Feature-Based Molecular Networking Facilitates the Comprehensive Identification of Differential Metabolites in Diabetic Cognitive Dysfunction Rats
Abstract
:1. Introduction
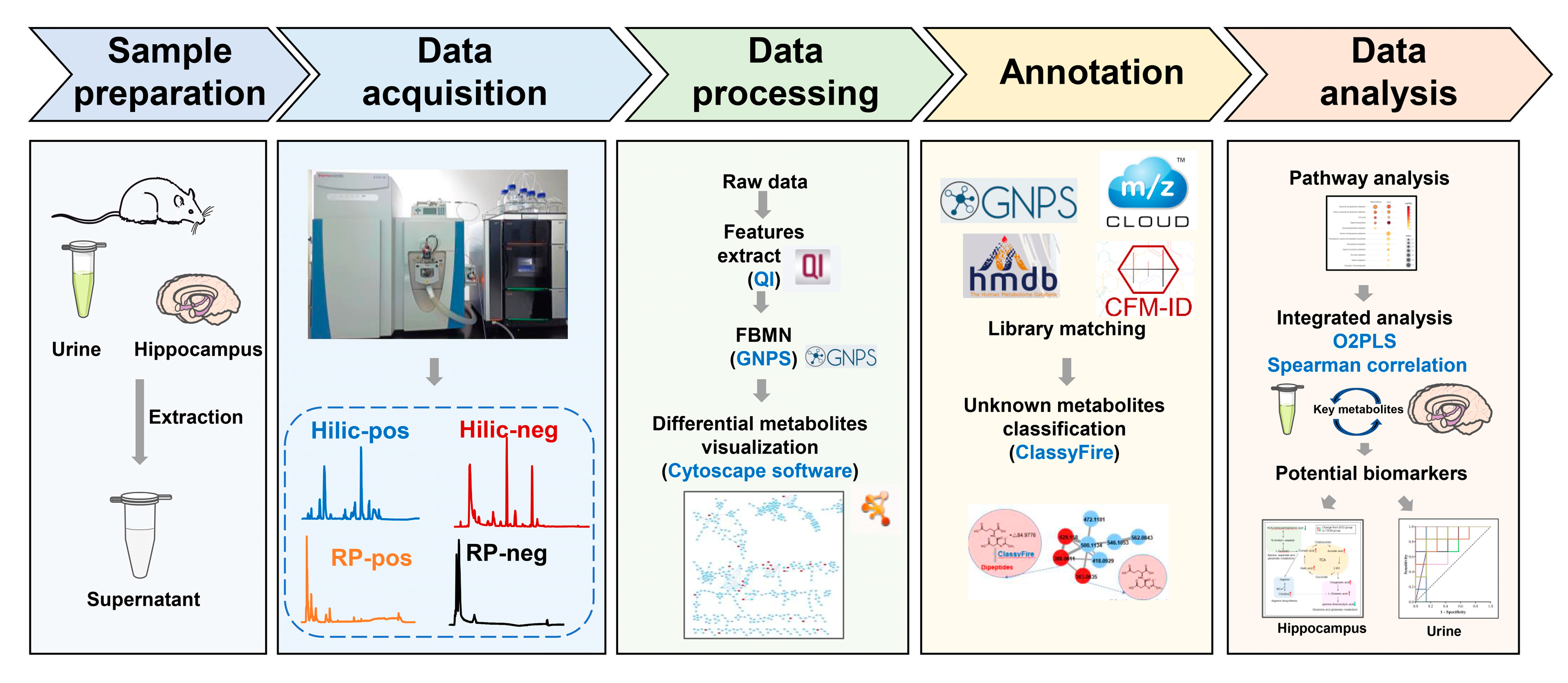
2. Materials and Methods
2.1. Reagents and Materials
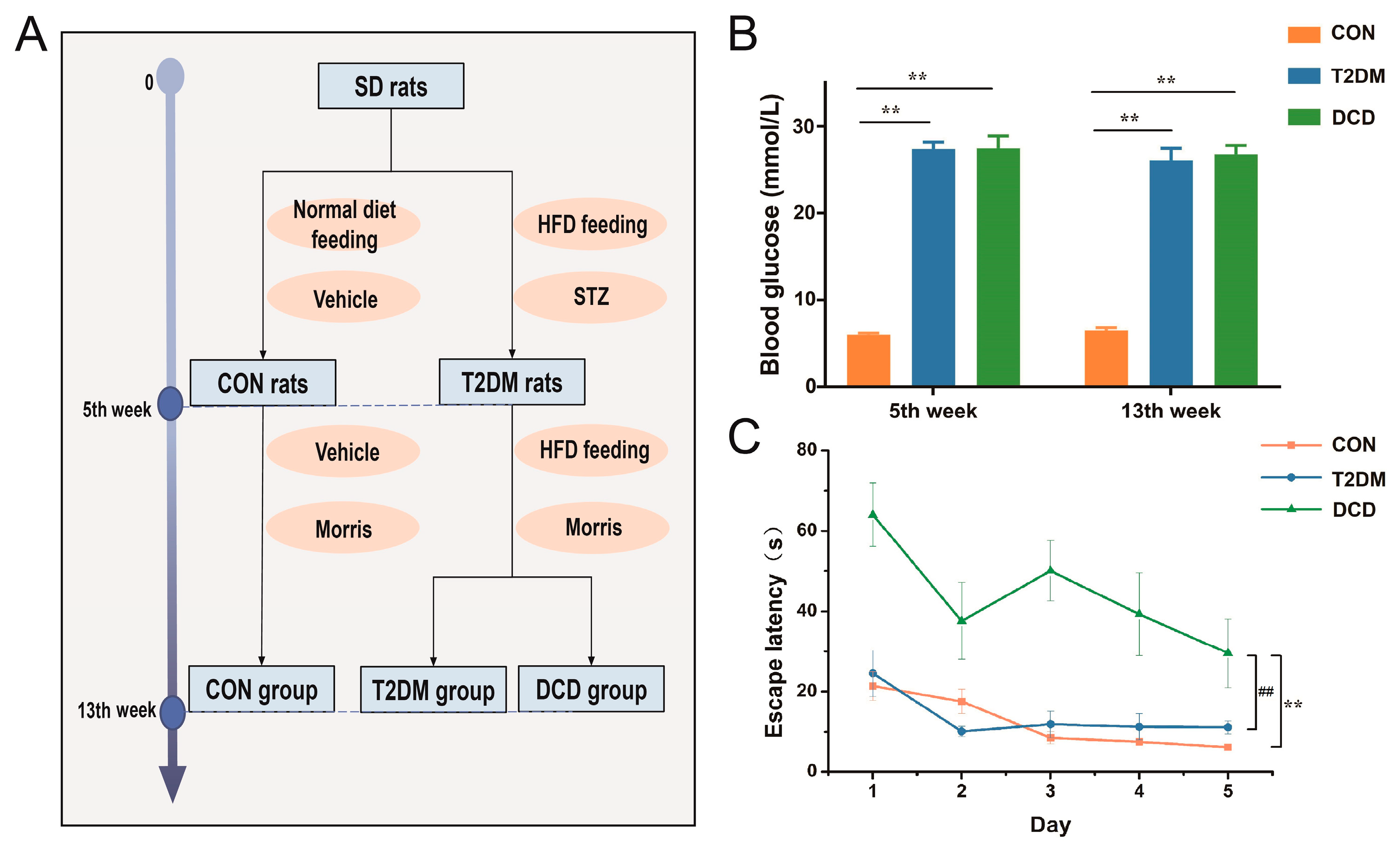
2.2. Animal Treatment
2.3. Morris Water Maze Test
2.4. Sample Preparation
2.4.1. Hippocampus Sample Preparation
2.4.2. Urine Sample Preparation
2.5. Data Acquisition for Untargeted Metabolomics Profiling
2.5.1. Chromatographic Conditions for the hippocampus Samples
2.5.2. Chromatographic Conditions of the Urine Samples
2.5.3. MS Conditions
2.6. Metabolomics Data Analysis
3. Results and Discussion
3.1. Establishment of the DCD Rat Model
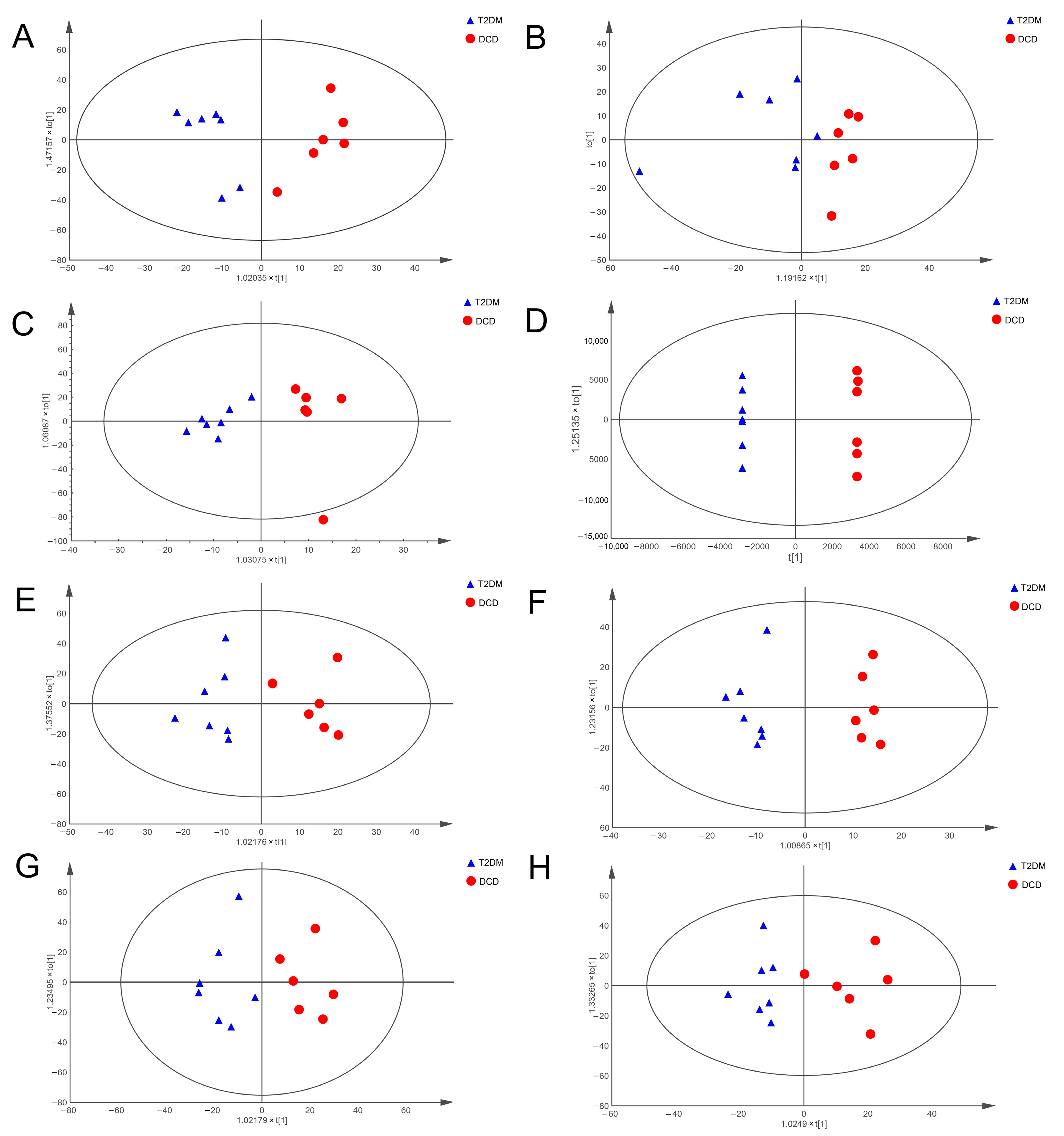
3.2. Altered Metabolic Profiles of Hippocampal Tissue and Urine in Rats with DCD
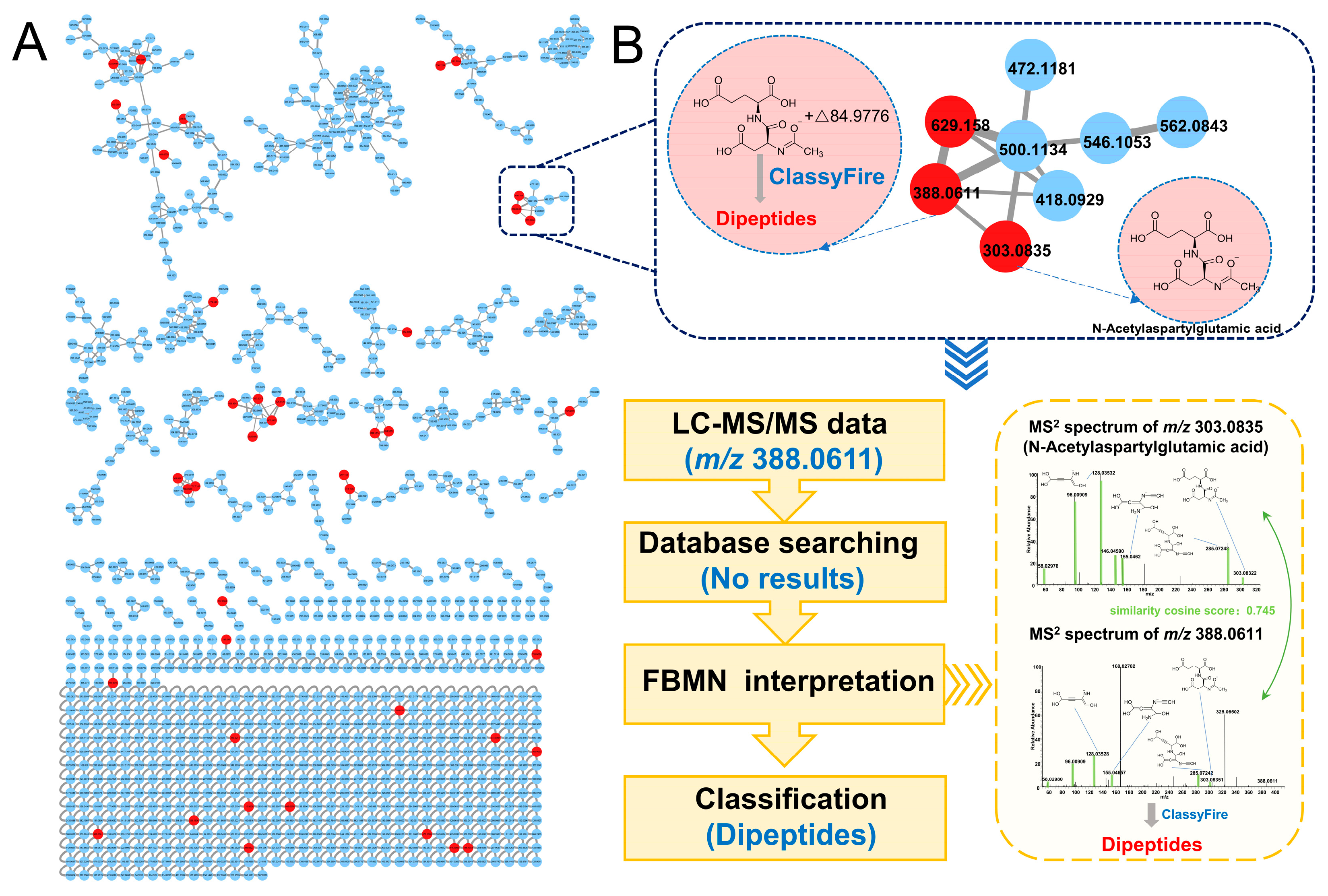
3.3. Identification of Differential Metabolites Based on FBMN
3.4. Enrichment of the Metabolic Pathways
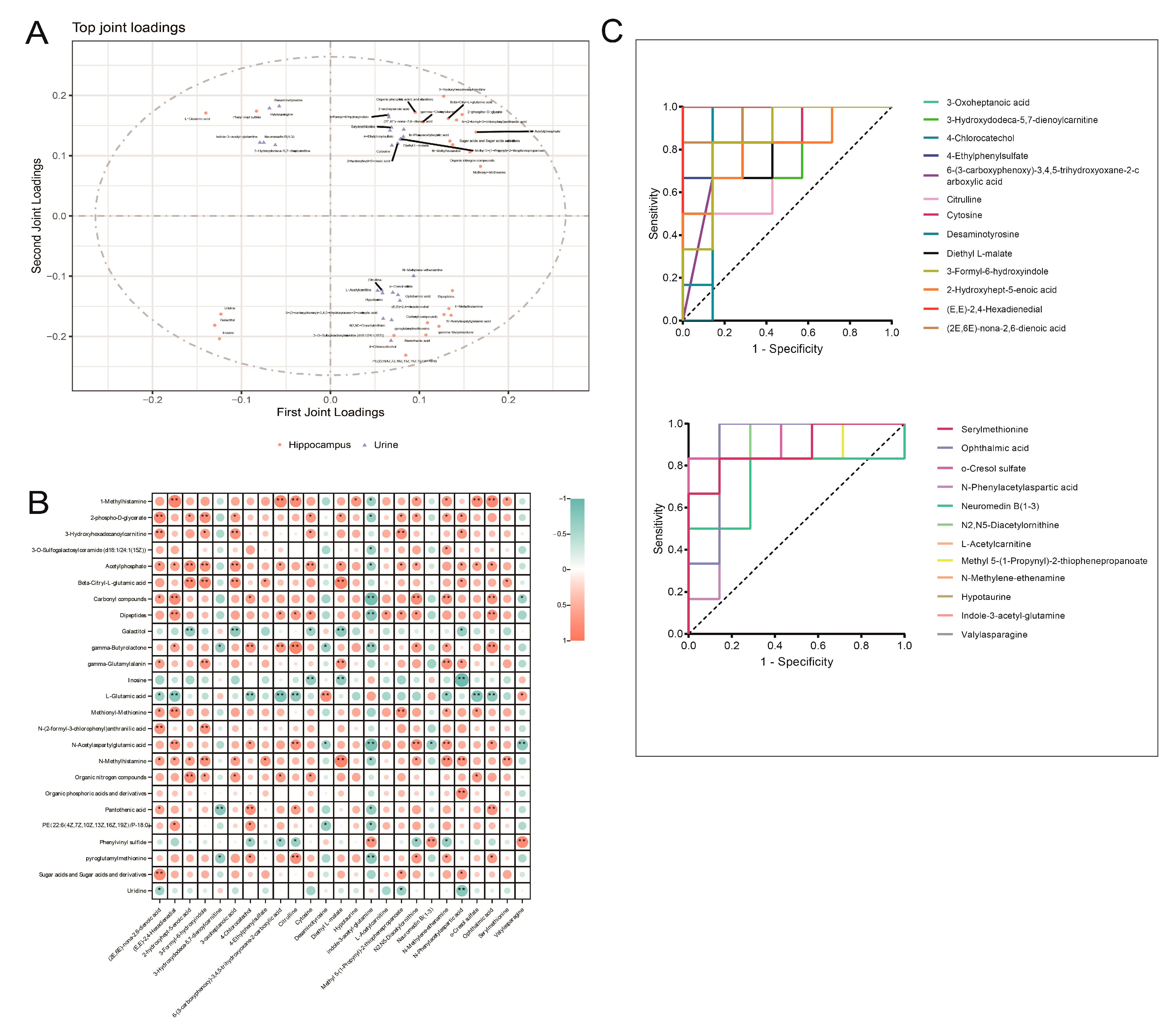
3.5. Integrated Analysis of Key Differential Metabolites in the Hippocampus and Urine of Rats with DCD
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. On behalf of the American Diabetes, A. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes-2023. Diabetes Care 2023, 46, S19–S40. [Google Scholar] [CrossRef]
