Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms
Abstract
:1. Introduction
2. Configuration and Expression of the KLF14 Gene
3. KLF14 Trans-Regulatory Network
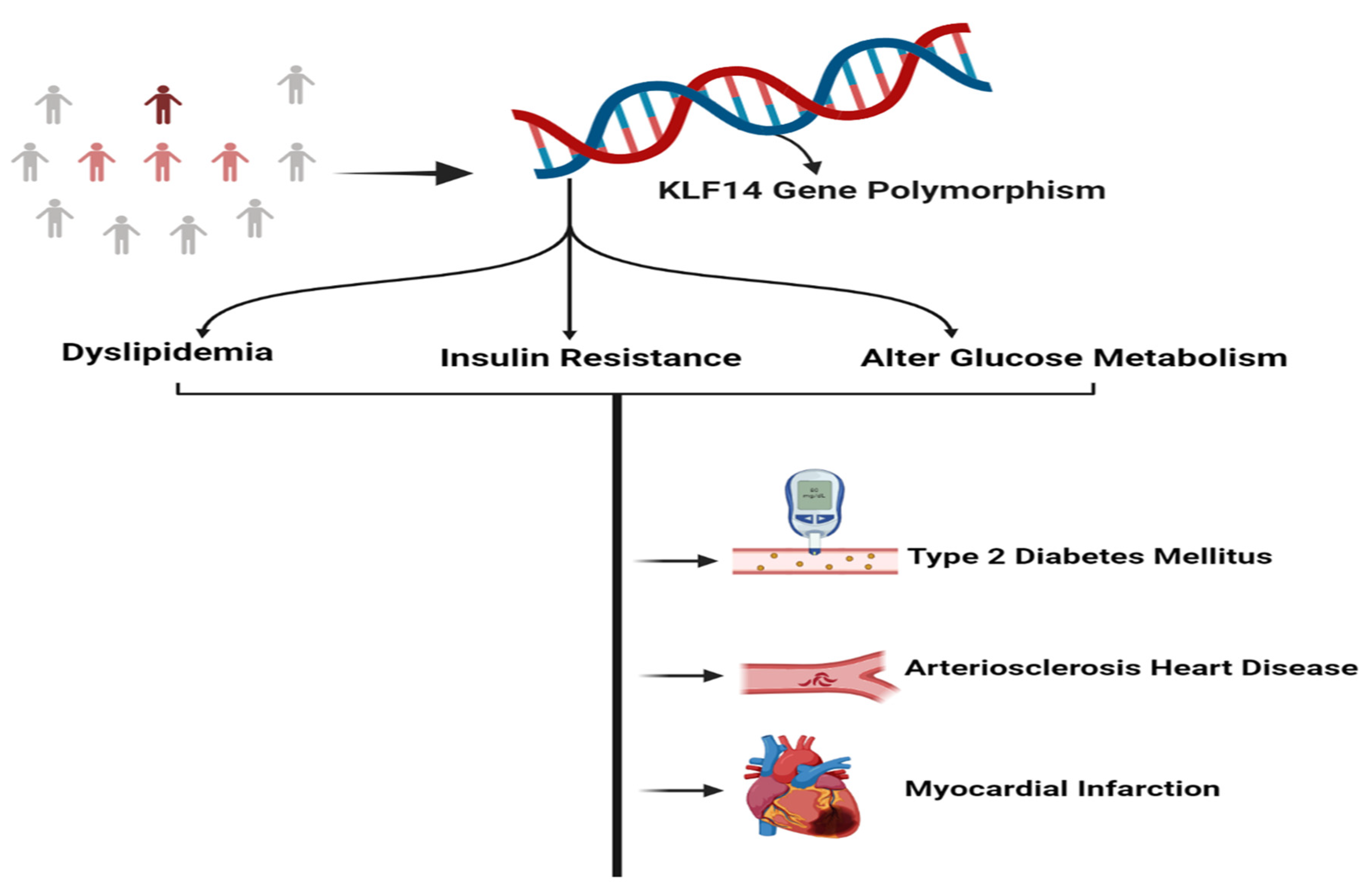
4. Role of KLF14 in the Progression of Metabolic Disorders
5. Role of KLF14 in Glucose Regulation
6. Role of KLF14 in Lipid Metabolism
7. KLF14 Role in Reverse Cholesterol Transport
8. Role of KLF14 in Insulin Sensitivity
9. Role of KLF14 in Human Age Prediction
10. Role of KLF14 in Cancer Suppression
11. Single Nucleotide Polymorphisms of KLF14
12. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- McConnell, B.B.; Yang, V.W.; Mallipattu, S.K.; Estrada, C.C.; He, J.C.; Talmasov, D.; Zhang, X.; Yu, B.; Nandan, M.O.; Bialkowska, A.B.; et al. Mammalian Krüppel-Like Factors in Health and Diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tetreault, M.P.; Yang, Y.; Katz, J.P. Krüppel-like factors in cancer. Nat. Rev. Cancer 2013, 13, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Kubo, A.; Liu, H.; Akita, K.; Laub, F.; Ramirez, F. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 2006, 107, 1357–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laub, F.; Lei, L.; Sumiyoshi, H.; Kajimura, D.; Dragomir, C.; Smaldone, S.; Puche, A.C.; Petros, T.J.; Mason, C.; Parada, L.F.; et al. Transcription Factor KLF7 Is Important for Neuronal Morphogenesis in Selected Regions of the Nervous System. Mol. Cells Biol. 2005, 25, 5699–5711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, M.; Kobayashi, A.; Yamashita, T.; Shimanuki, T.; Nakajima, O.; Takahashi, S.; Ikegami, S.; Inokuchi, K.; Yamashita, K.; Yamamoto, M.; et al. Functional Analysis of Basic Transcription Element Binding Protein by Gene Targeting Technology. Mol. Cells Biol. 2003, 23, 2489–2500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramaniam, M.; Gorny, G.; Johnsen, S.A.; Monroe, D.G.; Evans, G.L.; Fraser, D.G.; Rickard, D.J.; Rasmussen, K.; van Deursen, J.M.A.; Turner, R.T.; et al. TIEG1 Null Mouse-Derived Osteoblasts Are Defective in Mineralization and in Support of Osteoclast Differentiation In Vitro. Mol. Cells Biol. 2005, 25, 1191–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.-Z.; Gavriilidis, G.; Asano, H.; Stamatoyannopoulos, G. Functional study of transcription factor KLF11 by targeted gene inactivation. Blood Cells Mol. Dis. 2005, 34, 53–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; McPherson, L.; Feng, D.; Song, A.; Dong, C.; Lyu, S.C.; Krensky, A.M. Kruppel-like transcription factor 13 regulates T lymphocyte survival in vivo. J. Immunol. 2007, 178, 5496–5504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisch, S.; Gray, S.; Heymans, S.; Haldar, S.M.; Wang, B.; Pfister, O.; Jain, M.K. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. USA 2007, 104, 7074–7079. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Civelek, M. Transcription Factor KLF14 and Metabolic Syndrome. Front. Cardiovasc. Med. 2020, 7, 91. [Google Scholar] [CrossRef]
- Fokeng, M.G.; Atogho-Tiedeu, B.; Sobngwi, E. The Krüppel-like factor 14 (KLF14), master gene of multiple metabolic phenotypes: Putative TransRegulator Network. Transl. Biomed. 2016, 7, 2. [Google Scholar]
