Human Milk Lipids and Small Metabolites: Maternal and Microbial Origins
Abstract
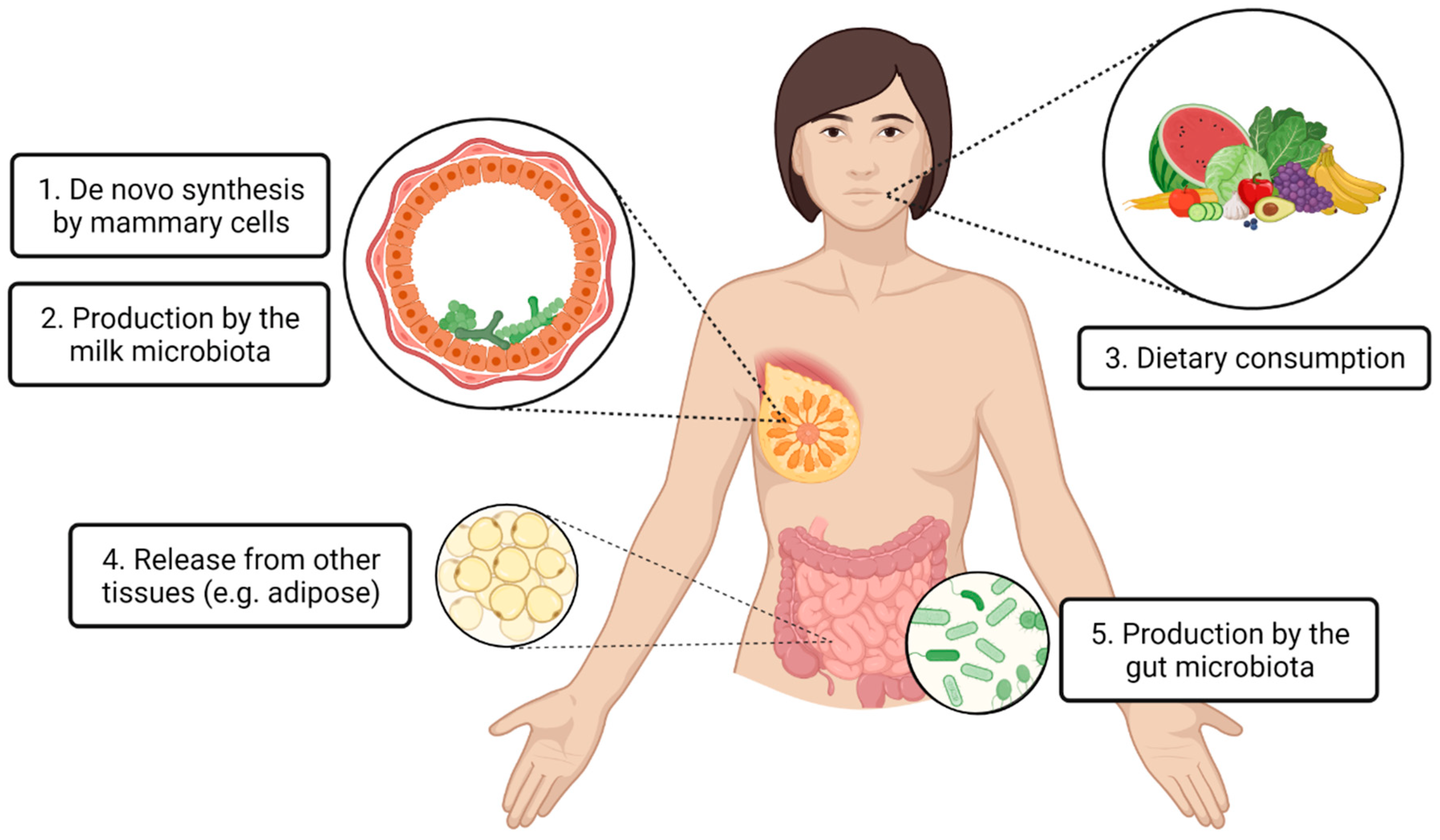
:1. Introduction
2. Local Production within the Mammary Gland
2.1. De Novo Synthesis by Mammary Cells
2.2. Production by the Milk Microbiota
3. Incorporation from Maternal Circulation
3.1. Dietary Consumption
3.2. Release from Other Tissues
3.3. Production by Gut Microbiota
4. Summary and Future Research Directions
4.1. Transcriptome Analysis of Human Milk Cells
4.2. Paired Sample Analyses
4.3. Dietary Studies
4.4. Bacterial Gene Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Victora, C.G.; Bahl, R.; Barros, A.J.; Franca, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef] [Green Version]
- Ladomenou, F.; Moschandreas, J.; Kafatos, A.; Tselentis, Y.; Galanakis, E. Protective effect of exclusive breastfeeding against infections during infancy: A prospective study. Arch. Dis. Child. 2010, 95, 1004–1008. [Google Scholar] [CrossRef]
- Vennemann, M.M.; Bajanowski, T.; Brinkmann, B.; Jorch, G.; Yucesan, K.; Sauerland, C.; Mitchell, E.A.; Ge, S.I.D.S.G. Does breastfeeding reduce the risk of sudden infant death syndrome? Pediatrics 2009, 123, e406–e410. [Google Scholar] [CrossRef] [Green Version]
- Lund-Blix, N.A.; Stene, L.C.; Rasmussen, T.; Torjesen, P.A.; Andersen, L.F.; Ronningen, K.S. Infant feeding in relation to islet autoimmunity and type 1 diabetes in genetically susceptible children: The MIDIA Study. Diabetes Care 2015, 38, 257–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horta, B.L.; Loret de Mola, C.; Victora, C.G. Long-term consequences of breastfeeding on cholesterol, obesity, systolic blood pressure and type 2 diabetes: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.; Raman, A.S.; Murch, S.H.; Rollins, N.C.; Gordon, J.I. Understanding the mother-breastmilk-infant “triad”. Science 2020, 367, 1070–1072. [Google Scholar] [CrossRef]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [Green Version]
- Bode, L.; McGuire, M.; Rodriguez, J.M.; Geddes, D.T.; Hassiotou, F.; Hartmann, P.E.; McGuire, M.K. It’s alive: Microbes and cells in human milk and their potential benefits to mother and infant. Adv. Nutr. 2014, 5, 571–573. [Google Scholar] [CrossRef] [Green Version]
- Geddes, D.T.; Gridneva, Z.; Perrella, S.L.; Mitoulas, L.R.; Kent, J.C.; Stinson, L.F.; Lai, C.T.; Sakalidis, V.; Twigger, A.-J.; Hartmann, P.E. 25 Years of Research in Human Lactation: From Discovery to Translation. Nutrients 2021, 13, 3071. [Google Scholar] [CrossRef]
- Garwolińska, D.; Namieśnik, J.; Kot-Wasik, A.; Hewelt-Belka, W. Chemistry of Human Breast Milk—A Comprehensive Review of the Composition and Role of Milk Metabolites in Child Development. J. Agric. Food Chem. 2018, 66, 11881–11896. [Google Scholar] [CrossRef] [PubMed]
- George, A.D.; Burugupalli, S.; Paul, S.; Mansell, T.; Burgner, D.; Meikle, P.J. The Role of Human Milk Lipids and Lipid Metabolites in Protecting the Infant against Non-Communicable Disease. Int. J. Mol. Sci. 2022, 23, 7490. [Google Scholar] [CrossRef]