- Kullmann, S.; Kleinridders, A.; Small, D.M.; Fritsche, A.; Häring, H.-U.; Preissl, H.; Heni, M. Central nervous pathways of insulin action in the control of metabolism and food intake. Lancet Diabetes Endocrinol. 2020, 8, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, V.; Sinclair, A.J.; Hill-Briggs, F.; Moran, C.; Biessels, G.J. Type 2 diabetes and cognitive dysfunction—Towards effective management of both comorbidities. Lancet Diabetes Endocrinol. 2020, 8, 535–545. [Google Scholar] [CrossRef]
- Biessels, G.J.; Nobili, F.; Teunissen, C.E.; Simó, R.; Scheltens, P. Understanding multifactorial brain changes in type 2 diabetes: A biomarker perspective. Lancet Neurol. 2020, 19, 699–710. [Google Scholar] [CrossRef]
- Pelimanni, E.; Jehkonen, M. Type 2 Diabetes and Cognitive Functions in Middle Age: A Meta-Analysis. J. Int. Neuropsychol. Soc. 2019, 25, 215–229. [Google Scholar] [CrossRef] [Green Version]
- Palta, P.; Schneider, A.L.; Biessels, G.J.; Touradji, P.; Hill-Briggs, F. Magnitude of cognitive dysfunction in adults with type 2 diabetes: A meta-analysis of six cognitive domains and the most frequently reported neuropsychological tests within domains. J. Int. Neuropsychol. Soc. 2014, 20, 278–291. [Google Scholar] [CrossRef] [Green Version]
- Morze, J.; Wittenbecher, C.; Schwingshackl, L.; Danielewicz, A.; Rynkiewicz, A.; Hu, F.B.; Guasch-Ferré, M. Metabolomics and Type 2 Diabetes Risk: An Updated Systematic Review and Meta-analysis of Prospective Cohort Studies. Diabetes Care 2022, 45, 1013–1024. [Google Scholar] [CrossRef]
- Liu, Z.; Dai, X.; Zhang, H.; Shi, R.; Hui, Y.; Jin, X.; Zhang, W.; Wang, L.; Wang, Q.; Wang, D.; et al. Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment. Nat. Commun. 2020, 11, 855. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Zhuang, P.; Lin, M.; Kang, M.; Liu, H.; Zhang, Y.; Yang, Z.; Chen, Y.; Zhang, Y. Urine Metabonomics Reveals Early Biomarkers in Diabetic Cognitive Dysfunction. J. Proteome Res. 2017, 16, 3180–3189. [Google Scholar] [CrossRef]