- Parker-Katiraee, L.; Carson, A.R.; Yamada, T.; Arnaud, P.; Feil, R.; Abu-Amero, S.N.; Scherer, S.W. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007, 3, e65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.; Li, L.; Zheng, X.-L.; Yin, W.-D.; Tang, C.-K. The role of Krüppel-like factor 14 in the pathogenesis of atherosclerosis. Atherosclerosis 2017, 263, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hsu, L.-A.; Teng, M.-S.; Chou, H.-H.; Ko, Y.-L. Differential Genetic and Epigenetic Effects of the KLF14 Gene on Body Shape Indices and Metabolic Traits. Int. J. Mol. Sci. 2022, 23, 4165. [Google Scholar] [CrossRef]
- Small, K.S.; Todorčević, M.; Civelek, M.; El-Sayed Moustafa, J.S.; Wang, X.; Simon, M.M.; McCarthy, M.I. Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition. Nat. Genet. 2018, 50, 572–580. [Google Scholar] [CrossRef]
- Chen, X.; Li, S.; Yang, Y.; Yang, X.; Liu, Y.; Hu, W.; Jin, L.; Wang, X. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J. Thromb. Haemost. 2012, 10, 1508–1514. [Google Scholar] [CrossRef]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef] [Green Version]
- Voight, B.F.; Scott, L.J.; Steinthorsdottir, V.; Morris, A.P.; Dina, C.; Welch, R.P.; Zeggini, E.; Huth, C.; Aulchenko, Y.S.; Thorleifsson, G.; et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010, 42, 579–589, Corrigendum on Nat. Genet. 2011, 43, 388. [Google Scholar] [CrossRef]
- Civelek, M.; Wu, Y.; Pan, C.; Raulerson, C.; Ko, A.; He, A.; Tilford, C.; Saleem, N.K.; Stančáková, A.; Scott, L.J.; et al. Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits. Am. J. Hum. Genet. 2017, 100, 428–443. [Google Scholar] [CrossRef] [Green Version]
- Mugatroyd, C.; Wu, Y.; Bockmühl, Y.; Spengler, D. The Janus face of DNA methylation in aging. Aging 2010, 2, 107–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, K.; Niaz, S.; Tahir, A.; Jabeen, K. Akash MSH. FTO, PPAR-γ and ABCC8 gene variation and hypertension as de-terminants of cardiometabolic risk in CVD patients. Metab. Clin. Exp. 2022, 128, 154972. [Google Scholar] [CrossRef]
- Jabeen, K.; Rehman, K.; Akash, M.S.H. Genetic mutations of APOEε4 carriers in cardiovascular patients lead to the de-velopment of insulin resistance and risk of Alzheimer’s disease. J. Biochem. Mol. Toxicol. 2022, 36, e22953. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Rehman, K.; Akash, M.S.H. Suhail S, Kamal S, Imran M, Assiri MA. Genetic polymorphism in angiotensinogen and its association with cardiometabolic diseases. Metabolites 2022, 12, 1291. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Rehman, K.; Akash, M.S.H. Suhail S, Rasheed S, Imran M, Assiri MA. Biochemical association between the prevalence of genetic polymorphism and myocardial infarction. Biocells 2023, 47, 473–484. [Google Scholar] [CrossRef]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef] [Green Version]
- Scohy, S.; Gabant, P.; Van Reeth, T.; Hertveldt, V.; Drèze, P.-L.; Van Vooren, P.; Rivière, M.; Szpirer, J.; Szpirer, C. Identification of KLF13 and KLF14 (SP6), Novel Members of the SP/XKLF Transcription Factor Family. Genomics 2000, 70, 93–101. [Google Scholar] [CrossRef]
