Metabolic and Transcriptomic Signatures of the Acute Psychological Stress Response in the Mouse Brain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
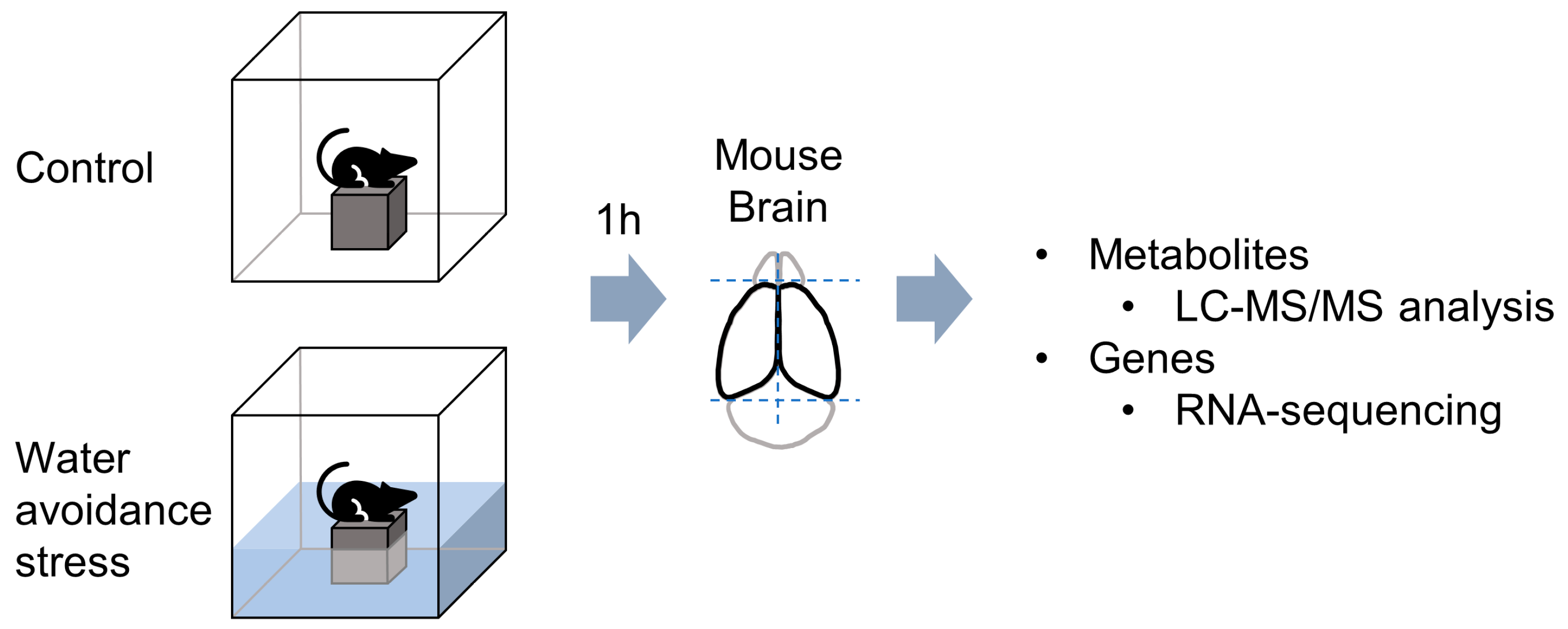
2.2. Water Avoidance Stress Procedure
2.3. Total RNA Extraction and RNA-Sequencing
2.4. Global Metabolite Profiling and Data Analysis
3. Results
3.1. Transcriptome Analysis of the Brain under Acute Stress
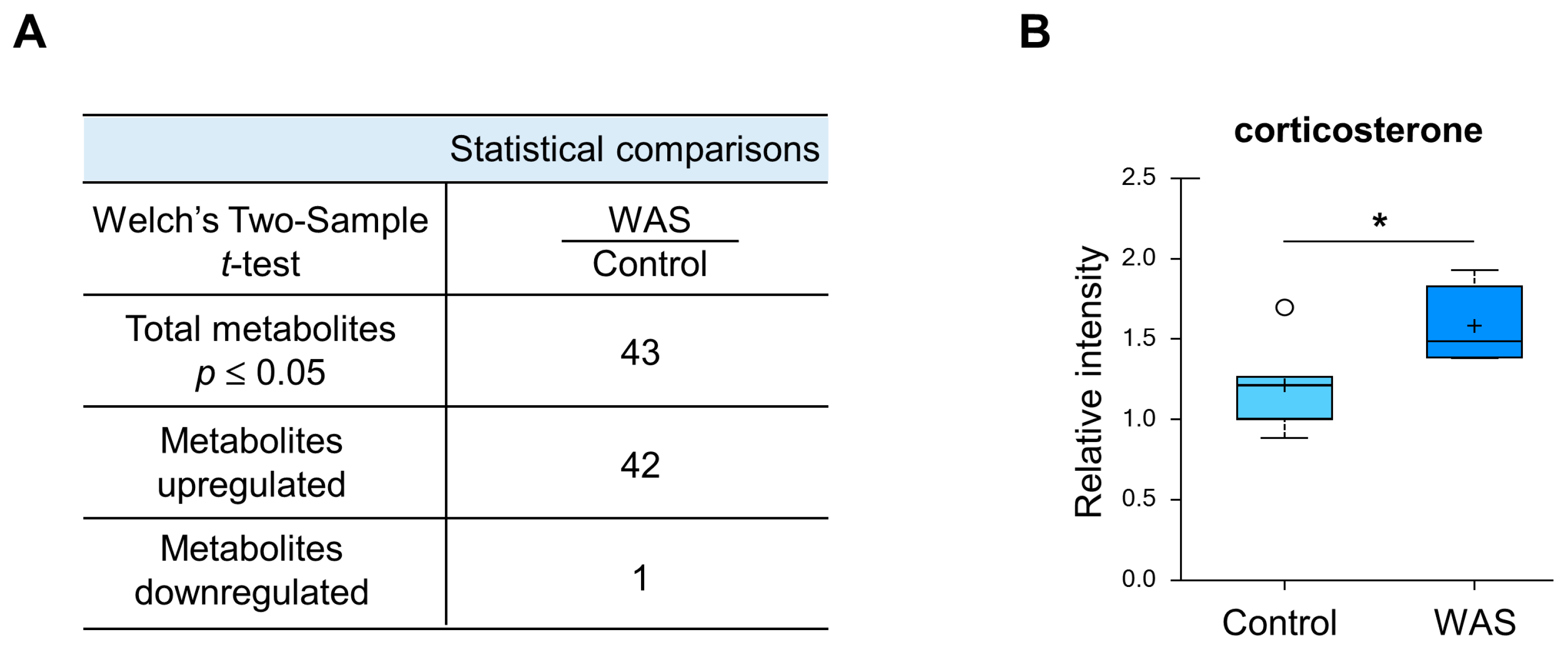
3.2. Unique Alterations in a Broad Range of Metabolites in the Mouse Brain Exposed to Acute Water Avoidance Stress
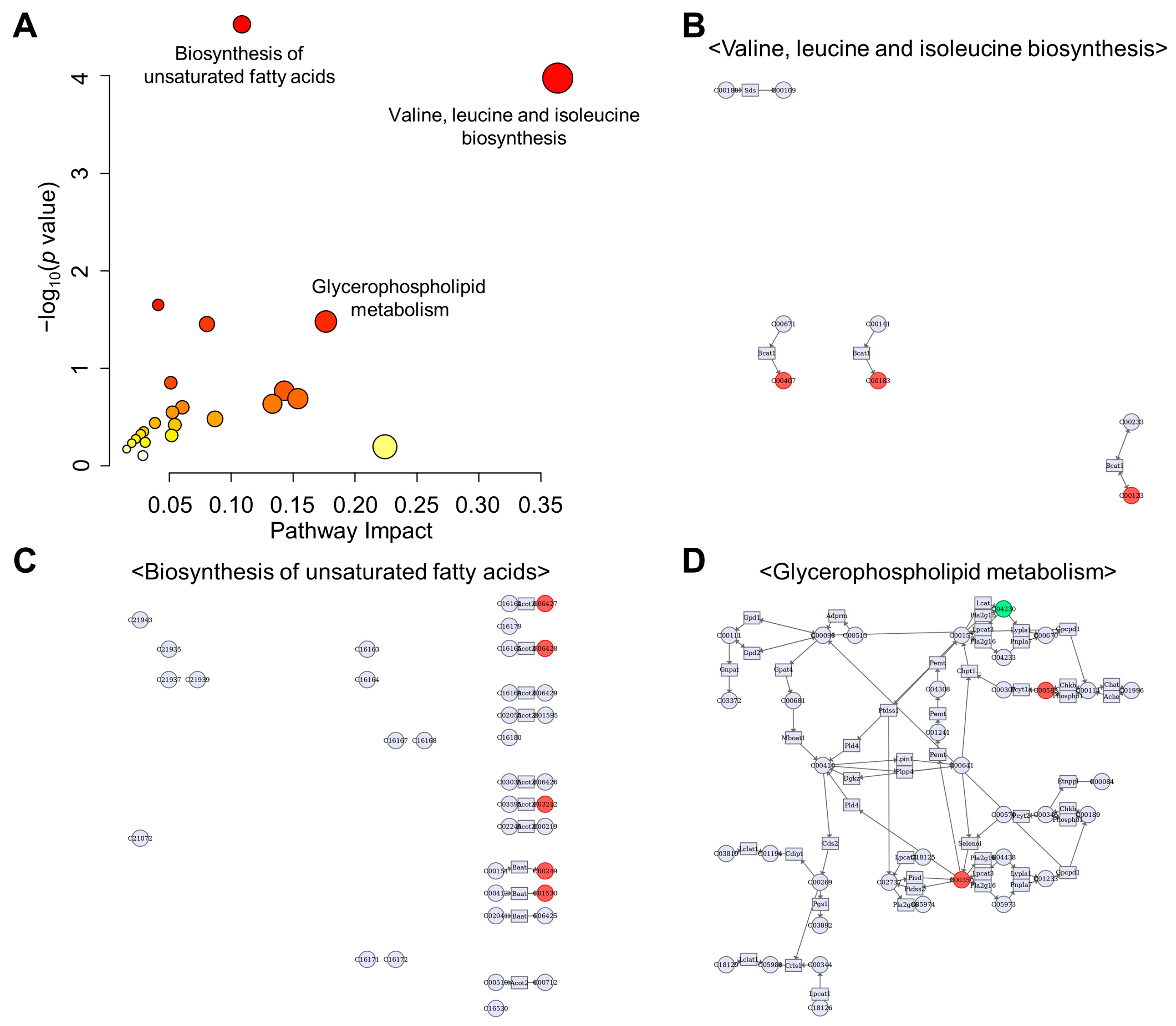
3.3. Activation of Lipolysis and Eicosanoid Metabolic Pathways
3.4. Acute Stress Elevates Endocannabinoid Levels in the Brain
3.5. Changes in Amino Acid Metabolism
3.6. Increased 4-Hydroxy-Nonenal-Glutathione Levels
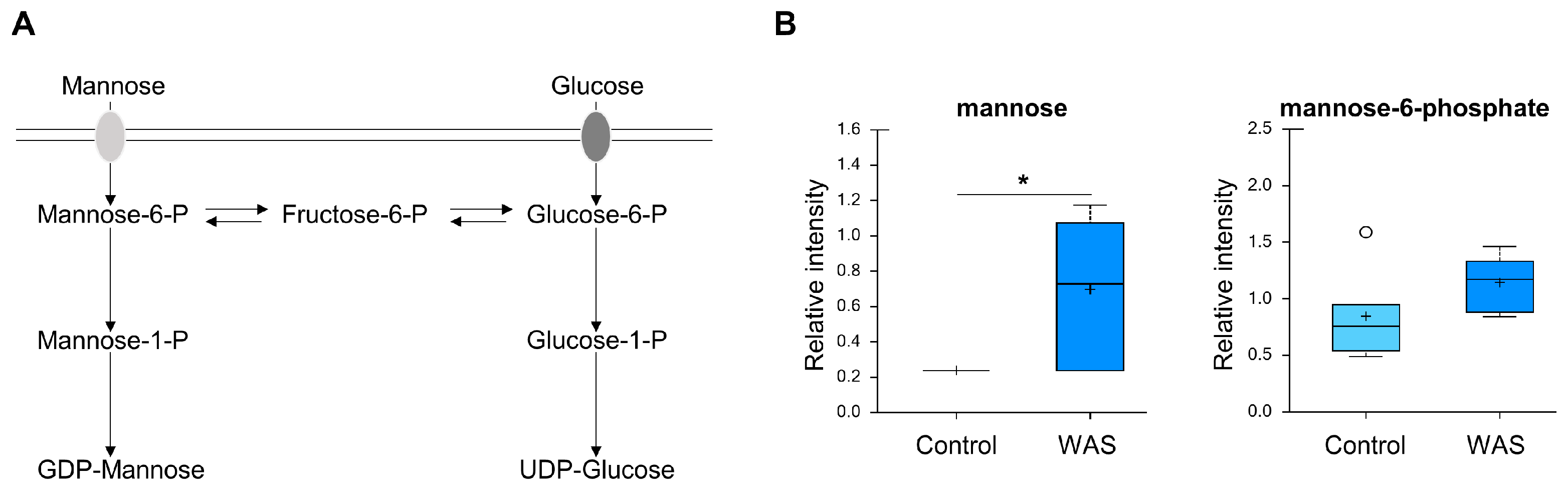
3.7. Increased Mannose Levels
3.8. Integration Analysis of Transcriptomic and Metabolomic Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selye, H. A Syndrome Produced by Diverse Nocuous Agents. Nature 1936, 138, 32. [Google Scholar] [CrossRef] [Green Version]
- Selye, H. The General Adaptation Syndrome and the Diseases of Adaptation. J. Clin. Endocrinol. 1946, 6, 117–230. [Google Scholar] [CrossRef]
- Selye, H. A Syndrome Produced by Diverse Nocuous Agents. J. Neuropsychiatry Clin. Neurosci. 1998, 10, 230a–231a. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulrich-Lai, Y.M.; Herman, J.P. Neural Regulation of Endocrine and Autonomic Stress Responses. Nat. Rev. Neurosci. 2009, 10, 397–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapolsky, R.M. Why Stress Is Bad for Your Brain. Science 1996, 273, 749–750. [Google Scholar] [CrossRef]
- McEwen, B.S. Protective and Damaging Effects of Stress Mediators. N. Engl. J. Med. 1998, 338, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Bonanno, G.A.; Westphal, M.; Mancini, A.D. Resilience to Loss and Potential Trauma. Annu. Rev. Clin. Psychol. 2011, 7, 511–535. [Google Scholar] [CrossRef] [Green Version]
- Kalisch, R.; Baker, D.G.; Basten, U.; Boks, M.P.; Bonanno, G.A.; Brummelman, E.; Chmitorz, A.; Fernàndez, G.; Fiebach, C.J.; Galatzer-Levy, I.; et al. The Resilience Framework as a Strategy to Combat Stress-Related Disorders. Nat. Hum. Behav. 2017, 1, 784–790. [Google Scholar] [CrossRef] [Green Version]
- Tost, H.; Champagne, F.A.; Meyer-Lindenberg, A. Environmental Influence in the Brain, Human Welfare and Mental Health. Nat. Neurosci. 2015, 18, 1421–1431. [Google Scholar] [CrossRef]