- Xiong, F.; Gong, K.; Xu, H.; Tu, Y.; Lu, J.; Zhou, Y.; He, W.; Li, W.; Li, C.; Zhao, L.; et al. Optimized integration of metabolomics and lipidomics reveals brain region-specific changes of oxidative stress and neuroinflammation in type 1 diabetic mice with cognitive decline. J. Adv. Res. 2023, 43, 233–245. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Keshari, K.R. Metabolic analysis as a driver for discovery, diagnosis, and therapy. Cell 2022, 185, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Teruya, T.; Chen, Y.J.; Kondoh, H.; Fukuji, Y.; Yanagida, M. Whole-blood metabolomics of dementia patients reveal classes of disease-linked metabolites. Proc. Natl. Acad. Sci. USA 2021, 118, e2022857118. [Google Scholar] [PubMed]
- Chen, R.; Zeng, Y.; Xiao, W.; Zhang, L.; Shu, Y. LC-MS-Based Untargeted Metabolomics Reveals Early Biomarkers in STZ-Induced Diabetic Rats With Cognitive Impairment. Front. Endocrinol. 2021, 12, 665309. [Google Scholar] [CrossRef]
- Tang, D.Q.; Zou, L.; Yin, X.X.; Ong, C.N. HILIC-MS for metabolomics: An attractive and complementary approach to RPLC-MS. Mass Spectrom. Rev. 2016, 35, 574–600. [Google Scholar]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Phelan, V.V. Feature-Based Molecular Networking for Metabolite Annotation. Methods Mol. Biol. 2020, 2104, 227–243. [Google Scholar]