- Nagai, R.; Friedman, S.L.; Kasuga, M. The Biology of Krüppel-Like Factors; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Pandya, A.Y.; Talley, L.I.; Frost, A.R.; Fitzgerald, T.J.; Trivedi, V.; Chakravarthy, M.; Chhieng, D.C.; Grizzle, W.E.; Engler, J.A.; Krontiras, H.; et al. Nuclear Localization of KLF4 Is Associated with an Aggressive Phenotype in Early-Stage Breast Cancer. Clin. Cancer Res. 2004, 10, 2709–2719. [Google Scholar] [CrossRef] [Green Version]
- Quadrini, K.J.; Bieker, J.J. Krüppel-like Zinc Fingers Bind to Nuclear Import Proteins and Are Required for Efficient Nuclear Localization of Erythroid Krüppel-like Factor. J. Biol. Chem. 2002, 277, 32243–32252. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Guo, Y.; Fan, Y.; Zhang, J.; Lomberk, G.A.; Zhou, Z.; Sun, L.; Chen, Y.E. Perhexiline activates KLF14 and reduces atherosclerosis by modulating ApoA-I production. J. Clin. Investig. 2015, 125, 3819–3830. [Google Scholar] [CrossRef] [Green Version]
- de Assuncao, T.M.; Lomberk, G.; Cao, S.; Yaqoob, U.; Mathison, A.; Simonetto, D.A.; Shah, V.H. New role for Kruppel-like factor 14 as a transcriptional activator involved in the generation of signaling lipids. J. Biol. Chem. 2014, 289, 15798–15809. [Google Scholar] [CrossRef]
- Sarmento, O.F.; Svingen, P.A.; Xiong, Y.; Xavier, R.J.; McGovern, D.; Smyrk, T.C. A novel role for KLF14 in T regulatory cell differentiation. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 188–202.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farris, W.; Mansourian, S.; Chang, Y.; Lindsley, L.; Eckman, E.A.; Frosch, M.P. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. USA 2003, 100, 4162–4167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakata, M.; Nagasaka, S.; Kusaka, I.; Matsuoka, H.; Ishibashi, S.; Yada, T. Effects of statins on the adipocyte maturation and expression of glucose transporter 4 (SLC2A4): Implications in glycaemic control. Diabetologia 2006, 49, 1881–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrat, G.R.; Hu, M.; Nguyen-Tu, M.-S.; Chabosseau, P.; Gaulton, K.J.; van de Bunt, M.; Siddiq, A.; Falchi, M.; Thurner, M.; Canouil, M.; et al. Decreased STARD10 Expression Is Associated with Defective Insulin Secretion in Humans and Mice. Am. J. Hum. Genet. 2017, 100, 238–256. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Gadue, P.; Rutter, G.A. Characterization of a type 2 diabetes–associated islet-specific enhancer cluster in STARD10 by genome editing of endoC-ßH1 cells. Diabetes 2018, 67 (Suppl. 1), 1708. [Google Scholar] [CrossRef]
- Wei, X.; Yang, R.; Wang, C.; Jian, X.; Li, L.; Liu, H.; Yang, G.; Li, Z. A novel role for the Krüppel-like factor 14 on macrophage inflammatory response and atherosclerosis development. Cardiovasc. Pathol. 2017, 27, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Steneberg, P.; Bernardo, L.; Edfalk, S.; Lundberg, L.; Backlund, F.; Östenson, C.-G. The Type 2 Diabetes–Associated Gene Ide Is Required for Insulin Secretion and Suppression of α-Synuclein Levels in β-Cells. Diabetes 2013, 62, 2004–2014. [Google Scholar] [CrossRef] [Green Version]
- Corrêa-Giannella, M.L.; Machado, U.F. SLC2A4 gene: A promising target for pharmacogenomics of insulin resistance. Pharmacogenomics 2013, 14, 847–850. [Google Scholar] [CrossRef] [Green Version]
