Association of Maternal Metabolites and Metabolite Networks with Newborn Outcomes in a Multi-Ancestry Cohort
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Sample Collection
2.2. Conventional Metabolites and Targeted Metabolomics Assays
2.3. Untargeted Metabolite Analyses
2.4. Statistical Analyses
3. Results
3.1. Study Population
3.2. Associations of Maternal Metabolites with Newborn Phenotypes
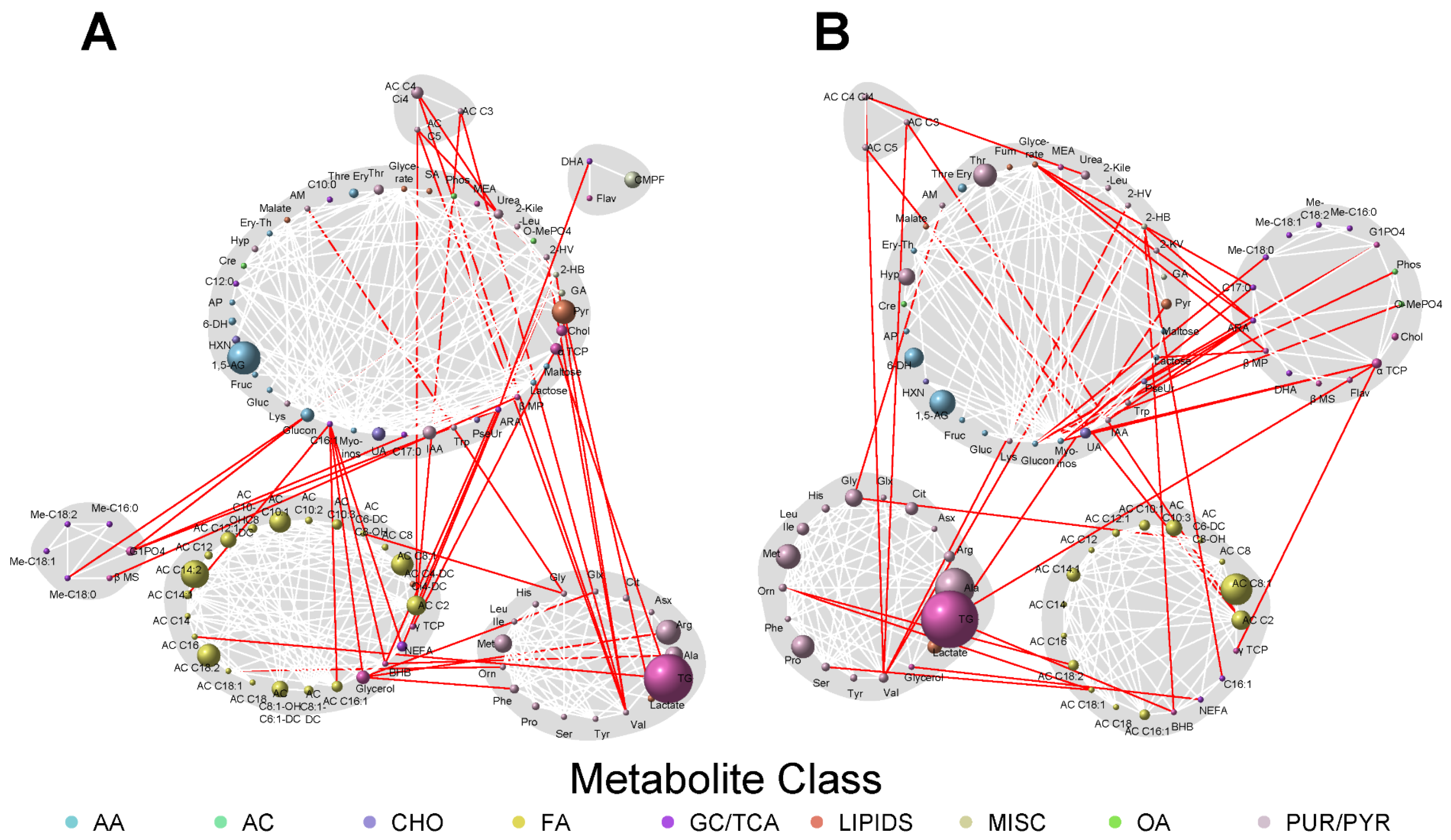
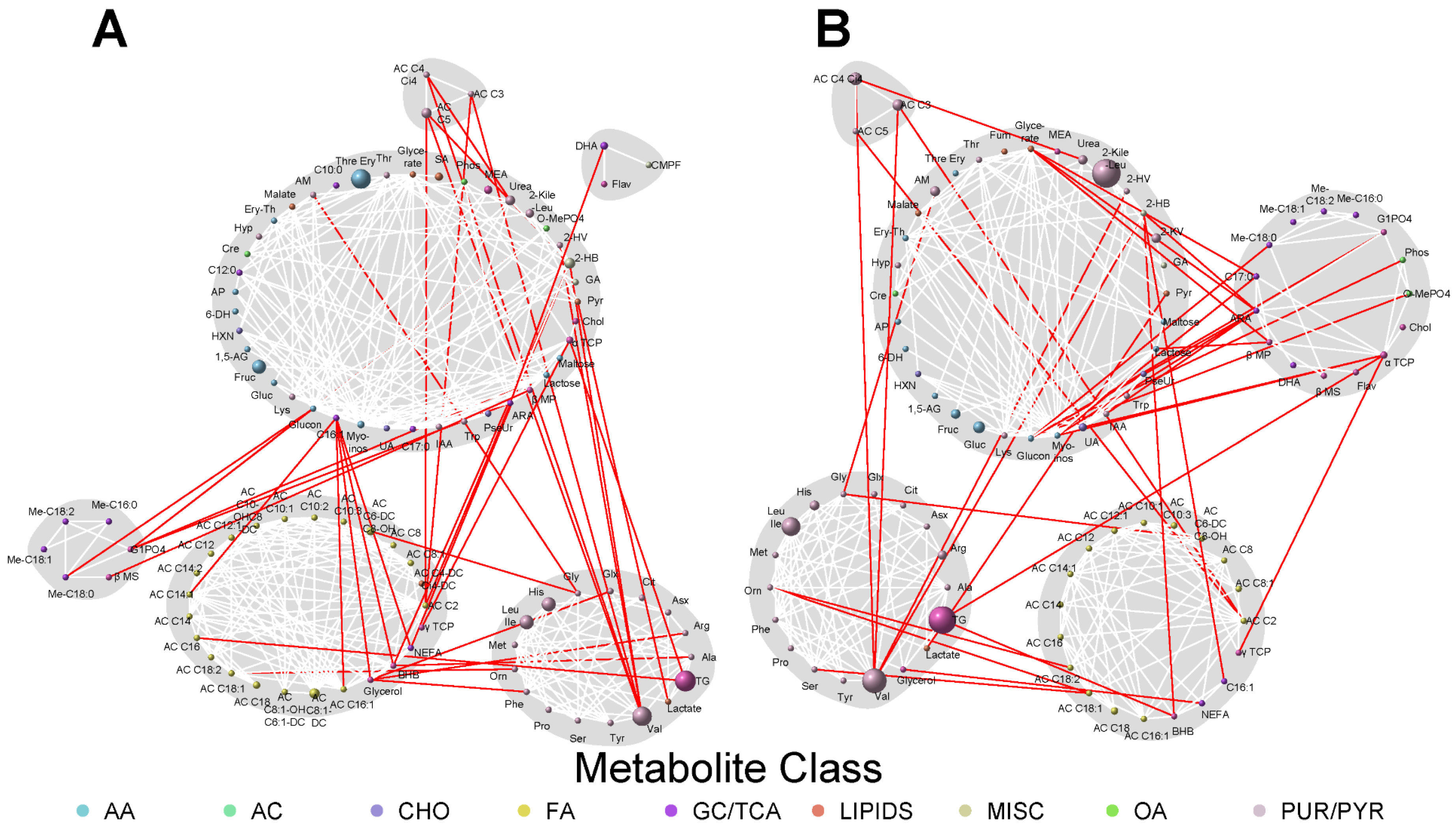
3.3. Network Analyses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hales, C.M.; Fryar, C.D.; Carroll, M.D.; Freedman, D.S.; Ogden, C.L. Trends in Obesity and Severe Obesity Prevalence in US Youth and Adults by Sex and Age, 2007–2008 to 2015–2016. JAMA 2018, 319, 1723–1725. [Google Scholar] [CrossRef]
- Josefson, J.L.; Scholtens, D.M.; Kuang, A.; Catalano, P.M.; Lowe, L.P.; Dyer, A.R.; Petito, L.C.; Lowe, W.L., Jr.; Metzger, B.E. Newborn Adiposity and Cord Blood C-Peptide as Mediators of the Maternal Metabolic Environment and Childhood Adiposity. Diabetes Care 2021, 44, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Kelly, A.S. Review of Childhood Obesity: From Epidemiology, Etiology, and Comorbidities to Clinical Assessment and Treatment. Mayo Clin. Proc. 2017, 92, 251–265. [Google Scholar] [CrossRef]
- Hadden, D.R.; McLaughlin, C. Normal and abnormal maternal metabolism during pregnancy. Semin. Fetal. Neonatal. Med. 2009, 14, 66–71. [Google Scholar] [CrossRef]
- Chen, Q.; Francis, E.; Hu, G.; Chen, L. Metabolomic profiling of women with gestational diabetes mellitus and their offspring: Review of metabolomics studies. J. Diabetes Complicat. 2018, 32, 512–523. [Google Scholar] [CrossRef]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef]
- Suhre, K.; Gieger, C. Genetic variation in metabolic phenotypes: Study designs and applications. Nat. Rev. Genet. 2012, 13, 759–769. [Google Scholar] [CrossRef]
- Josefson, J.L.; Catalano, P.M.; Lowe, W.L.; Scholtens, D.M.; Kuang, A.; Dyer, A.R.; Lowe, L.P.; Metzger, B.E. The Joint Associations of Maternal BMI and Glycemia with Childhood Adiposity. J. Clin. Endocrinol. Metab. 2020, 105, 2177–2188. [Google Scholar] [CrossRef]
- Catalano, P.M.; Shankar, K. Obesity and pregnancy: Mechanisms of short term and long term adverse consequences for mother and child. BMJ 2017, 356, j1. [Google Scholar] [CrossRef]
- Dabelea, D.; Crume, T. Maternal environment and the transgenerational cycle of obesity and diabetes. Diabetes 2011, 60, 1849–1855. [Google Scholar] [CrossRef]
- Catalano, P.M.; McIntyre, H.D.; Cruickshank, J.K.; McCance, D.R.; Dyer, A.R.; Metzger, B.E.; Lowe, L.P.; Trimble, E.R.; Coustan, D.R.; Hadden, D.R.; et al. The hyperglycemia and adverse pregnancy outcome study: Associations of GDM and obesity with pregnancy outcomes. Diabetes Care 2012, 35, 780–786. [Google Scholar] [CrossRef]