- Renai, L.; Ulaszewska, M.; Mattivi, F.; Bartoletti, R.; Del Bubba, M.; van der Hooft, J.J.J. Combining Feature-Based Molecular Networking and Contextual Mass Spectral Libraries to Decipher Nutrimetabolomics Profiles. Metabolites 2022, 12, 1005. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.F.; Nothias-Esposito, M.; da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J.; et al. Bioactivity-Based Molecular Networking for the Discovery of Drug Leads in Natural Product Bioassay-Guided Fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar]
- Kakumu, Y.; Thi Nguyen, M.T.; Mitsunaga, T. Molecular networking-based discovery of anti-inflammatory chromene dimers from Melicope pteleifolia. Phytochemistry 2022, 202, 113322. [Google Scholar] [CrossRef] [PubMed]
- Gomes, P.; Quirós-Guerrero, L.; Silva, C.; Pamplona, S.; Boutin, J.A.; Eberlin, M.; Wolfender, J.L.; Silva, M. Feature-Based Molecular Network-Guided Dereplication of Natural Bioactive Products from Leaves of Stryphnodendron pulcherrimum (Willd.) Hochr. Metabolites 2021, 11, 281. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Neto, F.C.; Raftery, D. Expanding Urinary Metabolite Annotation through Integrated Mass Spectral Similarity Networking. Anal. Chem. 2021, 93, 12001–12010. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jarmusch, A.K.; Vargas, F.; Aksenov, A.A.; Gauglitz, J.M.; Weldon, K.; Petras, D.; da Silva, R.; Quinn, R.; Melnik, A.V.; et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 2020, 38, 23–26. [Google Scholar] [CrossRef]