- Carrat, G.R.; Haythorne, E.; Tomas, A.; Haataja, L.; Müller, A.; Arvan, P.; Piunti, A.; Cheng, K.; Huang, M.; Pullen, T.J.; et al. The type 2 diabetes gene product STARD10 is a phosphoinositide-binding protein that controls insulin secretory granule biogenesis. Mol. Metab. 2020, 40, 101015. [Google Scholar] [CrossRef]
- Cunliffe, V.T. Eloquent silence: Developmental functions of Class I histone deacetylases. Curr. Opin. Genet. Dev. 2008, 18, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Kadosh, D.; Struhl, K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998, 12, 797–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cauchi, S.; Meyre, D.; Dina, C.; Choquet, H.; Samson, C.; Gallina, S.; Froguel, P. Transcription factor TCF7L2 genetic study in the French population: Expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes 2006, 55, 2903–2908. [Google Scholar] [CrossRef] [Green Version]
- Kong, A.; DIAGRAM Consortium; Steinthorsdottir, V.; Masson, G.; Thorleifsson, G.; Sulem, P.; Besenbacher, S.; Jonasdottir, A.; Sigurdsson, A.; Kristinsson, K.T.; et al. Parental origin of sequence variants associated with complex diseases. Nature 2009, 462, 868–874. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Ren, Y.; Lin, Z.; Tang, C.; Jia, Y.; Lai, Y.; Zhou, T.; Wu, S.; Liu, H.; Yang, G.; et al. Krüppel-like factor 14 increases insulin sensitivity through activation of PI3K/Akt signal pathway. Cells Signal. 2015, 27, 2201–2208. [Google Scholar] [CrossRef]
- Wang, J.; Badeanlou, L.; Bielawski, J.; Ciaraldi, T.P.; Samad, F. Sphingosine kinase 1 regulates adipose proinflammatory responses and insulin resistance. Am. J. Physiol. Metab. 2014, 306, E756–E768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front. Endocrinol. 2016, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Goossens, G.H. The role of adipose tissue dysfunction in the pathogenesis of obesity-related insulin resistance. Physiol. Behav. 2008, 94, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Goossens, G.H. The Metabolic Phenotype in Obesity: Fat Mass, Body Fat Distribution, and Adipose Tissue Function. Obes. Facts 2017, 10, 207–215. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. What We Talk About When We Talk About Fat. Cells 2014, 156, 20–44. [Google Scholar] [CrossRef] [Green Version]
- Iwaya, C.; Kitajima, H.; Yamamoto, K.; Maeda, Y.; Sonoda, N.; Shibata, H.; Inoguchi, T. DNA methylation of the Klf14 gene region in whole blood cells provides prediction for the chronic inflammation in the adipose tissue. Biochem. Biophys. Res. Commun. 2018, 497, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Bacos, K.; Gillberg, L.; Volkov, P.; Olsson, A.H.; Hansen, T.; Pedersen, O.; Gjesing, A.P.; Eiberg, H.; Tuomi, T.; Almgren, P.; et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat. Commun. 2016, 7, 11089. [Google Scholar] [CrossRef] [Green Version]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Lohoff, F.W. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar] [PubMed] [Green Version]
- Wang, L.; Tong, X.; Gu, F.; Zhang, L.; Chen, W.; Cheng, X.; Xie, L.; Chang, Y.; Zhang, H. The KLF14 transcription factor regulates hepatic gluconeogenesis in mice. J. Biol. Chem. 2017, 292, 21631–21642. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, P.N.; Fan, L.; Sweet, D.R.; Jain, M.K. The Krüppel-Like Factors and Control of Energy Homeostasis. Endocr. Rev. 2018, 40, 137–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergen, W.G.; Mersmann, H.J. Comparative Aspects of Lipid Metabolism: Impact on Contemporary Research and Use of Animal Models. J. Nutr. 2005, 135, 2499–2502. [Google Scholar] [CrossRef] [Green Version]