- Lowe, W.L., Jr.; Scholtens, D.M.; Lowe, L.P.; Kuang, A.; Nodzenski, M.; Talbot, O.; Catalano, P.M.; Linder, B.; Brickman, W.J.; Clayton, P.; et al. Association of Gestational Diabetes with Maternal Disorders of Glucose Metabolism and Childhood Adiposity. JAMA 2018, 320, 1005–1016. [Google Scholar] [CrossRef]
- Jacob, S.; Nodzenski, M.; Reisetter, A.C.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Ilkayeva, O.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; et al. Targeted Metabolomics Demonstrates Distinct and Overlapping Maternal Metabolites Associated With BMI, Glucose, and Insulin Sensitivity During Pregnancy Across Four Ancestry Groups. Diabetes Care 2017, 40, 911–919. [Google Scholar] [CrossRef]
- Kadakia, R.; Nodzenski, M.; Talbot, O.; Kuang, A.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Ilkayeva, O.R.; O’Neal, S.K.; Lowe, L.P.; et al. Maternal metabolites during pregnancy are associated with newborn outcomes and hyperinsulinaemia across ancestries. Diabetologia 2019, 62, 473–484. [Google Scholar] [CrossRef]
- The Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study. Int. J. Gynaecol. Obstet. 2002, 78, 69–77. [CrossRef]
- Metzger, B.E.; Lowe, L.P.; Dyer, A.R.; Trimble, E.R.; Chaovarindr, U.; Coustan, D.R.; Hadden, D.R.; McCance, D.R.; Hod, M.; McIntyre, H.D.; et al. Hyperglycemia and adverse pregnancy outcomes. N. Engl. J. Med. 2008, 358, 1991–2002. [Google Scholar]
- Scholtens, D.M.; Bain, J.R.; Reisetter, A.C.; Muehlbauer, M.J.; Nodzenski, M.; Stevens, R.D.; Ilkayeva, O.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; et al. Metabolic Networks and Metabolites Underlie Associations Between Maternal Glucose during Pregnancy and Newborn Size at Birth. Diabetes 2016, 65, 2039–2050. [Google Scholar] [CrossRef]
- Scholtens, D.M.; Muehlbauer, M.J.; Daya, N.R.; Stevens, R.D.; Dyer, A.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; Bain, J.R.; Lowe, W.L., Jr.; et al. Metabolomics reveals broad-scale metabolic perturbations in hyperglycemic mothers during pregnancy. Diabetes Care 2014, 37, 158–166. [Google Scholar] [CrossRef]
- Nodzenski, M.; Muehlbauer, M.J.; Bain, J.R.; Reisetter, A.C.; Lowe, W.L., Jr.; Scholtens, D.M. Metabomxtr: An R package for mixture-model analysis of non-targeted metabolomics data. Bioinformatics 2014, 30, 3287–3288. [Google Scholar] [CrossRef]
- Hayes, M.G.; Urbanek, M.; Hivert, M.F.; Armstrong, L.L.; Morrison, J.; Guo, C.; Lowe, L.P.; Scheftner, D.A.; Pluzhnikov, A.; Levine, D.M.; et al. Identification of HKDC1 and BACE2 as Genes Influencing Glycemic Traits During Pregnancy through Genome-Wide Association Studies. Diabetes 2013, 62, 3282–3291. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 2008, 9, 432–441. [Google Scholar] [CrossRef]
- Zhao, T.; Liu, H.; Roeder, K.; Lafferty, J.; Wasserman, L. The huge Package for High-dimensional Undirected Graph Estimation in R. J. Mach. Learn. Res. 2012, 13, 1059–1062. [Google Scholar]