- Wozniak, J.M.; Mills, R.H.; Olson, J.; Caldera, J.R.; Sepich-Poore, G.D.; Carrillo-Terrazas, M.; Tsai, C.M.; Vargas, F.; Knight, R.; Dorrestein, P.C.; et al. Mortality Risk Profiling of Staphylococcus aureus Bacteremia by Multi-omic Serum Analysis Reveals Early Predictive and Pathogenic Signatures. Cell 2020, 182, 1311–1327.e1314. [Google Scholar] [CrossRef]
- Jones, S.K.; McCarthy, D.M.; Vied, C.; Stanwood, G.D.; Schatschneider, C.; Bhide, P.G. Transgenerational transmission of aspartame-induced anxiety and changes in glutamate-GABA signaling and gene expression in the amygdala. Proc. Natl. Acad. Sci. USA 2022, 119, e2213120119. [Google Scholar] [CrossRef]
- Li, S.; Stern, A.M. Bioactive human Alzheimer brain soluble Aβ: Pathophysiology and therapeutic opportunities. Mol. Psychiatry 2022, 27, 3182–3191. [Google Scholar] [CrossRef]
- Ge, Y.; Chen, W.; Axerio-Cilies, P.; Wang, Y.T. NMDARs in Cell Survival and Death: Implications in Stroke Pathogenesis and Treatment. Trends Mol. Med. 2020, 26, 533–551. [Google Scholar] [CrossRef]
- Andersen, J.V.; Markussen, K.H.; Jakobsen, E.; Schousboe, A.; Waagepetersen, H.S.; Rosenberg, P.A.; Aldana, B.I. Glutamate metabolism and recycling at the excitatory synapse in health and neurodegeneration. Neuropharmacology 2021, 196, 108719. [Google Scholar] [CrossRef]
- Sloley, S.S.; Main, B.S.; Winston, C.N.; Harvey, A.C.; Kaganovich, A.; Korthas, H.T.; Caccavano, A.P.; Zapple, D.N.; Wu, J.Y.; Partridge, J.G.; et al. High-frequency head impact causes chronic synaptic adaptation and long-term cognitive impairment in mice. Nat. Commun. 2021, 12, 2613. [Google Scholar] [CrossRef]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef]
