 151, 125 | F/FB/S/L |
| 82 | Arbutoside Ⅰ | 7.60 | 697.2189 | C28H42O20 | Iridoids | [M−H]− | 535, 373, 355, 341 | F/FB/S/L |
| 83 | Arbutoside Ⅱ | 8.04 | 697.2189 | C28H42O20 | Iridoids | [M−H]− | 535, 373, 355, 341 | F/FB/S/L |
| 84 | Demethyl-strychoside A | 12.41 | 729.2239 | C32H42O19 | Iridoids | [M−H]− | 549, 505, 497, 453, 409 | F/FB/S/L |
| 85 | 7-O-Methyl morroniside | 9.86 | 419.1549 | C18H28O11 | Iridoids | [M−H]− | 239 | F/FB/S/L |
| 86 | Strychoside A Ⅰ | 14.15 | 743.2396 | C33H44O19 | Iridoids | [M−H]− | 581, 563, 511, 467 | F/FB/S/L |
| 87 | Strychoside A Ⅱ | 14.65 | 743.2396 | C33H44O19 | Iridoids | [M−H]− | 581, 563, 511, 467 | F/FB/S/L |
| 88 | Loganic acid-O-pentoside | 7.50 | 509.1868 | C21H32O14 | Iridoids | [M+H]+ | 377 | F/FB/S/L |
| 89 | Secoxyloganin Ⅰ | 5.50 | 405.1393 | C17H24O11 | Iridoids | [M+H]+ | 243, 211, 193, 167 | F/FB/S |
| 90 | Secoxyloganin Ⅱ | 7.10 | 405.1393 | C17H24O11 | Iridoids | [M+H]+ | 243, 211, 193, 167 | F/FB/S |
| 91 | Secoxyloganin Ⅲ | 10.50 | 405.1393 | C17H24O11 | Iridoids | [M+H]+ | 243, 211, 193, 167 | F/FB/S/L |
| 92 | Loganin Ⅰ | 8.90 | 391.1600 | C17H26O10 | Iridoids | [M+H]+ | 229. 211, 197, 193, 179, 167 | F/FB/S/L |
| 93 | Loganin Ⅱ | 10.20 | 391.1600 | C17H26O10 | Iridoids | [M+H]+ | 229. 211, 197, 193, 179, 167 | F/FB/S |
| 94 | 5α-Carboxystrictosidine Ⅰ | 12.80 | 575.2232 | C28H34N2O11 | Iridoids | [M+H]+ | 413, 395, 343 | F/FB |
| 95 | 5α-Carboxystrictosidine Ⅱ | 14.40 | 575.2232 | C28H34N2O11 | Iridoids | [M+H]+ | 413, 395, 343 | F/FB/L |
| 96 | 5α-Carboxystrictosidine Ⅲ | 15.60 | 575.2233 | C28H34N2O11 | Iridoids | [M+H]+ | 413, 395, 343 | F/FB/S |
| 97 | 5α-Carboxystrictosidine Ⅳ | 17.40 | 575.2232 | C28H34N2O11 | Iridoids | [M+H]+ | 413, 395, 343 | F/FB/S/L |
| 98 | Hydro-dimethyl lonijaposide C | 8.40 | 554.2234 | C26H35NO12 | Iridoids | [M+H]+ | 392, 374 | F/FB/S/L |
| 99 | Vogeloside Ⅰ | 11.20 | 389.1445 | C17H24O10 | Iridoids | [M+H]+ | 227, 209, 195, 177, 151 | F/FB/S/L |
| 100 | Vogeloside Ⅱ | 10.40 | 389.1445 | C17H24O10 | Iridoids | [M+H]+ | 227, 209, 195, 177, 151 | F/FB/S/L |
| 101 | 7-O-Ethylsweroside | 15.20 | 403.1596 | C18H26O10 | Iridoids | [M+H]+ | 241, 209, 177 | F/FB/S |
| 102 | Secologanic acid | 6.90 | 375.1287 | C16H22O10 | Iridoids | [M+H]+ | 213, 195, 151 | F/FB/S/L |
| 103 | Lonijaposide T | 10.50 | 594.2184 | C28H35NO13 | Iridoids | [M+H]+ | 362 | F/FB/S |
| 104 | Lonijaposide B | 12.10 | 538.1920 | C25H31NO12 | Iridoids | [M+H]+ | 376, 358, 344, 211 | F/FB/S/L |
| 105 | Dimethyl lonijaposide C | 8.90 | 552.2078 | C26H33NO12 | Iridoids | [M+H]+ | 390, 320,288 | F/FB/S |
| 106 | Sweroside | 8.70 | 359.1339 | C16H22O9 | Iridoids | [M+H]+ | 197, 179, 127 | F/FB/S/L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, L.; Ji, W.; Liu, S.; Fan, J.; Lu, H.; Wang, X. Metabolomics Analysis of Different Tissues of Lonicera japonica Thunb. Based on Liquid Chromatography with Mass Spectrometry. Metabolites 2023, 13, 186. https://doi.org/10.3390/metabo13020186
Wang Y, Li L, Ji W, Liu S, Fan J, Lu H, Wang X. Metabolomics Analysis of Different Tissues of Lonicera japonica Thunb. Based on Liquid Chromatography with Mass Spectrometry. Metabolites. 2023; 13(2):186. https://doi.org/10.3390/metabo13020186
Chicago/Turabian StyleWang, Yan, Lili Li, Wenhua Ji, Shuang Liu, Jiali Fan, Heng Lu, and Xiao Wang. 2023. "Metabolomics Analysis of Different Tissues of Lonicera japonica Thunb. Based on Liquid Chromatography with Mass Spectrometry" Metabolites 13, no. 2: 186. https://doi.org/10.3390/metabo13020186
APA StyleWang, Y., Li, L., Ji, W., Liu, S., Fan, J., Lu, H., & Wang, X. (2023). Metabolomics Analysis of Different Tissues of Lonicera japonica Thunb. Based on Liquid Chromatography with Mass Spectrometry. Metabolites, 13(2), 186. https://doi.org/10.3390/metabo13020186








