Development of a Urine Metabolomics Biomarker-Based Prediction Model for Preeclampsia during Early Pregnancy
Abstract
1. Introduction
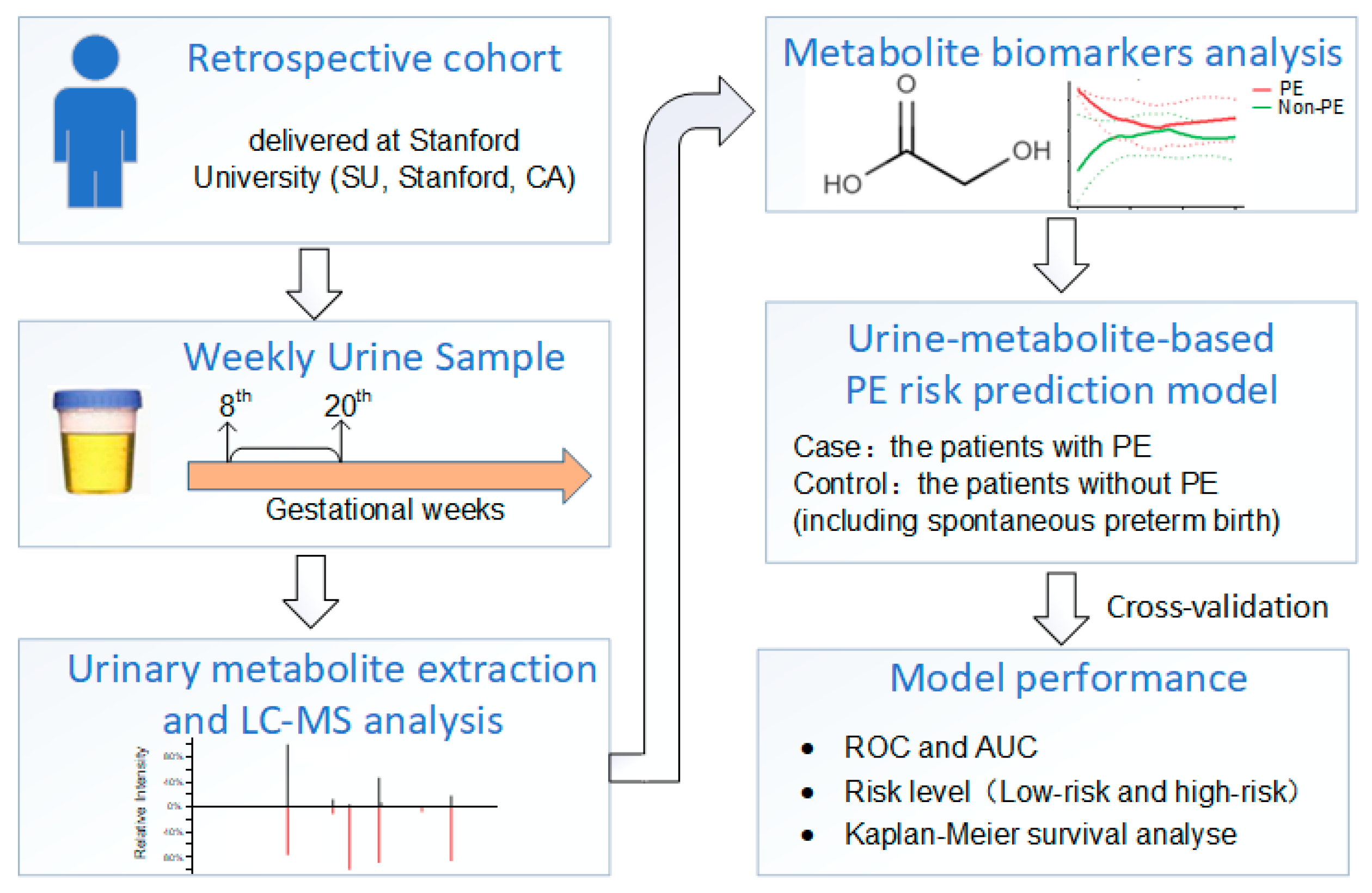
2. Method
2.1. PE Definition