- Reichardt, J.; Bornholdt, S. Statistical mechanics of community detection. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2006, 74, 016110. [Google Scholar] [CrossRef]
- Wahab, R.J.; Jaddoe, V.W.V.; Voerman, E.; Ruijter, G.J.G.; Felix, J.F.; Marchioro, L.; Uhl, O.; Shokry, E.; Koletzko, B.; Gaillard, R. Maternal Body Mass Index, Early-Pregnancy Metabolite Profile, and Birthweight. J. Clin. Endocrinol. Metab. 2022, 107, e315–e327. [Google Scholar] [CrossRef]
- Chia, A.R.; de Seymour, J.V.; Wong, G.; Sulek, K.; Han, T.L.; McKenzie, E.J.; Aris, I.M.; Godfrey, K.M.; Yap, F.; Tan, K.H.; et al. Maternal plasma metabolic markers of neonatal adiposity and associated maternal characteristics: The GUSTO study. Sci. Rep. 2020, 10, 9422. [Google Scholar] [CrossRef]
- Villar, J.; Ochieng, R.; Gunier, R.B.; Papageorghiou, A.T.; Rauch, S.; McGready, R.; Gauglitz, J.M.; Barros, F.C.; Vatish, M.; Fernandes, M.; et al. Association between fetal abdominal growth trajectories, maternal metabolite signatures early in pregnancy, and childhood growth and adiposity: Prospective observational multinational INTERBIO-21st fetal study. Lancet Diabetes Endocrinol. 2022, 10, 710–719. [Google Scholar] [CrossRef]
- Di Cianni, G.; Miccoli, R.; Volpe, L.; Lencioni, C.; Ghio, A.; Giovannitti, M.G.; Cuccuru, I.; Pellegrini, G.; Chatzianagnostou, K.; Boldrini, A.; et al. Maternal triglyceride levels and newborn weight in pregnant women with normal glucose tolerance. Diabet. Med. 2005, 22, 21–25. [Google Scholar] [CrossRef]
- Hashemipour, S.; Haji Seidjavadi, E.; Maleki, F.; Esmailzadehha, N.; Movahed, F.; Yazdi, Z. Level of maternal triglycerides is a predictor of fetal macrosomia in non-obese pregnant women with gestational diabetes mellitus. Pediatr. Neonatol. 2018, 59, 567–572. [Google Scholar] [CrossRef]
- Samsuddin, S.; Arumugam, P.A.; Md Amin, M.S.; Yahya, A.; Musa, N.; Lim, L.L.; Paramasivam, S.S.; Ratnasingam, J.; Ibrahim, L.; Chooi, K.C.; et al. Maternal lipids are associated with newborn adiposity, independent of GDM status, obesity and insulin resistance: A prospective observational cohort study. BJOG 2020, 127, 490–499. [Google Scholar] [CrossRef]
- Xi, F.; Chen, H.; Chen, Q.; Chen, D.; Chen, Y.; Sagnelli, M.; Chen, G.; Zhao, B.; Luo, Q. Second-trimester and third-trimester maternal lipid profiles significantly correlated to LGA and macrosomia. Arch. Gynecol. Obstet. 2021, 304, 885–894. [Google Scholar] [CrossRef]
- Barbour, L.A.; Farabi, S.S.; Friedman, J.E.; Hirsch, N.M.; Reece, M.S.; Van Pelt, R.E.; Hernandez, T.L. Postprandial Triglycerides Predict Newborn Fat More Strongly than Glucose in Women with Obesity in Early Pregnancy. Obesity (Silver Spring) 2018, 26, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Jousse, C.; Bruhat, A.; Ferrara, M.; Fafournoux, P. Physiological concentration of amino acids regulates insulin-like-growth-factor-binding protein 1 expression. Biochem. J. 1998, 334, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.S. The Emerging Role of Branched-Chain Amino Acids in Insulin Resistance and Metabolism. Nutrients 2016, 8, 405. [Google Scholar] [CrossRef]