- Godoy, L.D.; Rossignoli, M.T.; Delfino-Pereira, P.; Garcia-Cairasco, N.; de Lima Umeoka, E.H. A Comprehensive Overview on Stress Neurobiology: Basic Concepts and Clinical Implications. Front. Behav. Neurosci. 2018, 12, 127. [Google Scholar] [CrossRef] [Green Version]
- McEwen, B.S.; Akil, H. Revisiting the Stress Concept: Implications for Affective Disorders. J. Neurosci. 2020, 40, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, M.A.C.; Wand, G. Stress and the HPA Axis: Role of Glucocorticoids in Alcohol Dependence. Alcohol Res. Curr. Rev. 2012, 34, 468–483. [Google Scholar]
- Pagliaccio, D.; Luby, J.L.; Bogdan, R.; Agrawal, A.; Gaffrey, M.S.; Belden, A.C.; Botteron, K.N.; Harms, M.P.; Barch, D.M. Stress-System Genes and Life Stress Predict Cortisol Levels and Amygdala and Hippocampal Volumes in Children. Neuropsychopharmacology 2014, 39, 1245–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buss, C.; Davis, E.P.; Shahbaba, B.; Pruessner, J.C.; Head, K.; Sandman, C.A. Maternal Cortisol over the Course of Pregnancy and Subsequent Child Amygdala and Hippocampus Volumes and Affective Problems. Proc. Natl. Acad. Sci. USA 2012, 109, E1312–E1319. [Google Scholar] [CrossRef] [Green Version]
- McEwen, B.S.; Nasca, C.; Gray, J.D. Stress Effects on Neuronal Structure: Hippocampus, Amygdala, and Prefrontal Cortex. Neuropsychopharmacology 2016, 41, 3–23. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Ge, T.T.; Yin, G.; Cui, R.; Zhao, G.; Yang, W. Stress-Induced Functional Alterations in Amygdala: Implications for Neuropsychiatric Diseases. Front. Neurosci. 2018, 12, 367. [Google Scholar] [CrossRef] [Green Version]
- Lakshminarasimhan, H.; Chattarji, S. Stress Leads to Contrasting Effects on the Levels of Brain Derived Neurotrophic Factor in the Hippocampus and Amygdala. PLoS ONE 2012, 7, e30481. [Google Scholar] [CrossRef]
- Christoffel, D.J.; Golden, S.A.; Russo, S.J. Structural and Synaptic Plasticity in Stress-Related Disorders. Rev. Neurosci. 2011, 22, 535–549. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Li, C.; Wang, J.; Zhang, X.; Li, M.; Zhang, R.; Huang, Z.; Zhang, Y. Amygdala-Hippocampal Innervation Modulates Stress-Induced Depressive-like Behaviors through AMPA Receptors. Proc. Natl. Acad. Sci. USA 2021, 118, e2019409118. [Google Scholar] [CrossRef]
- Sumislawski, J.J.; Ramikie, T.S.; Patel, S. Reversible Gating of Endocannabinoid Plasticity in the Amygdala by Chronic Stress: A Potential Role for Monoacylglycerol Lipase Inhibition in the Prevention of Stress-Induced Behavioral Adaptation. Neuropsychopharmacology 2011, 36, 2750–2761. [Google Scholar] [CrossRef] [Green Version]
