Analysis of Metabolite Distribution in Rat Liver of High-Fat Model by Mass Spectrometry Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rat Model and Biochemistry Measurement
2.2. Liver Tissues Stained with Hematoxylin and Eosin (H&E)
2.3. Mass Spectrometry Imaging (MSI)
2.4. Data Processing and Drawing
3. Results
3.1. Pathological Results of Control and High-Lipid Models (HL)
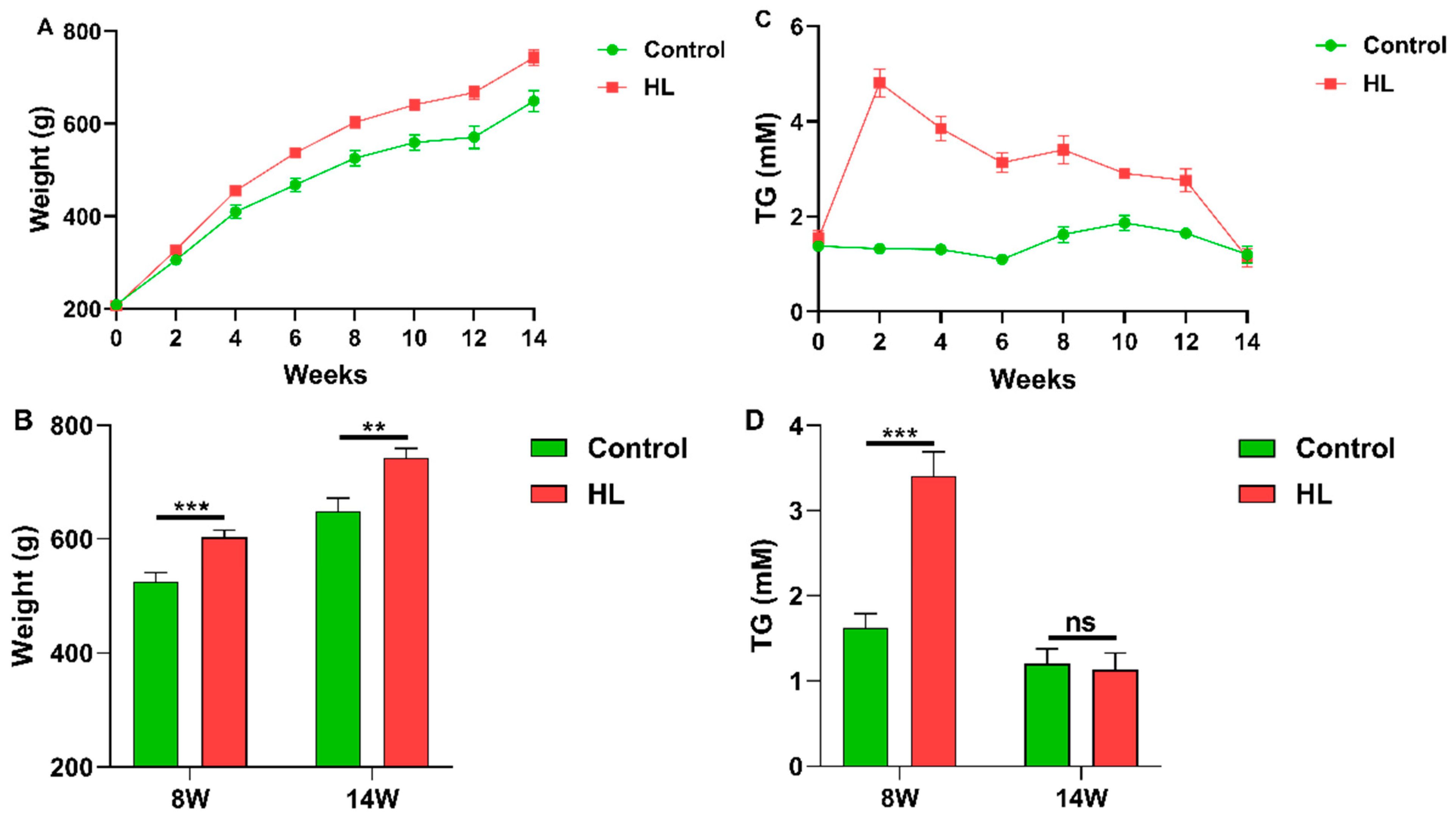
3.2. Changes of Body Weight and Blood TG Content with Time in Rats
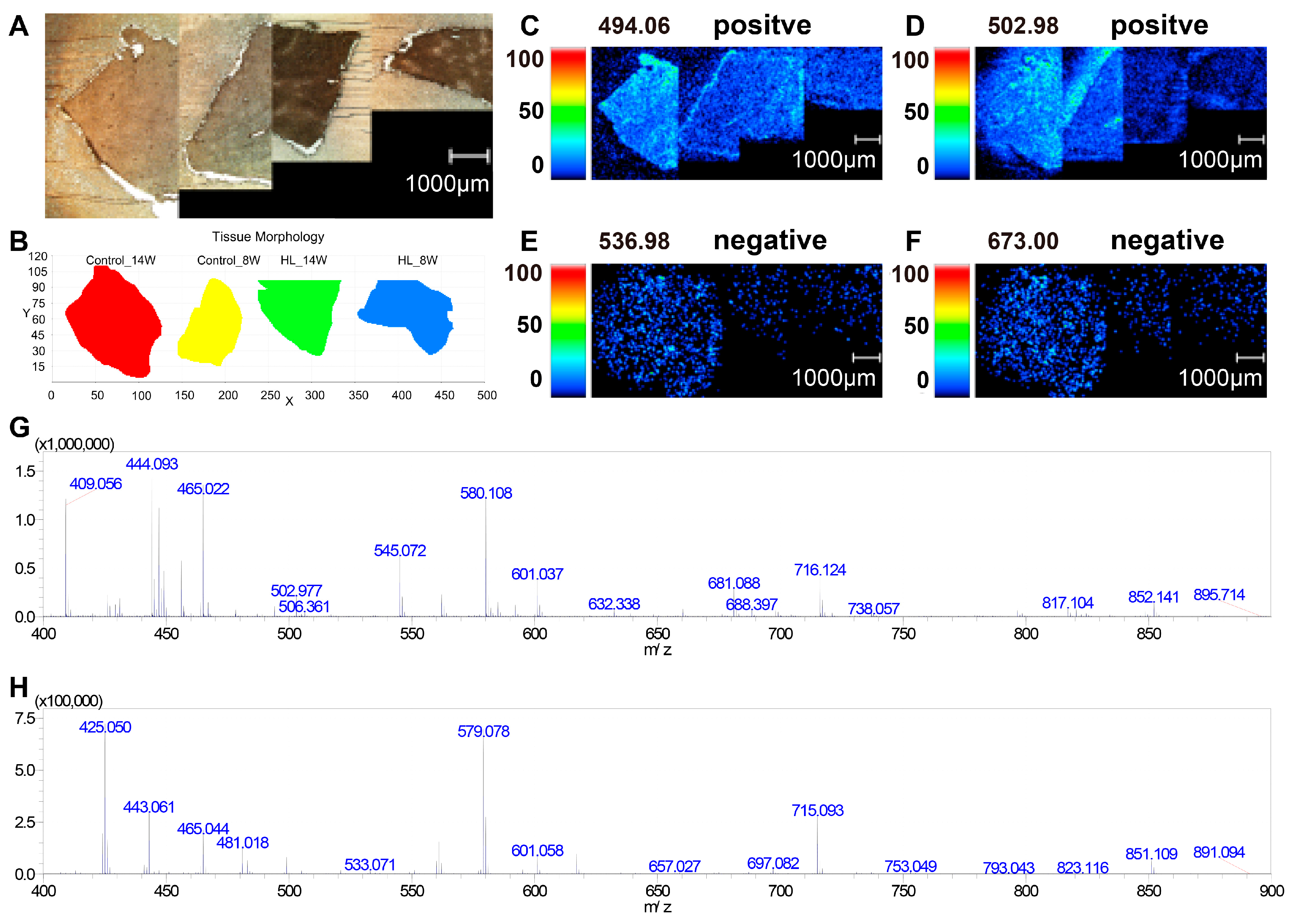
3.3. Photo of MSI and Mass Spectrum in Liver Tissues
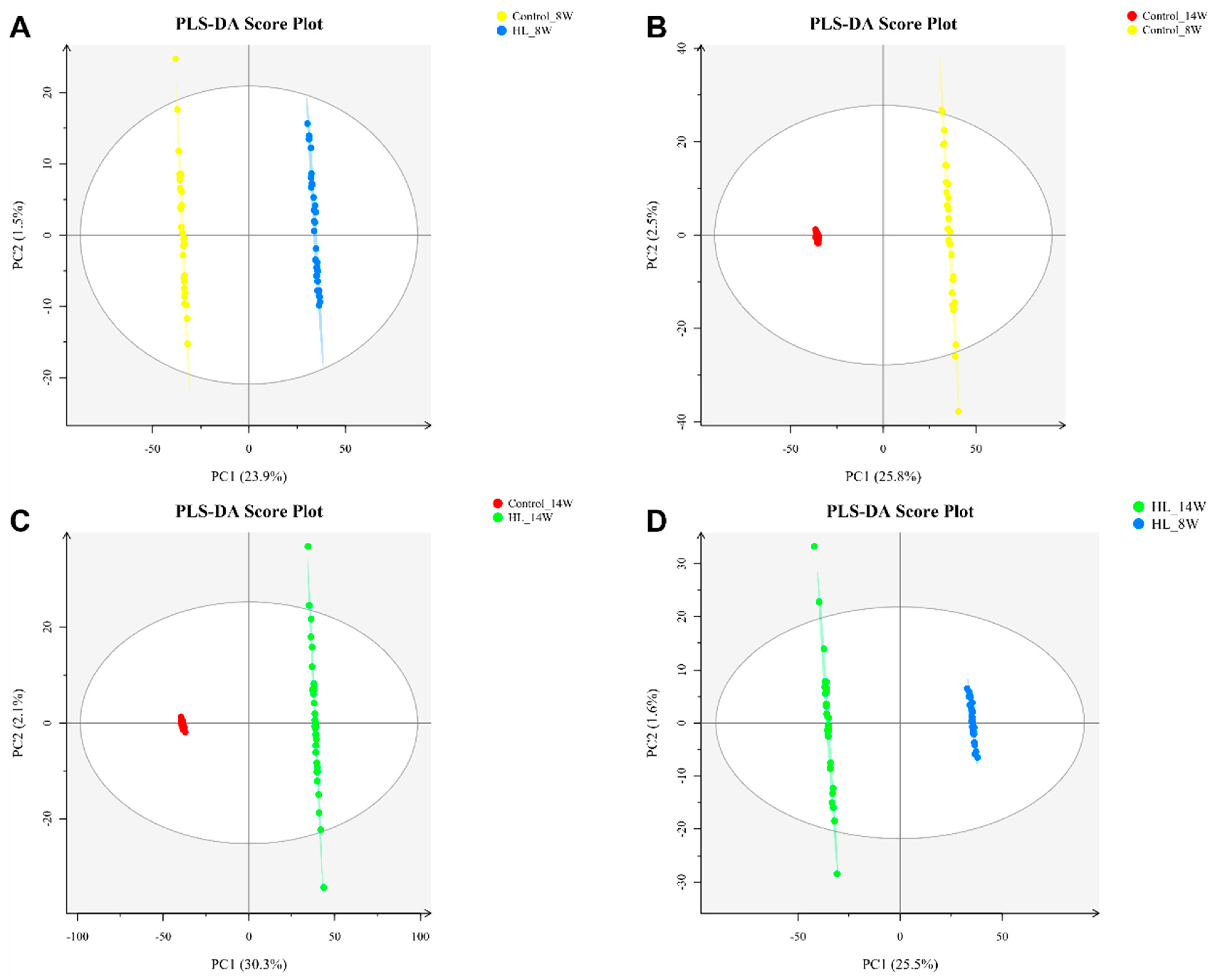
3.4. PLS-DA Analysis
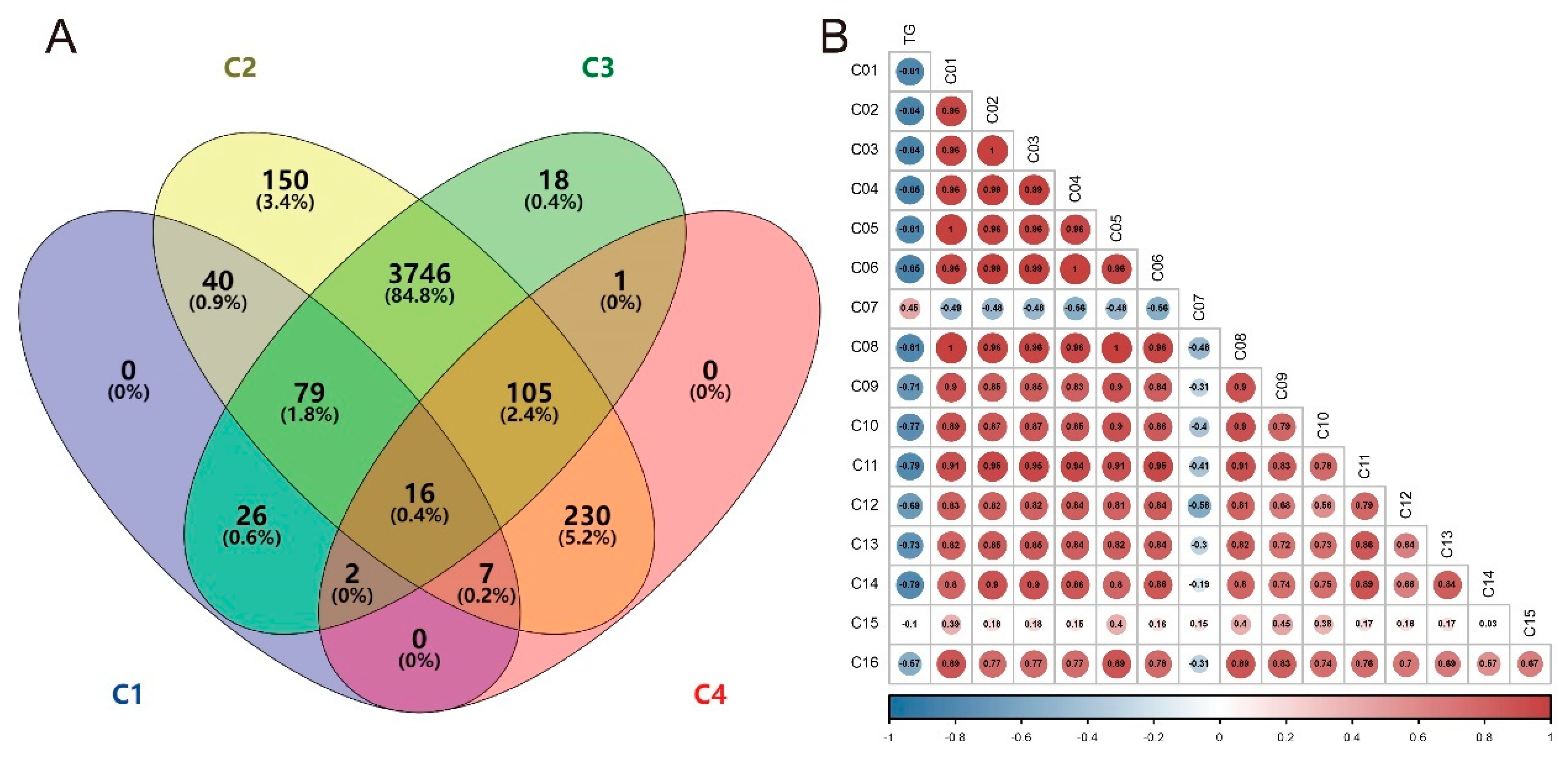
3.5. Differential Metabolites
3.6. MSI of Differential Metabolites
3.7. Correlation Analysis between TG and Differential Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HDL-C | High-density lipoprotein cholesterol |
| H&E | Hematoxylin and eosin |
| HFD | High-fat diet |
| HL | High-lipid or high-fat |
| LDL-C | Low-density lipoprotein cholesterol |
| MSI | Mass spectrometry imaging |
| PA | Phosphatidic acids |
| PG | Phosphatidylglycerols |
| PLS-DA | Partial least squares discriminant analysis |
| SE | Standard error |
| SM | Sphingomyelins |
| SPF | Specific pathogen-free |
| TC | Total cholesterol |
| TG | Triglyceride |
| VIP | Variable importance in the projection |
References