- Stinson, L.F.; Geddes, D.T. Microbial metabolites: The next frontier in human milk. Trends Microbiol. 2022, 30, 408–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Padhi, E.; Hasegawa, Y.; Larke, J.; Parenti, M.; Wang, A.; Hernell, O.; Lönnerdal, B.; Slupsky, C. Compositional Dynamics of the Milk Fat Globule and Its Role in Infant Development. Front. Pediatr. 2018, 6, 313. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wang, Y.; Yang, X.; Cheng, Y.; Zhang, H.; Xu, X.; Zhou, J.; Chen, H.; Su, M.; Yang, Y.; et al. Human Milk Lipid Profiles around the World: A Systematic Review and Meta-Analysis. Adv. Nutr. 2022, 13, 2519–2536. [Google Scholar] [CrossRef] [PubMed]
- Gardner, A.S.; Rahman, I.A.; Lai, C.T.; Hepworth, A.; Trengove, N.; Hartmann, P.E.; Geddes, D.T. Changes in Fatty Acid Composition of Human Milk in Response to Cold-Like Symptoms in the Lactating Mother and Infant. Nutrients 2017, 9, 1034. [Google Scholar] [CrossRef] [Green Version]
- George, A.D.; Gay, M.C.L.; Wlodek, M.E.; Trengove, R.D.; Murray, K.; Geddes, D.T. Untargeted lipidomics using liquid chromatography-ion mobility-mass spectrometry reveals novel triacylglycerides in human milk. Sci. Rep. 2020, 10, 9255. [Google Scholar] [CrossRef]
- Isaacs, C.E.; Kashyap, S.; Heird, W.C.; Thormar, H. Antiviral and antibacterial lipids in human milk and infant formula feeds. Arch. Dis. Child. 1990, 65, 861–864. [Google Scholar] [CrossRef] [Green Version]
- Meyer, D.M.; Brei, C.; Stecher, L.; Much, D.; Brunner, S.; Hauner, H. Associations between long-chain PUFAs in maternal blood, cord blood, and breast milk and offspring body composition up to 5 years: Follow-up from the INFAT study. Eur. J. Clin. Nutr. 2019, 73, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Ojo-Okunola, A.; Cacciatore, S.; Nicol, M.P.; du Toit, E. The Determinants of the Human Milk Metabolome and Its Role in Infant Health. Metabolites 2020, 10, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfs, D.; Lynes, M.D.; Tseng, Y.H.; Pierce, S.; Bussberg, V.; Darkwah, A.; Tolstikov, V.; Narain, N.R.; Rudolph, M.C.; Kiebish, M.A.; et al. Brown Fat-Activating Lipokine 12,13-diHOME in Human Milk Is Associated With Infant Adiposity. J. Clin. Endocrinol. Metab. 2021, 106, e943–e956. [Google Scholar] [CrossRef]
- Stinson, L.F.; Gay, M.C.L.; Koleva, P.T.; Eggesbø, M.; Johnson, C.C.; Wegienka, G.; du Toit, E.; Shimojo, N.; Munblit, D.; Campbell, D.E.; et al. Human Milk From Atopic Mothers Has Lower Levels of Short Chain Fatty Acids. Front. Immunol. 2020, 11, 1427. [Google Scholar] [CrossRef] [PubMed]
- Levan, S.R.; Stamnes, K.A.; Lin, D.L.; Panzer, A.R.; Fukui, E.; McCauley, K.; Fujimura, K.E.; McKean, M.; Ownby, D.R.; Zoratti, E.M.; et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 2019, 4, 1851–1861. [Google Scholar] [CrossRef]
- Prentice, P.M.; Schoemaker, M.H.; Vervoort, J.; Hettinga, K.; Lambers, T.T.; van Tol, E.A.F.; Acerini, C.L.; Olga, L.; Petry, C.J.; Hughes, I.A.; et al. Human Milk Short-Chain Fatty Acid Composition is Associated with Adiposity Outcomes in Infants. J. Nutr. 2019, 149, 716–722. [Google Scholar] [CrossRef]