- Douma, E.H.; de Kloet, E.R. Stress-Induced Plasticity and Functioning of Ventral Tegmental Dopamine Neurons. Neurosci. Biobehav. Rev. 2020, 108, 48–77. [Google Scholar] [CrossRef] [PubMed]
- Aceto, G.; Colussi, C.; Leone, L.; Fusco, S.; Rinaudo, M.; Scala, F.; Green, T.A.; Laezza, F.; D’Ascenzo, M.; Grassi, C. Chronic Mild Stress Alters Synaptic Plasticity in the Nucleus Accumbens through GSK3β-Dependent Modulation of Kv4. 2 Channels. Proc. Natl. Acad. Sci. USA 2020, 117, 8143–8153. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.W.; Wohleb, E.S. How Stress Shapes Neuroimmune Function: Implications for the Neurobiology of Psychiatric Disorders. Biol. Psychiatry 2021, 90, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Bagot, R.C.; Cates, H.M.; Purushothaman, I.; Lorsch, Z.S.; Walker, D.M.; Wang, J.; Huang, X.; Schlüter, O.M.; Maze, I.; Peña, C.J. Circuit-Wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron 2016, 90, 969–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, J.D.; Rubin, T.G.; Hunter, R.G.; McEwen, B.S. Hippocampal Gene Expression Changes Underlying Stress Sensitization and Recovery. Mol. Psychiatry 2014, 19, 1171–1178. [Google Scholar] [CrossRef] [Green Version]
- Misiewicz, Z.; Iurato, S.; Kulesskaya, N.; Salminen, L.; Rodrigues, L.; Maccarrone, G.; Martins, J.; Czamara, D.; Laine, M.A.; Sokolowska, E. Multi-Omics Analysis Identifies Mitochondrial Pathways Associated with Anxiety-Related Behavior. PLoS Genet. 2019, 15, e1008358. [Google Scholar] [CrossRef] [PubMed]
- Cerniauskas, I.; Winterer, J.; de Jong, J.W.; Lukacsovich, D.; Yang, H.; Khan, F.; Peck, J.R.; Obayashi, S.K.; Lilascharoen, V.; Lim, B.K. Chronic Stress Induces Activity, Synaptic, and Transcriptional Remodeling of the Lateral Habenula Associated with Deficits in Motivated Behaviors. Neuron 2019, 104, 899–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, P.J.; Chen, E.Y.; Tolstikov, V.; Peña, C.J.; Picone, J.A.; Shah, P.; Panagopoulos, K.; Strat, A.N.; Walker, D.M.; Lorsch, Z.S. Chronic Stress and Antidepressant Treatment Alter Purine Metabolism and Beta Oxidation within Mouse Brain and Serum. Sci. Rep. 2020, 10, 18134. [Google Scholar] [CrossRef] [PubMed]