- Goldberg, I.J. Diabetic Dyslipidemia: Causes and Consequences. J. Clin. Endocrinol. Metab. 2001, 86, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.C.; Proctor, S.D. Small Animal Models of Cardiovascular Disease: Tools for the Study of the Roles of Metabolic Syndrome, Dyslipidemia, and Atherosclerosis. Cardiovasc. Pathol. 2006, 15, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.S.; Kathiravan, M.K.; Somani, R.S.; Shishoo, C.J. The Biology and Chemistry of Hyperlipidemia. Bioorg. Med. Chem. 2007, 15, 4674–4699. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.H. Hyperlipidemia as a Risk Factor for Cardiovascular Disease. Prim. Care-Clin. Off. Pract. 2013, 40, 195–211. [Google Scholar] [CrossRef] [Green Version]
- Sudhakaran, S.; Bottiglieri, T.; Tecson, K.M.; Kluger, A.Y.; McCullough, P.A. Alteration of Lipid Metabolism in Chronic Kidney Disease, the Role of Novel Antihyperlipidemic Agents, and Future Directions. Rev. Cardiovasc. Med. 2018, 19, 77–88. [Google Scholar] [CrossRef]
- Babirak, S.P.; Brown, B.G.; Brunzell, J.D. Familial Combined Hyperlipidemia and Abnormal Lipoprotein Lipase. Arterioscler. Thromb. 1992, 12, 1176–1183. [Google Scholar] [CrossRef]
- Beaumont, J.L.; Carlson, L.A.; Cooper, G.R.; Fejfar, Z.; Fredrickson, D.S.; Strasser, T. Classification of Hyperlipidaemias and Hyperlipoproteinaemias. Bull. World Health Organ. 1970, 43, 891–915. [Google Scholar]
- Johnston, T.P.; Korolenko, T.A.; Sahebkar, A. P-407-Induced Mouse Model of Dose-Controlled Hyperlipidemia and Atherosclerosis: 25 Years Later. J. Cardiovasc. Pharmacol. 2017, 70, 339–352. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Zhang, H.; Ren, L.; Li, Q.; Li, N. Animal Models for the Atherosclerosis Research: A Review. Protein Cell 2011, 2, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Miao, H.; Chen, H.; Pei, S.; Bai, X.; Vaziri, N.D.; Zhao, Y.Y. Plasma Lipidomics Reveal Profound Perturbation of Glycerophospholipids, Fatty Acids, and Sphingolipids in Diet-Induced Hyperlipidemia. Chem. Biol. Interact. 