- Westbrook, R.; Zhang, C.; Yang, H.; Tian, J.; Guo, S.; Xue, Q.L.; Walston, J.; Le, A.; Abadir, P.M. Metabolomics-Based Identification of Metabolic Dysfunction in Frailty. J. Gerontol. A Biol. Sci. Med. Sci. 2022, 77, 2367–2372. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Jiang, L. Dysregulated Mitochondrial Dynamics and Metabolism in Obesity, Diabetes, and Cancer. Front. Endocrinol. 2019, 10, 570. [Google Scholar] [CrossRef] [Green Version]
- Nissen, J.D.; Pajęcka, K.; Stridh, M.H.; Skytt, D.M.; Waagepetersen, H.S. Dysfunctional TCA-Cycle Metabolism in Glutamate Dehydrogenase Deficient Astrocytes. Glia 2015, 63, 2313–2326. [Google Scholar] [CrossRef]
- Jo, S.L.; Yang, H.; Lee, S.R.; Heo, J.H.; Lee, H.W.; Hong, E.J. Curcumae Radix Decreases Neurodegenerative Markers through Glycolysis Decrease and TCA Cycle Activation. Nutrients 2022, 14, 1587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Tang, Y.B.; Dammer, E.; Liu, J.R.; Zhao, Y.W.; Zhu, L.; Ren, R.J.; Chen, H.Z.; Wang, G.; Cheng, Q. Dysregulated Urinary Arginine Metabolism in Older Adults With Amnestic Mild Cognitive Impairment. Front. Aging Neurosci. 2019, 11, 90. [Google Scholar] [CrossRef] [Green Version]
- Wiesinger, H. Arginine metabolism and the synthesis of nitric oxide in the nervous system. Prog. Neurobiol. 2001, 64, 365–391. [Google Scholar] [CrossRef]
- Yi, J.; Horky, L.L.; Friedlich, A.L.; Shi, Y.; Rogers, J.T.; Huang, X. L-arginine and Alzheimer′s disease. Int. J. Clin. Exp. Pathol. 2009, 2, 211–238. [Google Scholar]
- Virarkar, M.; Alappat, L.; Bradford, P.G.; Awad, A.B. L-arginine and nitric oxide in CNS function and neurodegenerative diseases. Crit. Rev. Food Sci. Nutr. 2013, 53, 1157–1167. [Google Scholar] [CrossRef]