- Lepack, A.E.; Bagot, R.C.; Peña, C.J.; Loh, Y.-H.E.; Farrelly, L.A.; Lu, Y.; Powell, S.K.; Lorsch, Z.S.; Issler, O.; Cates, H.M. Aberrant H3. 3 Dynamics in NAc Promote Vulnerability to Depressive-like Behavior. Proc. Natl. Acad. Sci. USA 2016, 113, 12562–12567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña, C.J.; Smith, M.; Ramakrishnan, A.; Cates, H.M.; Bagot, R.C.; Kronman, H.G.; Patel, B.; Chang, A.B.; Purushothaman, I.; Dudley, J. Early Life Stress Alters Transcriptomic Patterning across Reward Circuitry in Male and Female Mice. Nat. Commun. 2019, 10, 5098. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Damez-Werno, D.M.; Scobie, K.N.; Shao, N.-Y.; Dias, C.; Rabkin, J.; Koo, J.W.; Korb, E.; Bagot, R.C.; Ahn, F.H. ACF Chromatin-Remodeling Complex Mediates Stress-Induced Depressive-like Behavior. Nat. Med. 2015, 21, 1146–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Cheng, J.; Waddell, N.J.; Wang, Z.-J.; Pang, X.; Cao, Q.; Liu, A.; Chitaman, J.M.; Abreu, K.; Jasrotia, R.S. DNA Methyltransferase 3A Is Involved in the Sustained Effects of Chronic Stress on Synaptic Functions and Behaviors. Cereb. Cortex 2021, 31, 1998–2012. [Google Scholar] [CrossRef]
- von Ziegler, L.M.; Floriou-Servou, A.; Waag, R.; Das Gupta, R.R.; Sturman, O.; Gapp, K.; Maat, C.A.; Kockmann, T.; Lin, H.-Y.; Duss, S.N. Multiomic Profiling of the Acute Stress Response in the Mouse Hippocampus. Nat. Commun. 2022, 13, 1824. [Google Scholar] [CrossRef] [PubMed]
- Floriou-Servou, A.; von Ziegler, L.; Stalder, L.; Sturman, O.; Privitera, M.; Rassi, A.; Cremonesi, A.; Thöny, B.; Bohacek, J. Distinct Proteomic, Transcriptomic, and Epigenetic Stress Responses in Dorsal and Ventral Hippocampus. Biol. Psychiatry 2018, 84, 531–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roszkowski, M.; Manuella, F.; von Ziegler, L.; Durán-Pacheco, G.; Moreau, J.-L.; Mansuy, I.M.; Bohacek, J. Rapid Stress-Induced Transcriptomic Changes in the Brain Depend on Beta-Adrenergic Signaling. Neuropharmacology 2016, 107, 329–338. [Google Scholar] [CrossRef]
- Stankiewicz, A.M.; Goscik, J.; Majewska, A.; Swiergiel, A.H.; Juszczak, G.R. The Effect of Acute and Chronic Social Stress on the Hippocampal Transcriptome in Mice. PLoS ONE 2015, 10, e0142195. [Google Scholar] [CrossRef]
- Terenina, E.E.; Cavigelli, S.; Mormede, P.; Zhao, W.; Parks, C.; Lu, L.; Jones, B.C.; Mulligan, M.K. Genetic Factors Mediate the Impact of Chronic Stress and Subsequent Response to Novel Acute Stress. Front. Neurosci. 2019, 13, 438. [Google Scholar] [CrossRef] [Green Version]
- Tsolakidou, A.; Trümbach, D.; Panhuysen, M.; Pütz, B.; Deussing, J.; Wurst, W.; Sillaber, I.; Holsboer, F.; Rein, T. Acute Stress Regulation of Neuroplasticity Genes in Mouse Hippocampus CA3 Area—Possible Novel Signalling Pathways. Mol. Cell. Neurosci. 2008, 38, 444–452. [Google Scholar] [CrossRef]