2.2. Cohort Construction
2.3. Samples
2.4. Urinary Metabolite Extraction and Global Liquid Chromatography Mass Spectrometry (LC-MS/MS) Analysis
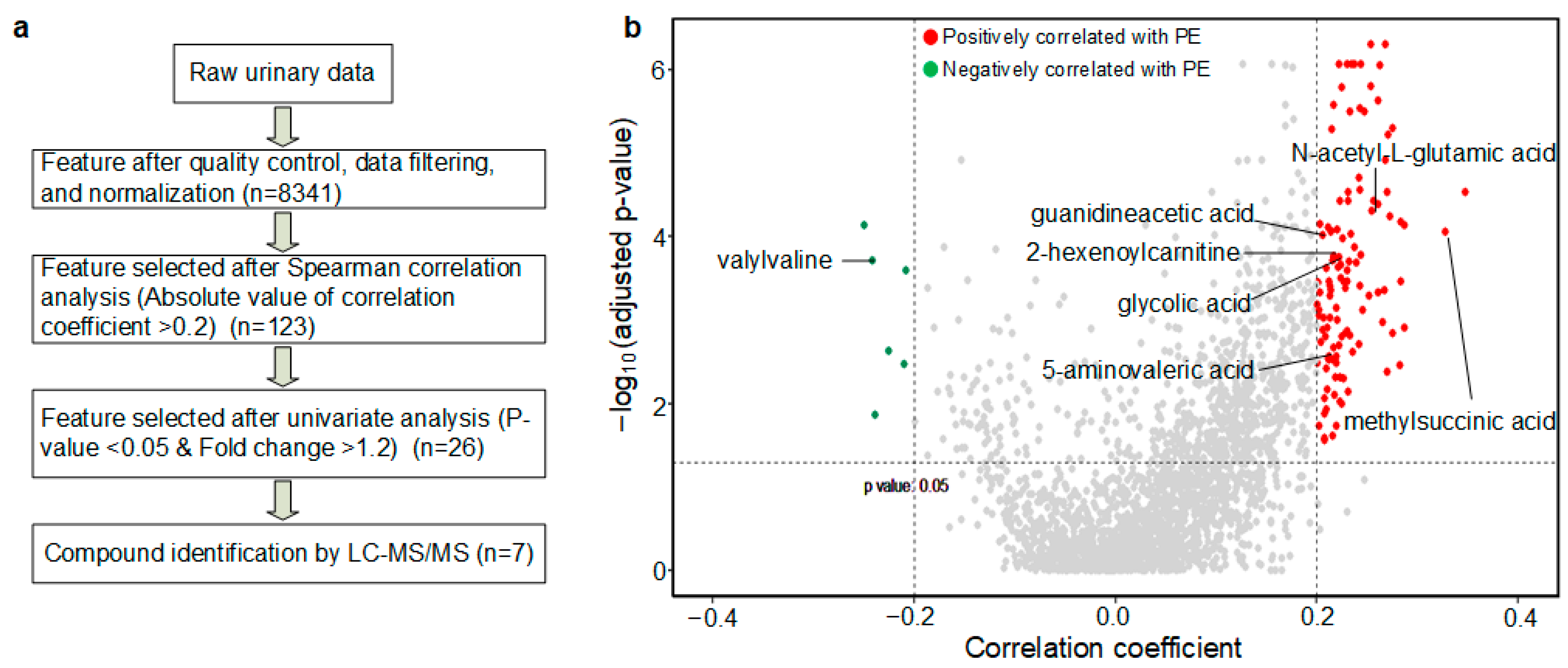
2.5. Data Preprocessing and Statistics