- Yoon, M.S. The Role of Mammalian Target of Rapamycin (mTOR) in Insulin Signaling. Nutrients 2017, 9, 1176. [Google Scholar] [CrossRef] [PubMed]
- Muta, K.; Morgan, D.A.; Rahmouni, K. The role of hypothalamic mTORC1 signaling in insulin regulation of food intake, body weight, and sympathetic nerve activity in male mice. Endocrinology 2015, 156, 1398–1407. [Google Scholar] [CrossRef]
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011, 12, 21–35. [Google Scholar] [CrossRef]
- Liu, Z.; Jeppesen, P.B.; Gregersen, S.; Bach Larsen, L.; Hermansen, K. Chronic Exposure to Proline Causes Aminoacidotoxicity and Impaired Beta-Cell Function: Studies In Vitro. Rev. Diabet. Stud. 2016, 13, 66–78. [Google Scholar] [CrossRef]
- Wu, G.; Bazer, F.W.; Datta, S.; Johnson, G.A.; Li, P.; Satterfield, M.C.; Spencer, T.E. Proline metabolism in the conceptus: Implications for fetal growth and development. Amino Acids 2008, 35, 691–702. [Google Scholar] [CrossRef]
- Hussain, T.; Tan, B.; Ren, W.; Rahu, N.; Kalhoro, D.H.; Yin, Y. Exploring polyamines: Functions in embryo/fetal development. Anim. Nutr. 2017, 3, 7–10. [Google Scholar] [CrossRef]
- Ryckman, K.K.; Berberich, S.L.; Dagle, J.M. Predicting gestational age using neonatal metabolic markers. Am. J. Obstet. Gynecol. 2016, 214, 515.e1–515.e13. [Google Scholar] [CrossRef]
- Song, Y.; Lyu, C.; Li, M.; Rahman, M.L.; Chen, Z.; Zhu, Y.; Hinkle, S.N.; Chen, L.; Mitro, S.D.; Li, L.J.; et al. Plasma Acylcarnitines during Pregnancy and Neonatal Anthropometry: A Longitudinal Study in a Multiracial Cohort. Metabolites 2021, 11, 885. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E. Alternative biomarkers for assessing glycemic control in diabetes: Fructosamine, glycated albumin, and 1,5-anhydroglucitol. Ann. Pediatr. Endocrinol. Metab. 2015, 20, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Saglam, B.; Uysal, S.; Sozdinler, S.; Dogan, O.E.; Onvural, B. Diagnostic value of glycemic markers HbA1c, 1,5-anhydroglucitol and glycated albumin in evaluating gestational diabetes mellitus. Ther. Adv. Endocrinol. Metab. 2017, 8, 161–167. [Google Scholar] [CrossRef]
- Crowther, C.A.; Hiller, J.E.; Moss, J.R.; McPhee, A.J.; Jeffries, W.S.; Robinson, J.S.; Australian Carbohydrate Intolerance Study in Pregnant Women Trial Group. Effect of treatment of gestational diabetes mellitus on pregnancy outcomes. N. Engl. J. Med. 2005, 352, 2477–2486. [Google Scholar] [CrossRef]