- Vitić, J.; Stevanović, J. Comparative studies of the serum lipoproteins and lipids in some domestic, laboratory and wild animals. Comp. Biochem. Physiol. Part B Comp. Biochem. 1993, 106, 223–229. [Google Scholar] [CrossRef]
- Xu, J.; Gu, Y.; Sun, J.; Zhu, H.; Lewis, D.F.; Wang, Y. Reduced CD200 expression is associated with altered Th1/Th2 cytokine production in placental trophoblasts from preeclampsia. Am. J. Reprod. Immunol. 2018, 79, 12763. [Google Scholar] [CrossRef] [PubMed]
- Vitali, C.; Khetarpal, S.A.; Rader, D.J. HDL Cholesterol Metabolism and the Risk of CHD: New Insights from Human Genetics. Curr. Cardiol. Rep. 2017, 19, 132. [Google Scholar] [CrossRef] [PubMed]
- Rader, D.J.; Alexander, E.T.; Weibel, G.L.; Billheimer, J.; Rothblat, G.H. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J. Lipid Res. 2009, 50, S189–S194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oishi, Y.; Manabe, I. Krüppel-Like Factors in Metabolic Homeostasis and Cardiometabolic Disease. Front. Cardiovasc. Med. 2018, 5, 69. [Google Scholar] [CrossRef]
- Pollak, N.M.; Hoffman, M.; Goldberg, I.J.; Drosatos, K. Krüppel-like factors: Crippling and un-crippling metabolic pathways. JACC Basic Transl. Sci. 2018, 3, 132–156. [Google Scholar] [CrossRef] [PubMed]
- Speliotes, E.K.; Yerges-Armstrong, L.M.; Wu, J.; Hernaez, R.; Kim, L.J.; Palmer, C.D. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011, 7, e1001324. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Vidaki, A.; Daniel, B.; Court, D.S. Forensic DNA methylation profiling—Potential opportunities and challenges. Forensic Sci. Int. Genet. 2013, 7, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Kader, F.; Ghai, M. DNA methylation and application in forensic sciences. Forensic Sci. Int. 2015, 249, 255–265. [Google Scholar] [CrossRef]
- Casillas, M.A., Jr.; Lopatina, N.; Andrews, L.G.; Tollefsbol, T.O. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol. Cells Biochem. 2003, 252, 33–43. [Google Scholar] [CrossRef]
- Silva, D.S.; Antunes, J.; Balamurugan, K.; Duncan, G.; Alho, C.S.; McCord, B. Developmental validation studies of epigenetic DNA methylation markers for the detection of blood, semen and saliva samples. Forensic Sci. Int. Genet. 2016, 23, 55–63. [Google Scholar] [CrossRef]
- Schilling, E.; Rehli, M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics 2007, 90, 314–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eipel, M.; Mayer, F.; Arent, T.; Ferreira, M.R.; Birkhofer, C.; Gerstenmaier, U.; Costa, I.G.; Ritz-Timme, S.; Wagner, W. Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures. Aging 2016, 8, 1034–1048. [Google Scholar] [CrossRef] [Green Version]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y.; et al. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2012, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Makowska, Ż.; Spas, A.; Parys-Proszek, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci. Int. Genet. 2014, 14, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Garagnani, P.; Bacalini, M.G.; Pirazzini, C.; Gori, D.; Giuliani, C.; Mari, D. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cells 2012, 11, 1132–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Lin, J.; Zhang, H.; Zhu, F.; Xie, R. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem. Biophys. Res. Commun. 2018, 503, 1848–1853. [Google Scholar] [CrossRef]