2015, 228, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Miao, H.; Zhao, Y.H.; Vaziri, N.D.; Tang, D.D.; Chen, H.; Chen, H.; Khazaeli, M.; Tarbiat-Boldaji, M.; Hatami, L.; Zhao, Y.Y. Lipidomics Biomarkers of Diet-Induced Hyperlipidemia and Its Treatment with Poria Cocos. J. Agric. Food Chem. 2016, 64, 969–979. [Google Scholar] [CrossRef]
- Bozkurt, B.; Aguilar, D.; Deswal, A.; Dunbar, S.B.; Francis, G.S.; Horwich, T.; Jessup, M.; Kosiborod, M.; Pritchett, A.M.; Ramasubbu, K.; et al. Contributory Risk and Management of Comorbidities of Hypertension, Obesity, Diabetes Mellitus, Hyperlipidemia, and Metabolic Syndrome in Chronic Heart Failure: A Scientific Statement from the American Heart Association. Circulation 2016, 134, e535–e578. [Google Scholar] [CrossRef]
- Yang, W.; Shi, H.; Zhang, J.; Shen, Z.; Zhou, G.; Hu, M. Effects of the Duration of Hyperlipidemia on Cerebral Lipids, Vessels and Neurons in Rats. Lipids Health Dis. 2017, 16, 26. [Google Scholar] [CrossRef] [Green Version]
- Heeren, J.; Scheja, L. Metabolic-Associated Fatty Liver Disease and Lipoprotein Metabolism. Mol. Metab. 2021, 50, 101238. [Google Scholar] [CrossRef]
- Kucera, O.; Cervinkova, Z. Experimental Models of Non-Alcoholic Fatty Liver Disease in Rats. World J. Gastroenterol. 2014, 20, 8364–8376. [Google Scholar] [CrossRef]