- Hartling, L.; Dryden, D.M.; Guthrie, A.; Muise, M.; Vandermeer, B.; Donovan, L. Benefits and harms of treating gestational diabetes mellitus: A systematic review and meta-analysis for the U.S. Preventive Services Task Force and the National Institutes of Health Office of Medical Applications of Research. Ann. Intern. Med. 2013, 159, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Landon, M.B. Is there a benefit to the treatment of mild gestational diabetes mellitus? Am. J. Obstet. Gynecol. 2010, 202, 649–653. [Google Scholar] [CrossRef]
- Landon, M.B.; Spong, C.Y.; Thom, E.; Carpenter, M.W.; Ramin, S.M.; Casey, B.; Wapner, R.J.; Varner, M.W.; Rouse, D.J.; Thorp, J.M., Jr.; et al. A multicenter, randomized trial of treatment for mild gestational diabetes. N. Engl. J. Med. 2009, 361, 1339–1348. [Google Scholar] [CrossRef]
- Vaarasmaki, M. Is it worth treating gestational diabetes: If so, when and how? Diabetologia 2016, 59, 1391–1395. [Google Scholar] [CrossRef]
- White, S.L.; Begum, S.; Vieira, M.C.; Seed, P.; Lawlor, D.L.; Sattar, N.; Nelson, S.M.; Welsh, P.; Pasupathy, D.; Poston, L.; et al. Metabolic phenotyping by treatment modality in obese women with gestational diabetes suggests diverse pathophysiology: An exploratory study. PLoS ONE 2020, 15, e0230658. [Google Scholar] [CrossRef]
- Mokkala, K.; Vahlberg, T.; Houttu, N.; Koivuniemi, E.; Laitinen, K. Distinct Metabolomic Profile Because of Gestational Diabetes and its Treatment Mode in Women with Overweight and Obesity. Obesity (Silver Spring) 2020, 28, 1637–1644. [Google Scholar] [CrossRef]
- Chen, L.W.; Tint, M.T.; Fortier, M.V.; Aris, I.M.; Shek, L.P.; Tan, K.H.; Chan, S.Y.; Gluckman, P.D.; Chong, Y.S.; Godfrey, K.M.; et al. Which anthropometric measures best reflect neonatal adiposity? Int. J. Obes. (Lond.) 2018, 42, 501–506. [Google Scholar] [CrossRef]
- Rodriguez-Cano, A.M.; Mier-Cabrera, J.; Munoz-Manrique, C.; Cardona-Perez, A.; Villalobos-Alcazar, G.; Perichart-Perera, O. Anthropometric and clinical correlates of fat mass in healthy term infants at 6 months of age. BMC Pediatr. 2019, 19, 60. [Google Scholar] [CrossRef]
- Liang, L.; Rasmussen, M.H.; Piening, B.; Shen, X.; Chen, S.; Rost, H.; Snyder, J.K.; Tibshirani, R.; Skotte, L.; Lee, N.C.; et al. Metabolic Dynamics and Prediction of Gestational Age and Time to Delivery in Pregnant Women. Cell 2020, 181, 1680–1692.e15. [Google Scholar] [CrossRef]

| Maternal Participants (n = 2337) | |
|---|---|
| Black | 663 (28.4%) |
| East Asian | 436 (18.7%) |
| Hispanic | 53 (2.3%) |
| South Asian | 629 (26.9%) |
| White | 556 (23.8%) |
| Maternal Characteristics | |
| Age at OGTT, years | 29.2 (5.8) |
| Height, cm | 161.0 (7.1) |
| BMI at OGTT, kg/m2 | 27.9 (5.1) |
| Mean arterial pressure, mmHg | 80.7 (7.8) |
| Fasting plasma glucose, mmol/L | 4.5 (0.4) |
| 1 h plasma glucose, mmol/L | 7.5 (1.7) |
| Gestational age at OGTT, weeks | 27.8 (1.8) |
| Gestational age at delivery, weeks | 39.8 (1.2) |