- Fan, G.; Sun, L.; Shan, P.; Zhang, X.; Huan, J.; Zhang, X.; Li, D.; Wang, T.; Wei, T.; Gu, X.; et al. Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat. Commun. 2015, 6, 8450. [Google Scholar] [CrossRef] [Green Version]
- Castedo, M.; Perfettini, J.-L.; Roumier, T.; Andreau, K.; Medema, R.; Kroemer, G. Cell death by mitotic catastrophe: A molecular definition. Oncogene 2004, 23, 2825–2837. [Google Scholar] [CrossRef] [Green Version]
- Gao, K.; Wang, J.; Li, L.; Zhai, Y.; Ren, Y.; You, H.; Wang, B.; Wu, X.; Li, J.; Liu, Z.; et al. Polymorphisms in Four Genes (KCNQ1 rs151290, KLF14 rs972283, GCKR rs780094 and MTNR1B rs10830963) and Their Correlation with Type 2 Diabetes Mellitus in Han Chinese in Henan Province, China. Int. J. Environ. Res. Public Health 2016, 13, 260. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, J.; Shen, J.; Hu, D.; Yan, G.; Liu, X.; Xu, X.; Pei, L.; Li, Y.; Sun, C. Association of KCNQ1 and KLF14 polymorphisms and risk of type 2 diabetes mellitus: A global meta-analysis. Hum. Immunol. 2014, 75, 342–347. [Google Scholar] [CrossRef]
- Shahvazian, E.; Mahmoudi, M.B.; Yazd, E.F.; Gharibi, S.; Moghimi, B.; HosseinNia, P.; Mirzaei, M. The KLF14 Variant is Associated with Type 2 Diabetes and HbA1C Level. Biochem. Genet. 2021, 59, 574–588. [Google Scholar] [CrossRef]
- Rees, S.D.; Hydrie, M.Z.; Shera, A.S.; Kumar, S.; O’Hare, J.P.; Barnett, A.H.; Basit, A.; Kelly, M.A. Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia 2011, 54, 1368–1374. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Yin, R.-X.; Huang, K.-K.; Zeng, X.-N.; Guo, T.; Lin, Q.-Z.; Wu, J.; Wu, D.-F.; Li, H.; Pan, S.-L. Association of the KLF14 rs4731702 SNP and Serum Lipid Levels in the Guangxi Mulao and Han Populations. BioMed Res. Int. 2013, 2013, 231515. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, M.F.; Gómez, M.E.V.; González, I.I.; Fernández, G.; Siewert, S. The association between the KLF14 rs4731702 SNP and the lipid profile in type 2 diabetes mellitus patients: A study in San Luis City, San Luis, Argentina. Open Access Libr. J. 2016, 3, 1–14. [Google Scholar] [CrossRef]
- Zabala, A.S.; Gomez, M.E.V.; Alvarez, M.F.; Siewert, S. Tetra Primer ARMS PCR Optimization to Detect Single Nucleotide Polymorphism of the KLF14 Gene. Oalib 2017, 4, 12. [Google Scholar] [CrossRef]
- Alvarez, M.F.; Gomez, M.E.V.; Siewert, S. Designing, Optimization and Validation of Tetra Primer ARMS-PCR Protocol for Genotyping Single Nucleotide Polymorphism rs4731702 (C/T) of KLF14 Gene Associated with Type 2 Diabetes Mellitus: A Study in San Luis, Argentina. Oalib 2016, 3, 1–11. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Chang, Y.-S.; Hsieh, C.-C.; Wang, R.-T.; Chang, J.-G.; Chen, C.-J.; Chang, S.-J. APOE and KLF14 genetic variants are sex-specific for low high-density lipoprotein cholesterol identified by a genome-wide association study. Genet. Mol. Biol. 2022, 45, e20210280. [Google Scholar] [CrossRef]