- Zhang, Q.; Fan, X.; Ye, R.; Hu, Y.; Zheng, T.; Shi, R.; Cheng, W.; Lv, X.; Chen, L.; Liang, P. The Effect of Simvastatin on Gut Microbiota and Lipid Metabolism in Hyperlipidemic Rats Induced by a High-Fat Diet. Front. Pharmacol. 2020, 11, 522. [Google Scholar] [CrossRef]
- Ge, S.; Liao, C.; Su, D.; Mula, T.; Gegen, Z.; Li, Z.; Tu, Y. Wuwei Qingzhuo San Ameliorates Hyperlipidemia in Mice Fed With HFD by Regulating Metabolomics and Intestinal Flora Composition. Front. Pharmacol. 2022, 13, 842671. [Google Scholar] [CrossRef]
- Wei, X.; Jia, X.; Liu, R.; Zhang, S.; Liu, S.; An, J.; Zhou, L.; Zhang, Y.; Mo, Y.; Li, X. Metabolic Pathway Analysis of Hyperuricaemia Patients with Hyperlipidaemia Based on High-Throughput Mass Spectrometry: A Case-Control Study. Lipids Health Dis. 2022, 21, 151. [Google Scholar] [CrossRef]
- Chen, H.; Miao, H.; Feng, Y.L.; Zhao, Y.Y.; Lin, R.C. Metabolomics in Dyslipidemia, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014; Volume 66, ISBN 9780128014011. [Google Scholar]
- Siemianowicz, K. Laboratory Diagnosis of Lipid Imbalance. Wiad. Lek. 1996, 49, 159–165. [Google Scholar]
- García-Heredia, A.; Kensicki, E.; Mohney, R.P.; Rull, A.; Triguero, I.; Marsillach, J.; Tormos, C.; MacKness, B.; MacKness, M.; Shih, D.M.; et al. Paraoxonase-1 Deficiency Is Associated with Severe Liver Steatosis in Mice Fed a High-Fat High-Cholesterol Diet: A Metabolomic Approach. J. Proteome Res. 2013, 12, 1946–1955. [Google Scholar] [CrossRef]