- Innis, S.M. Maternal Nutrition, Genetics, and Human Milk Lipids. Curr. Nutr. Rep. 2013, 2, 151–158. [Google Scholar] [CrossRef]
- Stinson, L.F.; Trevenen, M.L.; Geddes, D.T. Effect of Cold Storage on the Viable and Total Bacterial Populations in Human Milk. Nutrients 2022, 14, 1875. [Google Scholar] [CrossRef]
- Douglas, C.A.; Ivey, K.L.; Papanicolas, L.E.; Best, K.P.; Muhlhausler, B.S.; Rogers, G.B. DNA extraction approaches substantially influence the assessment of the human breast milk microbiome. Sci. Rep. 2020, 10, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garwolińska, D.; Hewelt-Belka, W.; Namieśnik, J.; Kot-Wasik, A. Rapid Characterization of the Human Breast Milk Lipidome Using a Solid-Phase Microextraction and Liquid Chromatography–Mass Spectrometry-Based Approach. J. Proteome Res. 2017, 16, 3200–3208. [Google Scholar] [CrossRef]
- Hewelt-Belka, W.; Garwolińska, D.; Belka, M.; Bączek, T.; Namieśnik, J.; Kot-Wasik, A. A new dilution-enrichment sample preparation strategy for expanded metabolome monitoring of human breast milk that overcomes the simultaneous presence of low- and high-abundance lipid species. Food Chem. 2019, 288, 154–161. [Google Scholar] [CrossRef]
- George, A.D.; Gay, M.C.L.; Trengove, R.D.; Geddes, D.T. Human Milk Lipidomics: Current Techniques and Methodologies. Nutrients 2018, 10, 1169. [Google Scholar] [CrossRef] [PubMed]
- Ganeshalingam, M.; Enstad, S.; Sen, S.; Cheema, S.; Esposito, F.; Thomas, R. Role of lipidomics in assessing the functional lipid composition in breast milk. Front. Nutr. 2022, 9, 899401. [Google Scholar] [CrossRef]
- Stinson, L.F.; Ma, J.; Sindi, A.S.; Geddes, D.T. Methodological approaches for studying the human milk microbiome. Nutr. Rev. 2022, nuac082. [Google Scholar] [CrossRef] [PubMed]
- Twigger, A.J.; Engelbrecht, L.K.; Bach, K.; Schultz-Pernice, I.; Pensa, S.; Stenning, J.; Petricca, S.; Scheel, C.H.; Khaled, W.T. Transcriptional changes in the mammary gland during lactation revealed by single cell sequencing of cells from human milk. Nat. Commun. 2022, 13, 562. [Google Scholar] [CrossRef] [PubMed]
- Christian, P.; Smith, E.R.; Lee, S.E.; Vargas, A.J.; Bremer, A.A.; Raiten, D.J. The need to study human milk as a biological system. Am. J. Clin. Nutr. 2021, 113, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Gay, M.C.L.; Koleva, P.T.; Slupsky, C.M.; Toit, E.D.; Eggesbo, M.; Johnson, C.C.; Wegienka, G.; Shimojo, N.; Campbell, D.E.; Prescott, S.L.; et al. Worldwide Variation in Human Milk Metabolome: Indicators of Breast Physiology and Maternal Lifestyle? Nutrients 2018, 10, 1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammad, M.A.; Haymond, M.W. Regulation of lipid synthesis genes and milk fat production in human mammary epithelial cells during secretory activation. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E700–E716. [Google Scholar] [CrossRef] [Green Version]