- Morris, A.P.; Voight, B.F.; Teslovich, T.M.; Ferreira, T.; Segrè, A.V.; Steinthorsdottir, V.; Strawbridge, R.J.; Khan, H.; Grallert, H.; Mahajan, A.; et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012, 44, 981–990. [Google Scholar] [CrossRef] [PubMed]


| Sr.# | Tans- Regulator Gene | Gene Encoded Protein | Gene Expression | KLF14 Interaction | Mutant KLF14 Impact on Gene Expression | Consequences | Ref. |
|---|---|---|---|---|---|---|---|
| 1. | IDE | Zinc metallopeptidase (Insulin degrading enzyme) | IDE degrades peptides, including insulin, glucagon, and amylin. Moreover, it regulates pancreatic β cells functions | KLF14 binds with promoter site of IDE gene | Impaired KLF14 induces irregularities in the normal IDE gene expression | Ultimate outcomes are glucose intolerance, impaired insulin degradation, insulin resistance, and type 2 diabetes mellitus inducement | [10,39] |
| 2. | SLC2A4 | GLUT4 | Regulates the insulin sensitive facilitative glucose transportation in fats and muscle cells | The promoter region of SLC2A4 gene has binding affinity for KLF14 | KLF14 regulates the SLC2A4 normal expression, any genetic variations in KLF14 lead toward compromised SLC2A4 gene expression in glucose regulation | Glucose intolerance which leads to type 2 diabetes mellitus development and obesity | [10,40] |
| 3. | STARD10 | StAR-related lipid transfer protein 10 | This phosphoinositide-binding protein regulates insulin secretion and synthesis | KLF14 has an affinity to bind with the promoter region of the STARD10 trans-gene | Mutant KLF14 binding on the promoter region of the gene increases the risk of type 2 diabetes mellitus by disrupting the normal gene expression | Glucose intolerance, impaired glucose-induced insulin secretion, and type 2 diabetes mellitus | [10,41] |
| Sr. # | Associated SNP | Ethnicity | Allele | Analysis Method | Sample Size | Study Design | Findings | Future Aspects | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1. | rs972283 | Argentina | A/G | Tetra-primer PCR | A total of 50 participants in which 25 were controls and 25 were diseased | Cross-sectional | KLF14 rs972283 (A/G) polymorphism has been found in the type 2 diabetic population of Argentina | Diabetic patients with the KLF14 rs972283 gene polymorphism are more prone to develop cardiac diseases in future | [84] |
| 2. | rs4731702 | San Luis, Argentina | C/T | Tetra Primer ARMS-PCR | A total of 60 in which 30 were controls and 30 diseased | Cross-sectional | KLF14 rs4731702 (C/T) polymorphism has been found in the type 2 diabetic population of Argentina | Diabetic patients with theKLF14 rs4731702 gene polymorphism are more prone to develop atherosclerotic and coronary artery diseases in the future | [85] |
| 3. | rs972283 | Han Chinese, Henan province | A/G | TaqMan PCR | A total of 1504 in which 768 were controls and 736 diseased | Large case control study | No association between KLF14 rs972283 and type 2 diabetes mellitus has been found | In the Henan province population, KLF14 rs972283 gene polymorphism has not prevailed. Hence, the KLF14 gene has no association with development of cardiac problems among the diabetic population | [78] |