- Sun, J.H.; Liu, X.; Cong, L.X.; Li, H.; Zhang, C.Y.; Chen, J.G.; Wang, C.M. Metabolomics Study of the Therapeutic Mechanism of Schisandra Chinensis Lignans in Diet-Induced Hyperlipidemia Mice. Lipids Health Dis. 2017, 16, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Yuan, B.; Miao, H.; Tan, Y.; Bai, X.; Zhao, Y.Y.; Wang, Y. Urine Metabolomics Reveals New Insights into Hyperlipidemia and the Therapeutic Effect of Rhubarb. Anal. Methods 2015, 7, 3113–3123. [Google Scholar] [CrossRef]
- Jiang, C.-Y.; Yang, K.-M.; Yang, L.; Miao, Z.-X.; Wang, Y.-H.; Zhu, H.-B. A 1H NMR-Based Metabonomic Investigation of Time-Related Metabolic Trajectories of the Plasma, Urine and Liver Extracts of Hyperlipidemic Hamsters. PLoS ONE 2013, 8, e0066786. [Google Scholar] [CrossRef] [Green Version]
- Amstalden van Hove, E.R.; Smith, D.F.; Heeren, R.M.A. A Concise Review of Mass Spectrometry Imaging. J. Chromatogr. A 2010, 1217, 3946–3954. [Google Scholar] [CrossRef] [PubMed]
- Spengler, B. Mass Spectrometry Imaging of Biomolecular Information. Anal. Chem. 2015, 87, 64–82. [Google Scholar] [CrossRef]
- Miura, D.; Fujimura, Y.; Wariishi, H. In Situ Metabolomic Mass Spectrometry Imaging: Recent Advances and Difficulties. J. Proteom. 2012, 75, 5052–5060. [Google Scholar] [CrossRef]
- Greer, T.; Sturm, R.; Li, L. Mass Spectrometry Imaging for Drugs and Metabolites. J. Proteom. 2011, 74, 2617–2631. [Google Scholar] [CrossRef] [Green Version]
- Buchberger, A.R.; DeLaney, K.; Johnson, J.; Li, L. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem. 2018, 90, 240–265. [Google Scholar] [CrossRef]
- Xia, H.; Tang, H.; Wang, F.; Yang, X.; Wang, Z.; Liu, H.; Pan, D.; Yang, C.; Wang, S.; Sun, G. An Untargeted Metabolomics Approach Reveals Further Insights of Lycium Barbarum Polysaccharides in High Fat Diet and Streptozotocin-Induced Diabetic Rats. Food Res. Int. 2019, 116, 20–29. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.H.; Noh, S.; Hur, H.J.; Sung, M.J.; Hwang, J.T.; Park, J.H.; Yang, H.J.; Kim, M.S.; Kwon, D.Y.; et al. Metabolomic Analysis of Livers and Serum from High-Fat Diet Induced Obese Mice. J. Proteome Res. 2011, 10, 722–731. [Google Scholar] [CrossRef]
- Kampa, J.M.; Sahin, M.; Slopianka, M.; Giampà, M.; Bednarz, H.; Ernst, R.; Riefke, B.; Niehaus, K.; Fatangare, A. Mass Spectrometry Imaging Reveals Lipid Upregulation and Bile Acid Changes Indicating Amitriptyline Induced Steatosis in a Rat Model. Toxicol. Lett. 2020, 325, 43–50. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [Green Version]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Smets, T.; Verbeeck, N.; Claesen, M.; Asperger, A.; Griffioen, G.; Tousseyn, T.; Waelput, W.; Waelkens, E.; de Moor, B. Evaluation of Distance Metrics and Spatial Autocorrelation in Uniform Manifold Approximation and Projection Applied to Mass Spectrometry Imaging Data. Anal. Chem. 2019, 91, 5706–5714. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Yin, R.; Brown, H.M.; Laskin, J. Spatial Segmentation of Mass Spectrometry Imaging Data by Combining Multivariate Clustering and Univariate Thresholding. Anal. Chem. 2021, 93, 3477–3485. [Google Scholar] [CrossRef]
- Smets, T.; Waelkens, E.; de Moor, B. Prioritization of m/z-Values in Mass Spectrometry Imaging Profiles Obtained Using Uniform Manifold Approximation and Projection for Dimensionality Reduction. Anal. Chem. 2020, 92, 5240–5248. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Heijs, B.; Kostidis, S.; Mahfouz, A.; Rietjens, R.G.J.; Bijkerk, R.; Koudijs, A.; van der Pluijm, L.A.K.; van den Berg, C.W.; Dumas, S.J.; et al. Analyzing Cell-Type-Specific Dynamics of Metabolism in Kidney Repair. Nat. Metab. 2022, 4, 1109–1118. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2018, 8, 1–22. [Google Scholar] [CrossRef]
- Han, X.; Ye, H. Overview of Lipidomic Analysis of Triglyceride Molecular Species in Biological Lipid Extracts. J. Agric. Food Chem. 2021, 69, 8895–8909. [Google Scholar] [CrossRef]
- Maruyama, C.; Imamura, K.; Teramoto, T. Assessment of LDL Particle Size by Triglyceride/HDL-Cholesterol Ratio in Non-Diabetic, Healthy Subjects without Prominent Hyperlipidemia. J. Atheroscler. Thromb. Orig. Artic. 2003, 10, 186–191. [Google Scholar] [CrossRef] [Green Version]
- Tirosh, A.; Shai, I.; Bitzur, R.; Kochba, I.; Tekes-Manova, D.; Israeli, E.; Shochat, T.; Rudich, A. Changes in Triglyceride Levels over Time and Risk of Type 2 Diabetes in Young Men. Diabetes Care 2008, 31, 2032–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czech, M.P.; Tencerova, M.; Pedersen, D.J.; Aouadi, M. Insulin Signalling Mechanisms for Triacylglycerol Storage. Diabetologia 2013, 56, 949–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Zhang, Y.; Wei, F.; Song, J.; Cao, Z.; Chen, C.; Zhang, K.; Feng, S.; Wang, Y.; Li, W.D. Triglyceride Is an Independent Predictor of Type 2 Diabetes among Middle-Aged and Older Adults: A Prospective Study with 8-Year Follow-Ups in Two Cohorts. J. Transl. Med. 2019, 17, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Wang, N.; Ahn, D.U.; Ma, M. Long Term Egg Yolk Consumption Alters Lipid Metabolism and Attenuates Hyperlipidemia in Mice Fed a High-Fat Diet Based on Lipidomics Analysis. Eur. J. Lipid Sci. Technol. 2019, 121, 1800496. [Google Scholar] [CrossRef]