- Fan, L.; Yang, L.; Li, X.; Teng, T.; Xiang, Y.; Liu, X.; Jiang, Y.; Zhu, Y.; Zhou, X.; Xie, P. Proteomic and Metabolomic Characterization of Amygdala in Chronic Social Defeat Stress Rats. Behav. Brain Res. 2021, 412, 113407. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, S.; Pu, J.; Yang, L.; Zhou, X.; Liu, L.; Jiang, X.; Zhang, H.; Teng, T.; Tian, L. Integrated Metabolomics and Proteomics Analysis of Hippocampus in a Rat Model of Depression. Neuroscience 2018, 371, 207–220. [Google Scholar] [CrossRef]
- Dulka, B.N.; Bourdon, A.K.; Clinard, C.T.; Muvvala, M.B.K.; Campagna, S.R.; Cooper, M.A. Metabolomics Reveals Distinct Neurochemical Profiles Associated with Stress Resilience. Neurobiol. Stress 2017, 7, 103–112. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, M.; Gu, S.; Zhang, X.; Feng, G.; Ma, X.; Xu, S.; Wu, E.; Huang, J.H.; Wang, F. Metabolomics Changes in Brain-Gut Axis after Unpredictable Chronic Mild Stress. Psychopharmacology 2022, 239, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Mu, J.; Xu, T.; Linghu, T.; Zhao, H.; Tian, J.; Qin, X. Metabolomic Analysis of the Hippocampus in a Rat Model of Chronic Mild Unpredictable Stress-Induced Depression Based on a Pathway Crosstalk and Network Module Approach. J. Pharm. Biomed. Anal. 2021, 193, 113755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, W.; Huang, Y.; Zeng, Z.; Yang, X.; Huang, H.; Wen, J.; Cao, Y.; Sun, H. Hippocampal Metabolic Alteration in Rat Exhibited Susceptibility to Prenatal Stress. J. Affect. Disord. 2019, 259, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Tian, J.; Xiang, H.; Guo, X.; Zhang, L.; Du, G.; Qin, X. A 1H-NMR Plasma Metabonomic Study of Acute and Chronic Stress Models of Depression in Rats. Behav. Brain Res. 2013, 241, 86–91. [Google Scholar] [CrossRef]
- Lo Iacono, L.; Valzania, A.; Visco-Comandini, F.; Viscomi, M.T.; Felsani, A.; Puglisi-Allegra, S.; Carola, V. Regulation of Nucleus Accumbens Transcript Levels in Mice by Early-Life Social Stress and Cocaine. Neuropharmacology 2016, 103, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, X.; Teng, T.; Fan, L.; Liu, X.; Xiang, Y.; Jiang, Y.; Xie, P.; Zhu, D. Multi-Omics Analysis of the Amygdala in a Rat Chronic Unpredictable Mild Stress Model of Depression. Neuroscience 2021, 463, 174–183. [Google Scholar] [CrossRef]