- Suburu, J.; Shi, L.; Wu, J.; Wang, S.; Samuel, M.; Thomas, M.J.; Kock, N.D.; Yang, G.; Kridel, S.; Chen, Y.Q. Fatty acid synthase is required for mammary gland development and milk production during lactation. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1132–E1143. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.; Pasco, D.; Pawlak, J.; Thompson, B.J.; Stampfer, M.; Nandi, S. Thioesterase II, a new marker enzyme for human cells of breast epithelial origin. J. Natl. Cancer Inst. 1984, 73, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Gagné, H.T.; Pitelka, D.R.; Abraham, S. The effect of dietary fat on lipogenesis in mammary gland and liver from lactating and virgin mice. Biochem. J. 1969, 115, 807–815. [Google Scholar] [CrossRef]
- Hachey, D.L.; Thomas, M.R.; Emken, E.A.; Garza, C.; Brown-Booth, L.; Adlof, R.O.; Klein, P.D. Human lactation: Maternal transfer of dietary triglycerides labeled with stable isotopes. J. Lipid Res. 1987, 28, 1185–1192. [Google Scholar] [CrossRef]
- Chatterjee, D.; Subba Reddy, K.V.; Ray, T.K. Lipid synthesis in lactating mammary gland. Trends Biochem. Sci. 1979, 4, 278–280. [Google Scholar] [CrossRef]
- Boutinaud, M.; Herve, L.; Lollivier, V. Mammary epithelial cells isolated from milk are a valuable, non-invasive source of mammary transcripts. Front. Genet. 2015, 6, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stinson, L.F.; Sindi, A.S.M.; Cheema, A.S.; Lai, C.T.; Muhlhausler, B.S.; Wlodek, M.E.; Payne, M.S.; Geddes, D.T. The human milk microbiome: Who, what, when, where, why, and how? Nutr. Rev. 2020, 79, 529–543. [Google Scholar] [CrossRef]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 25, 324–335.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheema, A.S.; Trevenen, M.L.; Turlach, B.A.; Furst, A.J.; Roman, A.S.; Bode, L.; Gridneva, Z.; Lai, C.T.; Stinson, L.F.; Payne, M.S.; et al. Exclusively Breastfed Infant Microbiota Develops over Time and Is Associated with Human Milk Oligosaccharide Intakes. Int. J. Mol. Sci. 2022, 23, 2804. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; O’Sullivan, A.; Barile, D.; German, J.B.; Lonnerdal, B.; Slupsky, C.M. The human milk metabolome reveals diverse oligosaccharide profiles. J. Nutr. 2013, 143, 1709–1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulsen, K.O.; Meng, F.; Lanfranchi, E.; Young, J.F.; Stanton, C.; Ryan, C.A.; Kelly, A.L.; Sundekilde, U.K. Dynamic Changes in the Human Milk Metabolome Over 25 Weeks of Lactation. Front. Nutr. 2022, 9, 917659. [Google Scholar] [CrossRef]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [Green Version]
- Lentner, C.; West Cadwell, N.J. (Eds.) Geigy Scientific Tables, 8th ed.; Medical Education Div., Ciba-Geigy Corp: Basel, Switzerland, 1992; pp. 165–177. [Google Scholar]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Jia, K.; Wu, M.; Chen, M. Unveiling the influence of ultrasonic-assisted lipolysis: A pilot study of short-chain fatty acid profiling in human milk based on mass spectrometry. Food Chem. Adv. 2022, 1, 100097. [Google Scholar] [CrossRef]