- Sacks, F.M.; Bray, G.A.; Carey, V.J.; Smith, S.R.; Ryan, D.H.; Anton, S.D.; Mcmanus, K.; Champagne, C.M.; Bishop, L.M.; Laranjo, N.; et al. Comparison of Weight-Loss Diets with Different Compositions of Fat, Protein, and Carbohydrates. N. Engl. J. Med. 2009, 360, 859–873. [Google Scholar] [CrossRef] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, H.; Wang, W.; Xiang, X.; Li, Y.; Zhao, J.; Huang, Y.; Di, S.; Zhuo, Q.; Nie, H. Analysis of Metabolite Distribution in Rat Liver of High-Fat Model by Mass Spectrometry Imaging. Metabolites 2023, 13, 411. https://doi.org/10.3390/metabo13030411
Mao H, Wang W, Xiang X, Li Y, Zhao J, Huang Y, Di S, Zhuo Q, Nie H. Analysis of Metabolite Distribution in Rat Liver of High-Fat Model by Mass Spectrometry Imaging. Metabolites. 2023; 13(3):411. https://doi.org/10.3390/metabo13030411
Chicago/Turabian StyleMao, Hongmei, Wenjun Wang, Xuesong Xiang, Yan Li, Jinpeng Zhao, Yin Huang, Shuangshuang Di, Qin Zhuo, and Honggang Nie. 2023. "Analysis of Metabolite Distribution in Rat Liver of High-Fat Model by Mass Spectrometry Imaging" Metabolites 13, no. 3: 411. https://doi.org/10.3390/metabo13030411
APA StyleMao, H., Wang, W., Xiang, X., Li, Y., Zhao, J., Huang, Y., Di, S., Zhuo, Q., & Nie, H. (2023). Analysis of Metabolite Distribution in Rat Liver of High-Fat Model by Mass Spectrometry Imaging. Metabolites, 13(3), 411. https://doi.org/10.3390/metabo13030411