- Sillivan, S.E.; Jones, M.E.; Jamieson, S.; Rumbaugh, G.; Miller, C.A. Bioinformatic Analysis of Long-Lasting Transcriptional and Translational Changes in the Basolateral Amygdala Following Acute Stress. PLoS ONE 2019, 14, e0209846. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, A.M.; Bridgewater, B.R.; Liu, Q.; Mitchell, M.W.; Robinson, R.J.; Dai, H.; Stewart, S.J.; DeHaven, C.D.; Miller, L.A.D. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. Metabolomics 2014, 4, 1. [Google Scholar]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef]
- Dhama, K.; Latheef, S.K.; Dadar, M.; Samad, H.A.; Munjal, A.; Khandia, R.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Bhatt, P. Biomarkers in Stress Related Diseases/Disorders: Diagnostic, Prognostic, and Therapeutic Values. Front. Mol. Biosci. 2019, 6, 91. [Google Scholar] [CrossRef]
- Tan, S.T.; Ramesh, T.; Toh, X.R.; Nguyen, L.N. Emerging Roles of Lysophospholipids in Health and Disease. Prog. Lipid Res. 2020, 80, 101068. [Google Scholar] [CrossRef]
- Dennis, E.A. Phospholipase A2 in Eicosanoid Generation. Am. J. Respir. Crit. Care Med. 2000, 161, S32–S35. [Google Scholar] [CrossRef]
- Mechoulam, R.; Parker, L.A. The Endocannabinoid System and the Brain. Annu. Rev. Psychol. 2013, 64, 21–47. [Google Scholar] [CrossRef] [Green Version]
- Raso, G.M.; Russo, R.; Calignano, A.; Meli, R. Palmitoylethanolamide in CNS Health and Disease. Pharmacol. Res. 2014, 86, 32–41. [Google Scholar] [CrossRef]
- Connor, M.; Vaughan, C.W.; Vandenberg, R.J. N-Acyl Amino Acids and N-acyl Neurotransmitter Conjugates: Neuromodulators and Probes for New Drug Targets. Br. J. Pharmacol. 2010, 160, 1857–1871. [Google Scholar] [CrossRef]
- Csala, M.; Kardon, T.; Legeza, B.; Lizák, B.; Mandl, J.; Margittai, É.; Puskás, F.; Száraz, P.; Szelényi, P.; Bánhegyi, G. On the Role of 4-Hydroxynonenal in Health and Disease. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2015, 1852, 826–838. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS Spectra Processing, Multi-Omics Integration and Covariate Adjustment of Global Metabolomics Data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Farooqui, A.A.; Yang, H.; Rosenberger, T.A.; Horrocks, L.A. Phospholipase A2 and Its Role in Brain Tissue. J. Neurochem. 1997, 69, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Tassoni, D.; Kaur, G.; Weisinger, R.; Sinclair, A. The Role of Eicosanoids in the Brain. Asia Pac. J. Clin. Nutr. 2008, 17, 220–228. [Google Scholar] [PubMed]
- Gaire, B.P.; Choi, J.-W. Critical Roles of Lysophospholipid Receptors in Activation of Neuroglia and Their Neuroinflammatory Responses. Int. J. Mol. Sci. 2021, 22, 7864. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.G.; Chan, R.B.; Bravo, F.V.; Miranda, A.; Silva, R.R.; Zhou, B.; Marques, F.; Pinto, V.; Cerqueira, J.J.; Di Paolo, G. The Impact of Chronic Stress on the Rat Brain Lipidome. Mol. Psychiatry 2016, 21, 80–88. [Google Scholar] [CrossRef] [Green Version]