- Gomez-Gallego, C.; Kumar, H.; Garcia-Mantrana, I.; du Toit, E.; Suomela, J.P.; Linderborg, K.M.; Zhang, Y.; Isolauri, E.; Yang, B.; Salminen, S.; et al. Breast Milk Polyamines and Microbiota Interactions: Impact of Mode of Delivery and Geographical Location. Ann. Nutr. Metab. 2017, 70, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Duranton, F.; Cohen, G.; De Smet, R.; Rodriguez, M.; Jankowski, J.; Vanholder, R.; Argiles, A.; European Uremic Toxin Work Group. Normal and pathologic concentrations of uremic toxins. J. Am. Soc. Nephrol. 2012, 23, 1258–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Gouveia-Figueira, S.; Domellof, M.; Zivkovic, A.M.; Nording, M.L. Oxylipins, endocannabinoids, and related compounds in human milk: Levels and effects of storage conditions. Prostagland. Other Lipid Mediat. 2016, 122, 28–36. [Google Scholar] [CrossRef]
- Stanford, K.I.; Lynes, M.D.; Takahashi, H.; Baer, L.A.; Arts, P.J.; May, F.J.; Lehnig, A.C.; Middelbeek, R.J.W.; Richard, J.J.; So, K.; et al. 12,13-diHOME: An Exercise-Induced Lipokine that Increases Skeletal Muscle Fatty Acid Uptake. Cell Metab. 2018, 27, 1111–1120.e3. [Google Scholar] [CrossRef] [Green Version]
- Lichtenberger, L.M.; Gardner, J.W.; Barreto, J.C.; Morriss, F.H., Jr. Evidence for a role of volatile amines in the development of neonatal hypergastrinemia. J. Pediatr. Gastroenterol. Nutr. 1991, 13, 342–346. [Google Scholar] [CrossRef]
- Bain, M.A.; Faull, R.; Fornasini, G.; Milne, R.W.; Evans, A.M. Accumulation of trimethylamine and trimethylamine-N-oxide in end-stage renal disease patients undergoing haemodialysis. Nephrol. Dial. Transplant. 2006, 21, 1300–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trabado, S.; Al-Salameh, A.; Croixmarie, V.; Masson, P.; Corruble, E.; Feve, B.; Colle, R.; Ripoll, L.; Walther, B.; Boursier-Neyret, C.; et al. The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS ONE 2017, 12, e0173615. [Google Scholar] [CrossRef] [Green Version]
- Asnicar, F.; Manara, S.; Zolfo, M.; Truong, D.T.; Scholz, M.; Armanini, F.; Ferretti, P.; Gorfer, V.; Pedrotti, A.; Tett, A.; et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems 2017, 2, e00164-16. [Google Scholar] [CrossRef] [Green Version]
- Parsons, J.B.; Rock, C.O. Bacterial lipids: Metabolism and membrane homeostasis. Prog. Lipid Res. 2013, 52, 249–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cronan, J.E.; Thomas, J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009, 459, 395–433. [Google Scholar] [CrossRef] [Green Version]
- Neville, M.C. The physiological basis of milk secretion. Ann. N. Y. Acad. Sci. 1990, 586, 1–11. [Google Scholar] [CrossRef]
- Shennan, D.B.; Boyd, C.A. The functional and molecular entities underlying amino acid and peptide transport by the mammary gland under different physiological and pathological conditions. J. Mammary Gland Biol. Neoplasia 2014, 19, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Scow, R.O.; Chernick, S.S.; Fleck, T.R. Lipoprotein lipase and uptake of triacylglycerol, cholesterol and phosphatidylcholine from chylomicrons by mammary and adipose tissue of lactating rats in vivo. Biochim. Biophys. Acta 1977, 487, 297–306. [Google Scholar] [CrossRef]