| Smoking (continued smoking in pregnancy) | 61 (2.6%) |
| Alcohol (continued alcohol consumption in pregnancy) | 103 (4.4%) |
| Parity (nulliparous) | 1245 (53.3%) |
| Newborn Participants (n = 2337) | |
| Black | 559 (23.9%) |
| East Asian | 654 (28.0%) |
| Hispanic | 451 (19.3%) |
| South Asian | 50 (2.1%) |
| White | 623 (26.7%) |
| Newborn Characteristics | |
| Birthweight, g | 3391.5 (485.0) |
| SSF | 12.5 (2.7) |
| Cord blood C-peptide, nmol/L | 0.3 (0.2) |
| Male sex | 1175 (50.3%) |
| Female sex | 1162 (49.7%) |
| Model 1 Beta (CI, p) | Model 2 Beta (CI, p) | |
|---|---|---|
| Birthweight | ||
| Fasting metabolites | ||
| Arginine | −12.92 (−31.86–6.03, 0.35) | −33.07 (−51.80–14.35, 1.18 × 10−2) |
| Triglycerides | 52.13 (34.53–69.73, 4.81 × 10−7) * | 44.80 (27.58–62.03, 2.43 × 10−5) |
| Alanine | 30.31 (12.91–47.70, 1.86 × 10−2) | 25.26 (7.97–42.55, 3.06 × 10−2) |
| Methionine | 22.07 (4.96–39.18, 8.13 × 10−2) | 24.51 (7.86–41.17, 3.06 × 10−2) |
| Pyruvate | 32.57 (17.77–47.37, 8.90 × 10−4) | 25.16 (10.64–39.67, 1.84 × 10−2) |
| AC C2 | −20.61 (−38.00–−3.22, 0.11) | −26.21 (−43.22–−9.20, 2.37 × 10−2) |
| AC C8:1 | −10.74 (−28.18–6.69, 0.39) | −28.54 (−45.81–−11.27, 1.58 × 10−2) |
| AC C8:1-OH/C6:1-DC | −21.06 (−38.84–−3.29, 0.11) | −24.14 (−41.45–−6.82, 4.12 × 10−2) |
| AC C10:1 | −22.05 (−39.08–−5.02, 8.13 × 10−2) | −26.973 (−43.58–−10.36, 1.60 × 10−2) |
| AC C12:1 | −18.87 (−36.12–−1.63, 0.12) | −23.01 (−39.93–−6.10, 4.57 × 10−2) |
| AC C14:2 | −29.05 (−46.12–−11.99, 1.86 × 10−2 | −31.70 (−48.42–−14.98, 6.76 × 10−3) |
| AC C18:2 | −28.45 (−46.14–−10.76, 2.67 × 10−2) | −29.35 (−46.65–−12.06, 1.45 × 10−2) |
| 1,5-Anhydroglucitol | −33.31 (−49.46–−17.17, 1.43 × 10−3) | −33.06 (−48.88–−17.23, 2.32 × 10−3) |
| One-hour metabolites | ||
| Leucine/Isoleucine | 38.92 (21.57–56.26, 1.88 × 10−4) | 20.33 (3.10–37.55, 8.13 × 10−2) |
| Valine | 34.77 (17.04–52.50, 1.16 × 10−3) | 17.36 (−0.178–34.90, 0.13) |
| Triglycerides | 59.48 (41.75–77.20, 3.93 × 10−9) | 49.66 (32.270–67.06, 1.61 × 10−6) |
| Alanine | 42.74 (25.18–60.30, 6.35 × 10−5) | 39.81 (22.68–56.94, 1.79 × 10−4) |
| Glutamine/Glutamic acid | 33.36 (15.40–51.33, 2.27 × 10−3) | 11.24 (−6.69–29.17, 0.36) |
| Methionine | 35.44 (18.39–52.48, 5.20 × 10−4) | 29.51 (12.90–46.11, 8.21 × 10−3) |
| Proline | 36.23 (19.17–53.30, 4.28 × 10−4) | 28.53 (11.89–45.18, 1.03 × 10−2) |
| Hydroxyprolines | 21.47 (9.88–33.07, 5.23 × 10−3) | 16.22 (4.91–27.53, 5.40 × 10−2) |
| Threonine | 23.25 (11.70–34.80, 2.64 × 10−3) | 19.44 (8.19–30.68, 1.93 × 10−2) |
| Tyrosine | 30.36 (13.20–47.51, 3.86 × 10−3) | 14.23 (−2.73–31.18, 0.20) |
| Glycine | 11.89 (−5.77–29.56, 0.46) | 25.29 (8.02–42.56, 3.85 × 10−2) |
| Pyruvate | 22.56 (9.52–35.60, 8.06 × 10−3) | 13.52 (0.73–26.31, 0.30) |
| AC C2 | −4.05 (−21.81–13.71, 0.85) | −27.04 (−44.66–9.43, 2.87 × 10−2) |