- Morena, M.; Patel, S.; Bains, J.S.; Hill, M.N. Neurobiological Interactions between Stress and the Endocannabinoid System. Neuropsychopharmacology 2016, 41, 80–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, M.N.; Patel, S.; Campolongo, P.; Tasker, J.G.; Wotjak, C.T.; Bains, J.S. Functional Interactions between Stress and the Endocannabinoid System: From Synaptic Signaling to Behavioral Output. J. Neurosci. 2010, 30, 14980–14986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, J.M.; Vecchiarelli, H.A.; Morena, M.; Lee, T.T.Y.; Hermanson, D.J.; Kim, A.B.; McLaughlin, R.J.; Hassan, K.I.; Kühne, C.; Wotjak, C.T.; et al. Corticotropin-Releasing Hormone Drives Anandamide Hydrolysis in the Amygdala to Promote Anxiety. J. Neurosci. 2015, 35, 3879 LP–3892 LP. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Kingsley, P.J.; Mackie, K.; Marnett, L.J.; Winder, D.G. Repeated Homotypic Stress Elevates 2-Arachidonoylglycerol Levels and Enhances Short-Term Endocannabinoid Signaling at Inhibitory Synapses in Basolateral Amygdala. Neuropsychopharmacology 2009, 34, 2699–2709. [Google Scholar] [CrossRef] [Green Version]
- Rademacher, D.J.; Meier, S.E.; Shi, L.; Ho, W.-S.V.; Jarrahian, A.; Hillard, C.J. Effects of Acute and Repeated Restraint Stress on Endocannabinoid Content in the Amygdala, Ventral Striatum, and Medial Prefrontal Cortex in Mice. Neuropharmacology 2008, 54, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Zhang, C.; Kilicarslan, M.; Piening, B.D.; Bjornson, E.; Hallström, B.M.; Groen, A.K.; Ferrannini, E.; Laakso, M.; Snyder, M. Integrated Network Analysis Reveals an Association between Plasma Mannose Levels and Insulin Resistance. Cell Metab. 2016, 24, 172–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardinoglu, A.; Stančáková, A.; Lotta, L.A.; Kuusisto, J.; Boren, J.; Blüher, M.; Wareham, N.J.; Ferrannini, E.; Groop, P.H.; Laakso, M. Plasma Mannose Levels Are Associated with Incident Type 2 Diabetes and Cardiovascular Disease. Cell Metab. 2017, 26, 281–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernstrom, J.D. Branched-Chain Amino Acids and Brain Function. J. Nutr. 2005, 135, 1539S–1546S. [Google Scholar] [CrossRef] [Green Version]
- Yudkoff, M. Interactions in the Metabolism of Glutamate and the Branched-Chain Amino Acids and Ketoacids in the CNS. Neurochem. Res. 2017, 42, 10–18. [Google Scholar] [CrossRef] [Green Version]