- Bescós, R.; Sureda, A.; Tur, J.A.; Pons, A. The effect of nitric-oxide-related supplements on human performance. Sports Med. 2012, 42, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Bahadoran, Z.; Mirmiran, P.; Kashfi, K.; Ghasemi, A. Endogenous flux of nitric oxide: Citrulline is preferred to Arginine. Acta Physiol. 2021, 231, e13572. [Google Scholar] [CrossRef] [PubMed]
- Tewari, D.; Sah, A.N.; Bawari, S.; Nabavi, S.F.; Dehpour, A.R.; Shirooie, S.; Braidy, N.; Fiebich, B.L.; Vacca, R.A.; Nabavi, S.M. Role of Nitric Oxide in Neurodegeneration: Function, Regulation, and Inhibition. Curr. Neuropharmacol. 2021, 19, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.R. Nitric oxide neurons and neurotransmission. Prog. Neurobiol. 2010, 90, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, M.K.; Kartawy, M.; Amal, H. The role of nitric oxide in brain disorders: Autism spectrum disorder and other psychiatric, neurological, and neurodegenerative disorders. Redox Biol. 2020, 34, 101567. [Google Scholar] [CrossRef]
- Chen, P.; Guo, Y.; Jia, L.; Wan, J.; He, T.; Fang, C.; Li, T. Interaction Between Functionally Activate Endometrial Microbiota and Host Gene Regulation in Endometrial Cancer. Front. Cell Dev. Biol. 2021, 9, 727286. [Google Scholar] [CrossRef]
- Teixeira, F.C.; Soares, M.S.P.; Blödorn, E.B.; Domingues, W.B.; Reichert, K.P.; Zago, A.M.; Carvalho, F.B.; Gutierres, J.M.; Gonçales, R.A.; da Cruz Fernandes, M.; et al. Investigating the Effect of Inosine on Brain Purinergic Receptors and Neurotrophic and Neuroinflammatory Parameters in an Experimental Model of Alzheimer′s Disease. Mol. Neurobiol. 2022, 59, 841–855. [Google Scholar] [CrossRef]