| AC C6 | 29.21 (10.17–48.25, 1.59 × 10−2) | 11.72 (−7.01–30.46, 0.36) |
| AC C8:1 | −11.02 (−28.43–6.39, 0.50) | −35.39 (−52.68–−18.09, 1.36 × 10−3) |
| Lactate | 30.03 (11.80–48.26, 8.22 × 10−3) | 23.08 (5.11–41.06, 7.04 × 10−2) |
| 3-Hydroxybutyrate | 39.80 (22.19–57.40, 1.88 × 10−4) | 3.83 (−14.60–22.25, 0.71) |
| Glucose and other aldohexoses | 27.01 (11.33–42.68, 8.06 × 10−3) | −0.56 (−16.90–15.79, 0.95) |
| 6-Deoxyhexose | 17.07 (5.98–28.17, 2.33 × 10−2) | 16.67 (5.89–27.45, 4.46 × 10−2) |
| 3-Carboxy-4-methyl-5-propyl-2-furanpropanoic acid | −16.99 (−28.95–−5.03, 4.04 × 10−2) | −16.65 (−28.26–−5.04, 5.40 × 10−2) |
| 1,5-Anhydroglucitol | −31.51 (−47.33–−15.69, 2.64 × 10−3) | −28.51 (−44.07–−12.95, 1.82 × 10−2) |
| Palmitoleic acid | 11.63 (3.35–19.92, 4.04 × 10−2) | 3.96 (−4.23–12.15, 0.71) |
| Sum of Skinfolds | ||
| Fasting metabolites | ||
| Leucine/Isoleucine | 0.15 (0.05–0.26, 4.09 × 10−2) | 0.09 (−0.01–0.19, 0.25) |
| Valine | 0.21 (0.10–0.32, 4.26 × 10−3) | 0.12 (0.01–0.22, 0.16) |
| Triglycerides | 0.25 (0.15–0.36, 1.70 × 10−4) | 0.21 (0.11–0.31, 4.84 × 10−3) |
| Alanine | 0.15 (0.05–0.25, 4.09 × 10−2) | 0.10 (−0.01–0.20, 0.23) |
| Proline | 0.16 (0.06–0.26, 3.57 × 10−2) | 0.09 (−0.02–0.19, 0.28) |
| Glycine | −0.15 (−0.26–−0.05, 4.09 × 10−2) | −0.11 (−0.21–−0.00, 0.17) |
| Pyruvate | 0.20 (0.11–0.29, 5.39 × 10−4) | 0.16 (0.07–0.25, 2.05 × 10−2) |
| AC C5:1 | 0.18 (0.07–0.28, 1.68 × 10−2) | 0.17 (0.07–0.27, 2.33 × 10−2) |
| AC C14:2 | −0.16 (−0.26–−0.06, 3.45 × 10−2) | −0.17 (−0.26–−0.07, 2.74 × 10−2) |
| 1,5-Anhydroglucitol | −0.17 (−0.26–−0.07, 1.72 × 10−2) | −0.16 (−0.25–−0.06, 2.70 × 10−2) |
| One-hour metabolites | ||
| Leucine/Isoleucine | 0.23 (0.12–0.34, 2.72 × 10−4) | 0.14 (0.04–0.24, 4.73 × 10−2) |
| Valine | 0.25 (0.14–0.36, 1.41 × 10−4) | 0.17 (0.06–0.27, 1.85 × 10−2) |
| Triglycerides | 0.29 (0.18–0.39, 1.32 × 10−5) | 0.23 (0.12–0.33, 1.68 × 10−3) |
| Alanine | 0.19 (0.09–0.30, 5.10 × 10−3) | 0.17 (0.07–0.28, 1.85 × 10−2) |
| Proline | 0.21 (0.10–0.31, 1.81 × 10−3) | 0.16 (0.06–0.26, 1.85 × 10−2) |
| Hydroxyprolines | 0.15 (0.08–0.21, 1.92 × 10−3) | 0.12 (0.05–0.19, 1.21 × 10−2) |
| Creatinine | 0.10 (0.03–0.17, 3.57 × 10−2) | 0.08 (0.02–0.15, 0.15) |
| Threonine | 0.14 (0.07–0.21, 2.28 × 10−3) | 0.12 (0.05–0.19, 1.21 × 10−2) |
| Glycolic acid | −0.07 (−0.11–−0.02, 4.87 × 10−2) | −0.07 (−0.11–−0.02, 5.03 × 10−2) |
| Pyruvate | 0.15 (0.07–0.23, 5.68 × 10−3) | 0.10 (0.02–0.18, 0.12) |
| AC C5:1 | 0.16 (0.06–0.27, 2.13 × 10−2) | 0.16 (0.06–0.26, 1.85e × 10−2) |
| AC C2 | −0.02 (−0.13–0.08, 0.80) | −0.14 (−0.25–−0.04, 4.73 × 10−2) |
| AC C10-OH/C8-DC | −0.07 (−0.17–0.04, 0.55) | −0.16 (−0.27–−0.06, 1.85 × 10−2) |
| AC C14:1 | −0.04 (−0.14–0.07, 0.77) | −0.18 (−0.28–−0.08, 1.85 × 10−2) |
| AC C14:1-OH | −0.10 (−0.21–−0.00, 0.21) | −0.15 (−0.25–−0.05, 2.55 × 10−2) |
| AC C14:2 | −0.04 (−0.15–0.06, 0.77) | −0.17 (−0.27–−0.06, 1.85 × 10−2) |