| 4. | rs972283 | Globally | A/G | Fixed and random effects meta-analysis | A total of 5 studies with 106,535 controls and 50,552 diseased | Global meta-analysis | KLF14 rs972283 (A/G) polymorphism association with the increased risk of type 2 diabetes has been found throughout the world | Globally, diabetic patients are at high risk of developing cardiac illnesses | [79] |
| 5. | rs4731702 | Han and Guangxi Mulao | C/T | RFLP-PCR | A total of 1467 subjects, 740 from the Han and 727 from the Guangxi population | Cross-sectional study | Both of the populations show the same allele frequency pattern for KLF14 rs4731702 but there is a higher allele frequency in females than males | In the Han and Guangxi Mulao populations, females have more KLF14 rs4731702 frequency differences, thus they are more prone to develop cardiac problems and atherosclerosis | [82] |
| 6. | rs1364422 | Taiwan | T/C | Affymetrix Axiom genotyping array | A total of 41,526 participants | Genome-wide association study | The number of genes including the KLF14 gene was examined to determine the gene polymorphism frequency and results found that KLF14 rs1364422 has a strong association with the risk of type 2 diabetes especially in females | In females of the Taiwan population, KLF14 rs1364422 gene polymorphism altered the metabolic factors including HDL-C, which can lead to the development of cardiac diseases, especially in diabetic female patients | [86] |
| 7. | rs76603546 | Iranian population | C/T | RFLP-PCR | A total of 2064 participants | Case control study | In the Iranian population, KLF14 rs76603546 gene polymorphism has been found | KLF14 rs76603546 has been associated with an increased risk of type 2 diabetes mellitus and altered levels of Hb1AC and BMI | [80] |
| 8. | rs13233731 | European population | G/A | Large scale genotyping using metabochip | A total of 149821 participants, in which 56862 were controls and 12171 were diseased | Large scale meta-analysis | In the European population, KLF14 rs13233731 has been found as a risk of type 2 diabetes mellitus | The meta-analysis study has found KLF14 rs13233731 SNP in the European population, which can induce metabolic disorders especially diabetes and cardiac disorders | [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akash, M.S.H.; Rasheed, S.; Rehman, K.; Ibrahim, M.; Imran, M.; Assiri, M.A. Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms. Metabolites 2023, 13, 199. https://doi.org/10.3390/metabo13020199
Akash MSH, Rasheed S, Rehman K, Ibrahim M, Imran M, Assiri MA. Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms. Metabolites. 2023; 13(2):199. https://doi.org/10.3390/metabo13020199
Chicago/Turabian StyleAkash, Muhammad Sajid Hamid, Sumbal Rasheed, Kanwal Rehman, Muhammad Ibrahim, Muhammad Imran, and Mohammed A. Assiri. 2023. "Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms" Metabolites 13, no. 2: 199. https://doi.org/10.3390/metabo13020199
APA StyleAkash, M. S. H., Rasheed, S., Rehman, K., Ibrahim, M., Imran, M., & Assiri, M. A. (2023). Biochemical Activation and Regulatory Functions of Trans-Regulatory KLF14 and Its Association with Genetic Polymorphisms. Metabolites, 13(2), 199. https://doi.org/10.3390/metabo13020199