- Freed, L.M.; Berkow, S.E.; Hamosh, P.; York, C.M.; Mehta, N.R.; Hamosh, M. Lipases in human milk: Effect of gestational age and length of lactation on enzyme activity. J. Am. Coll. Nutr. 1989, 8, 143–150. [Google Scholar] [CrossRef]
- Walker, R.E.; Harvatine, K.J.; Ross, A.C.; Wagner, E.A.; Riddle, S.W.; Gernand, A.D.; Nommsen-Rivers, L.A. Fatty Acid Transfer from Blood to Milk is Disrupted in Mothers with Low Milk Production, Obesity, and Inflammation. J. Nutr. 2022, 152, 2716–2726. [Google Scholar] [CrossRef] [PubMed]
- Aumeistere, L.; Ciproviča, I.; Zavadska, D.; Andersons, J.; Volkovs, V.; Ceļmalniece, K. Impact of Maternal Diet on Human Milk Composition Among Lactating Women in Latvia. Medicina 2019, 55, 173. [Google Scholar] [CrossRef] [Green Version]
- Moutsioulis, A.A.; Rule, D.C.; Murrieta, C.M.; Bauman, D.E.; Lock, A.L.; Barbano, D.M.; Carey, G.B. Human breast milk enrichment in conjugated linoleic acid after consumption of a conjugated linoleic acid-rich food product: A pilot study. Nutr. Res. 2008, 28, 437–442. [Google Scholar] [CrossRef]
- Juber, B.A.; Jackson, K.H.; Johnson, K.B.; Harris, W.S.; Baack, M.L. Breast milk DHA levels may increase after informing women: A community-based cohort study from South Dakota USA. Int. Breastfeed. J. 2016, 12, 7. [Google Scholar] [CrossRef] [Green Version]
- Fidler, N.; Sauerwald, T.; Pohl, A.; Demmelmair, H.; Koletzko, B. Docosahexaenoic acid transfer into human milk after dietary supplementation: A randomized clinical trial. J. Lipid Res. 2000, 41, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, F.; Fleith, M.; Goyer, A.; Samuel, T.M.; Elmelegy-Masserey, I.; Fontannaz, P.; Cruz-Hernandez, C.; Thakkar, S.K.; Monnard, C.; De Castro, C.A.; et al. Human milk fatty acid composition and its association with maternal blood and adipose tissue fatty acid content in a cohort of women from Europe. Eur. J. Nutr. 2022, 61, 2167–2182. [Google Scholar] [CrossRef]
- Del Prado, M.; Villalpando, S.; Elizondo, A.; Rodríguez, M.; Demmelmair, H.; Koletzko, B. Contribution of dietary and newly formed arachidonic acid to human milk lipids in women eating a low-fat diet. Am. J. Clin. Nutr. 2001, 74, 242–247. [Google Scholar] [CrossRef] [Green Version]
- Bzikowska-Jura, A.; Czerwonogrodzka-Senczyna, A.; Jasińska-Melon, E.; Mojska, H.; Olędzka, G.; Wesołowska, A.; Szostak-Węgierek, D. The Concentration of Omega-3 Fatty Acids in Human Milk Is Related to Their Habitual but Not Current Intake. Nutrients 2019, 11, 1585. [Google Scholar] [CrossRef] [Green Version]
- Dixon, J.B. Mechanisms of chylomicron uptake into lacteals. Ann. N. Y. Acad. Sci. 2010, 1207 (Suppl. 1), E52–E57. [Google Scholar] [CrossRef] [Green Version]