| AC C16:1 | −0.03 (−0.14–0.07, 0.77) | −0.16 (−0.26–−0.05, 2.11 × 10−2) |
| AC C20-OH/C18-DC | −0.12 (−0.22–0.01, 1.69 × 10−1) | −0.14 (−0.24–−0.03, 4.86 × 10−2) |
| Lactate | 0.17 (0.06–0.28, 2.13 × 10−2) | 0.13 (0.02–0.23, 9.99 × 10−2) |
| 3-Hydroxybutyrate | 0.17 (0.06–0.27, 2.13 × 10−2) | −0.03 (−0.14–0.09, 0.72) |
| Glucose and other aldohexoses | 0.15 (0.05–0.24, 2.54 × 10−2) | 0.00 (−0.01–0.10, 0.99) |
| 1,5-Anhydroglucitol | −0.17 (−0.27–−0.08, 5.68 × 10−3) | −0.15 (−0.25–−0.06, 2.55 × 10−2) |
| Cord C-Peptide | ||
| Fasting metabolites | ||
| Arginine | 0.04 (0.02–0.07, 2.67 × 10−2) | 0.019 (−0.01–0.04, 0.64) |
| Leucine/Isoleucine | 0.04 (0.02–0.07, 6.96 × 10−3) | 0.029 (0.01–0.05, 0.14) |
| Valine | 0.06 (0.03–0.08, 1.67 × 10−4) | 0.04 (0.01–0.06, 0.11) |
| Triglycerides | 0.05 (0.02–0.07, 4.28 × 10−3) | 0.04 (0.01–0.06, 0.11) |
| AC C5 | 0.04 (0.01–0.06, 2.67 × 10−2) | 0.02 (0.00–0.05, 0.33) |
| AC C5-OH/C3-DC | 0.04 (0.02–0.07, 7.94 × 10−3) | 0.03 (0.01–0.06, 0.11) |
| One-hour metabolites | ||
| Arginine | 0.04 (0.01–0.06, 4.26 × 10−2) | 0.02 (−0.00–0.05, 0.47) |
| Leucine/Isoleucine | 0.05 (0.03–0.08, 3.49 × 10−4) | 0.04 (0.01–0.06, 6.34 × 10−2) |
| Valine | 0.06 (0.03–0.08, 1.92 × 10−4) | 0.04 (0.02–0.07, 2.31 × 10−2) |
| Triglycerides | 0.06 (0.03–0.08, 1.92 × 10−4) | 0.05 (0.02–0.07, 1.86 × 10−2) |
| 3-Hydroxybutyrate | 0.04 (0.02–0.07, 1.01 × 10−2) | 0.01 (−0.02–0.03, 0.85) |
| Glucose and other aldohexoses | 0.05 (0.03–0.07, 1.41 × 10−4) | 0.03 (0.00–0.05, 0.56) |
| 2-Ketoleucine/ketoisoleucine | 0.03 (0.02–0.04, 8.03 × 10−4) | 0.03 (0.01–0.04, 1.17 × 10−2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gleason, B.; Kuang, A.; Bain, J.R.; Muehlbauer, M.J.; Ilkayeva, O.R.; Scholtens, D.M.; Lowe, W.L., Jr. Association of Maternal Metabolites and Metabolite Networks with Newborn Outcomes in a Multi-Ancestry Cohort. Metabolites 2023, 13, 505. https://doi.org/10.3390/metabo13040505
Gleason B, Kuang A, Bain JR, Muehlbauer MJ, Ilkayeva OR, Scholtens DM, Lowe WL Jr. Association of Maternal Metabolites and Metabolite Networks with Newborn Outcomes in a Multi-Ancestry Cohort. Metabolites. 2023; 13(4):505. https://doi.org/10.3390/metabo13040505
Chicago/Turabian StyleGleason, Brooke, Alan Kuang, James R. Bain, Michael J. Muehlbauer, Olga R. Ilkayeva, Denise M. Scholtens, and William L. Lowe, Jr. 2023. "Association of Maternal Metabolites and Metabolite Networks with Newborn Outcomes in a Multi-Ancestry Cohort" Metabolites 13, no. 4: 505. https://doi.org/10.3390/metabo13040505
APA StyleGleason, B., Kuang, A., Bain, J. R., Muehlbauer, M. J., Ilkayeva, O. R., Scholtens, D. M., & Lowe, W. L., Jr. (2023). Association of Maternal Metabolites and Metabolite Networks with Newborn Outcomes in a Multi-Ancestry Cohort. Metabolites, 13(4), 505. https://doi.org/10.3390/metabo13040505