- Nascimento, F.P.; Macedo-Júnior, S.J.; Lapa-Costa, F.R.; Cezar-Dos-Santos, F.; Santos, A.R.S. Inosine as a Tool to Understand and Treat Central Nervous System Disorders: A Neglected Actor? Front. Neurosci. 2021, 15, 703783. [Google Scholar] [CrossRef]
- Burnstock, G. Physiology and pathophysiology of purinergic neurotransmission. Physiol. Rev. 2007, 87, 659–797. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, K.; Zhai, C.; Li, X.; Gang, H.; Gao, X. Feature-Based Molecular Networking Facilitates the Comprehensive Identification of Differential Metabolites in Diabetic Cognitive Dysfunction Rats. Metabolites 2023, 13, 538. https://doi.org/10.3390/metabo13040538
Du K, Zhai C, Li X, Gang H, Gao X. Feature-Based Molecular Networking Facilitates the Comprehensive Identification of Differential Metabolites in Diabetic Cognitive Dysfunction Rats. Metabolites. 2023; 13(4):538. https://doi.org/10.3390/metabo13040538
Chicago/Turabian StyleDu, Ke, Chuanjia Zhai, Xuejiao Li, Hongchuan Gang, and Xiaoyan Gao. 2023. "Feature-Based Molecular Networking Facilitates the Comprehensive Identification of Differential Metabolites in Diabetic Cognitive Dysfunction Rats" Metabolites 13, no. 4: 538. https://doi.org/10.3390/metabo13040538
APA StyleDu, K., Zhai, C., Li, X., Gang, H., & Gao, X. (2023). Feature-Based Molecular Networking Facilitates the Comprehensive Identification of Differential Metabolites in Diabetic Cognitive Dysfunction Rats. Metabolites, 13(4), 538. https://doi.org/10.3390/metabo13040538