- Sakwinska, O.; Bosco, N. Host Microbe Interactions in the Lactating Mammary Gland. Front. Microbiol. 2019, 10, 1863. [Google Scholar] [CrossRef] [PubMed]
- Lynes, M.D.; Leiria, L.O.; Lundh, M.; Bartelt, A.; Shamsi, F.; Huang, T.L.; Takahashi, H.; Hirshman, M.F.; Schlein, C.; Lee, A.; et al. The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nat. Med. 2017, 23, 631–637. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.I.N.; Rackaityte, E.; Magnaye, K.; Porsche, C.; Özçam, M.; Levan, S.; Lynch, S. Gut bacterial-derived 12,13-diHOME promotes inflammatory macrophage polarization and epigenetic modifications. J. Allergy Clin. Immunol. 2022, 149, AB231. [Google Scholar] [CrossRef]
- Descamps, H.C.; Herrmann, B.; Wiredu, D.; Thaiss, C.A. The path toward using microbial metabolites as therapies. EBioMedicine 2019, 44, 747–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorburn, A.N.; Macia, L.; Mackay, C.R. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 2014, 40, 833–842. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B.K.; Morisseau, C.; Garen, G.; Cherney, M.M.; Garen, C.; Niu, C.; Hammock, B.D.; James, M.N. The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J. Mol. Biol. 2008, 381, 897–912. [Google Scholar] [CrossRef] [Green Version]
- Decker, M.; Arand, M.; Cronin, A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch. Toxicol. 2009, 83, 297–318. [Google Scholar] [CrossRef] [Green Version]
- Morisseau, C. Role of epoxide hydrolases in lipid metabolism. Biochimie 2013, 95, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [Green Version]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [Green Version]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Backhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glowacki, R.W.P.; Martens, E.C. In sickness and health: Effects of gut microbial metabolites on human physiology. PLoS Pathog. 2020, 16, e1008370. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Maher, A.D.; Hayes, B.; Cocks, B.; Marett, L.; Wales, W.J.; Rochfort, S.J. Latent biochemical relationships in the blood-milk metabolic axis of dairy cows revealed by statistical integration of 1H NMR spectroscopic data. J. Proteome Res. 2013, 12, 1428–1435. [Google Scholar] [CrossRef]
- Nyquist, S.K.; Gao, P.; Haining, T.K.J.; Retchin, M.R.; Golan, Y.; Drake, R.S.; Kolb, K.; Mead, B.E.; Ahituv, N.; Martinez, M.E.; et al. Cellular and transcriptional diversity over the course of human lactation. Proc. Natl. Acad. Sci. USA 2022, 119, e2121720119. [Google Scholar] [CrossRef]
- Lemay, D.G.; Ballard, O.A.; Hughes, M.A.; Morrow, A.L.; Horseman, N.D.; Nommsen-Rivers, L.A. RNA sequencing of the human milk fat layer transcriptome reveals distinct gene expression profiles at three stages of lactation. PLoS ONE 2013, 8, e67531. [Google Scholar] [CrossRef] [Green Version]
- Tanweer, A.; Khan, S.; Mustafa, F.N.; Imran, S.; Humayun, A.; Hussain, Z.-U.-N. Improving dietary data collection tools for better nutritional assessment—A systematic review. Comput. Methods Programs Biomed. Update 2022, 2, 100067. [Google Scholar] [CrossRef]