- Baranyi, A.; Amouzadeh-Ghadikolai, O.; von Lewinski, D.; Rothenhäusler, H.-B.; Theokas, S.; Robier, C.; Mangge, H.; Reicht, G.; Hlade, P.; Meinitzer, A. Branched-Chain Amino Acids as New Biomarkers of Major Depression-a Novel Neurobiology of Mood Disorder. PLoS ONE 2016, 11, e0160542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranyi, A.; Meinitzer, A.; Rothenhäusler, H.-B.; Amouzadeh-Ghadikolai, O.; Lewinski, D.V.; Breitenecker, R.J.; Herrmann, M. Metabolomics Approach in the Investigation of Depression Biomarkers in Pharmacologically Induced Immune-Related Depression. PLoS ONE 2018, 13, e0208238. [Google Scholar] [CrossRef]
- Fellendorf, F.T.; Platzer, M.; Pilz, R.; Rieger, A.; Kapfhammer, H.-P.; Mangge, H.; Dalkner, N.; Zelzer, S.; Meinitzer, A.; Birner, A. Branched-Chain Amino Acids Are Associated with Metabolic Parameters in Bipolar Disorder. World J. Biol. Psychiatry 2019, 20, 821–826. [Google Scholar] [CrossRef]
- Webhofer, C.; Gormanns, P.; Tolstikov, V.; Zieglgänsberger, W.; Sillaber, I.; Holsboer, F.; Turck, C.W. Metabolite Profiling of Antidepressant Drug Action Reveals Novel Drug Targets beyond Monoamine Elevation. Transl. Psychiatry 2011, 1, e58. [Google Scholar] [CrossRef] [Green Version]
- Callery, P.S.; Geelhaar, L.A. 1-Piperideine as an In Vivo Precursor of the Γ-Aminobutyric Acid Homologue 5-Aminopentanoic Acid. J. Neurochem. 1985, 45, 946–948. [Google Scholar] [CrossRef]
- Kalecký, K.; German, D.C.; Montillo, A.A.; Bottiglieri, T. Targeted Metabolomic Analysis in Alzheimer’s Disease Plasma and Brain Tissue in Non-Hispanic Whites. J. Alzheimer’s Dis. 2022, 86, 1875. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Park, J.; Kim, S. Metabolic and Transcriptomic Signatures of the Acute Psychological Stress Response in the Mouse Brain. Metabolites 2023, 13, 453. https://doi.org/10.3390/metabo13030453
Lee H, Park J, Kim S. Metabolic and Transcriptomic Signatures of the Acute Psychological Stress Response in the Mouse Brain. Metabolites. 2023; 13(3):453. https://doi.org/10.3390/metabo13030453
Chicago/Turabian StyleLee, Haein, Jina Park, and Seyun Kim. 2023. "Metabolic and Transcriptomic Signatures of the Acute Psychological Stress Response in the Mouse Brain" Metabolites 13, no. 3: 453. https://doi.org/10.3390/metabo13030453
APA StyleLee, H., Park, J., & Kim, S. (2023). Metabolic and Transcriptomic Signatures of the Acute Psychological Stress Response in the Mouse Brain. Metabolites, 13(3), 453. https://doi.org/10.3390/metabo13030453