- Demmelmair, H.; Kuhn, A.; Dokoupil, K.; Hegele, V.; Sauerwald, T.; Koletzko, B. Human lactation: Oxidation and maternal transfer of dietary 13C-labelled α-linolenic acid into human milk. Isot. Environ. Health Stud. 2016, 52, 270–280. [Google Scholar] [CrossRef]
- Ruiz, L.; Garcia-Carral, C.; Rodriguez, J.M. Unfolding the Human Milk Microbiome Landscape in the Omics Era. Front. Microbiol. 2019, 10, 1378. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Gomez, K.; Flores, N.; Castaneda, H.M.; Martinez-Batallar, G.; Hernandez-Chavez, G.; Ramirez, O.T.; Gosset, G.; Encarnacion, S.; Bolivar, F. New insights into Escherichia coli metabolism: Carbon scavenging, acetate metabolism and carbon recycling responses during growth on glycerol. Microb. Cell Factories 2012, 11, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanGuilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008, 44, 619–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Metabolite | Origin |
| Mean Level in Circulation | Mean Level in Milk | References |
|---|---|---|---|---|---|
| Short chain fatty acids (acetate, butyrate, formate) | Bacterial fermentation of polysaccharides |
| Acetate: 41.9 ± 15.1 Butyrate: 1.0 (0.3–1.5) Formate: 32.8 ± 13.3 | Acetate: 27.5 ± 28.2 Butyrate: 192 ± 149 Formate: 12.2 ± 14.8 | Milk: [46] Blood: [48,49] |
| Polyamines (putrescine, spermidine, spermine) | Arginine derivative produced by host and bacterial cells |
| Putrescine: 0.05 ± 0.03 Spermidine: 0.07 ± 0.04 Spermine: 0.03 ± 0.04 | Putrescine: 0.21 (0–1.14) Spermidine: 3.53 (1.43–6.47) Spermine: 5.08 (0.76–5.94) | Milk: [52] Blood: [53] |
| 12,13-DiHOME | End metabolic product of linoleic acid produced by host, fungal, and bacterial cells |
| 0.0012 ± 0.00018 | 0.0056 ± 0.0004 | Milk: [54] Blood: [55] |
| Trimethylamine | Bacterial metabolism of choline |
| 0.418 ± 0.124 | <5 ppm | Milk: [56] Blood: [57] |
| Hippurate | Bacterial metabolism of polyphenols and host conjugation from glycine and microbial benzoate |
| 16.74 ± 11.16 | 7.8 ± 4.2 | Milk: [46] Blood: [53] |
| Taurine | Bacterial deconjugation of taurine from host taurine-bound bile acids |
| 93.0 ± 35.7 | 190 ± 66.6 | Milk: [46] Blood: [58] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stinson, L.F.; George, A.D. Human Milk Lipids and Small Metabolites: Maternal and Microbial Origins. Metabolites 2023, 13, 422. https://doi.org/10.3390/metabo13030422
Stinson LF, George AD. Human Milk Lipids and Small Metabolites: Maternal and Microbial Origins. Metabolites. 2023; 13(3):422. https://doi.org/10.3390/metabo13030422
Chicago/Turabian StyleStinson, Lisa F., and Alexandra D. George. 2023. "Human Milk Lipids and Small Metabolites: Maternal and Microbial Origins" Metabolites 13, no. 3: 422. https://doi.org/10.3390/metabo13030422
APA StyleStinson, L. F., & George, A. D. (2023). Human Milk Lipids and Small Metabolites: Maternal and Microbial Origins. Metabolites, 13(3), 422. https://doi.org/10.3390/metabo13030422