2.6. Metabolite Identification and Metabolite-Based Modeling
3. Results
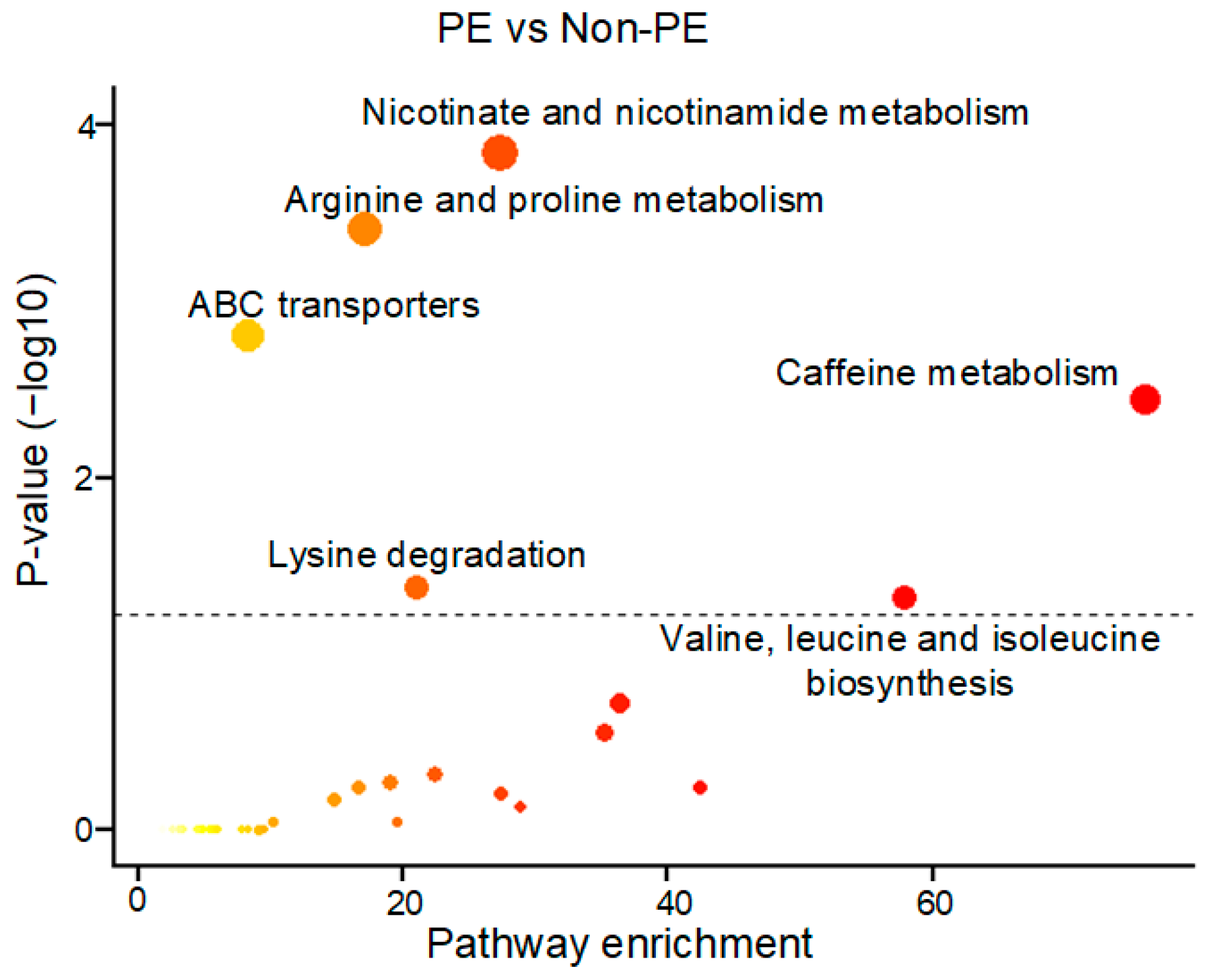
3.1. A Unique PE-Associated Metabolomics Pattern and Metabolic Pathway Analyses
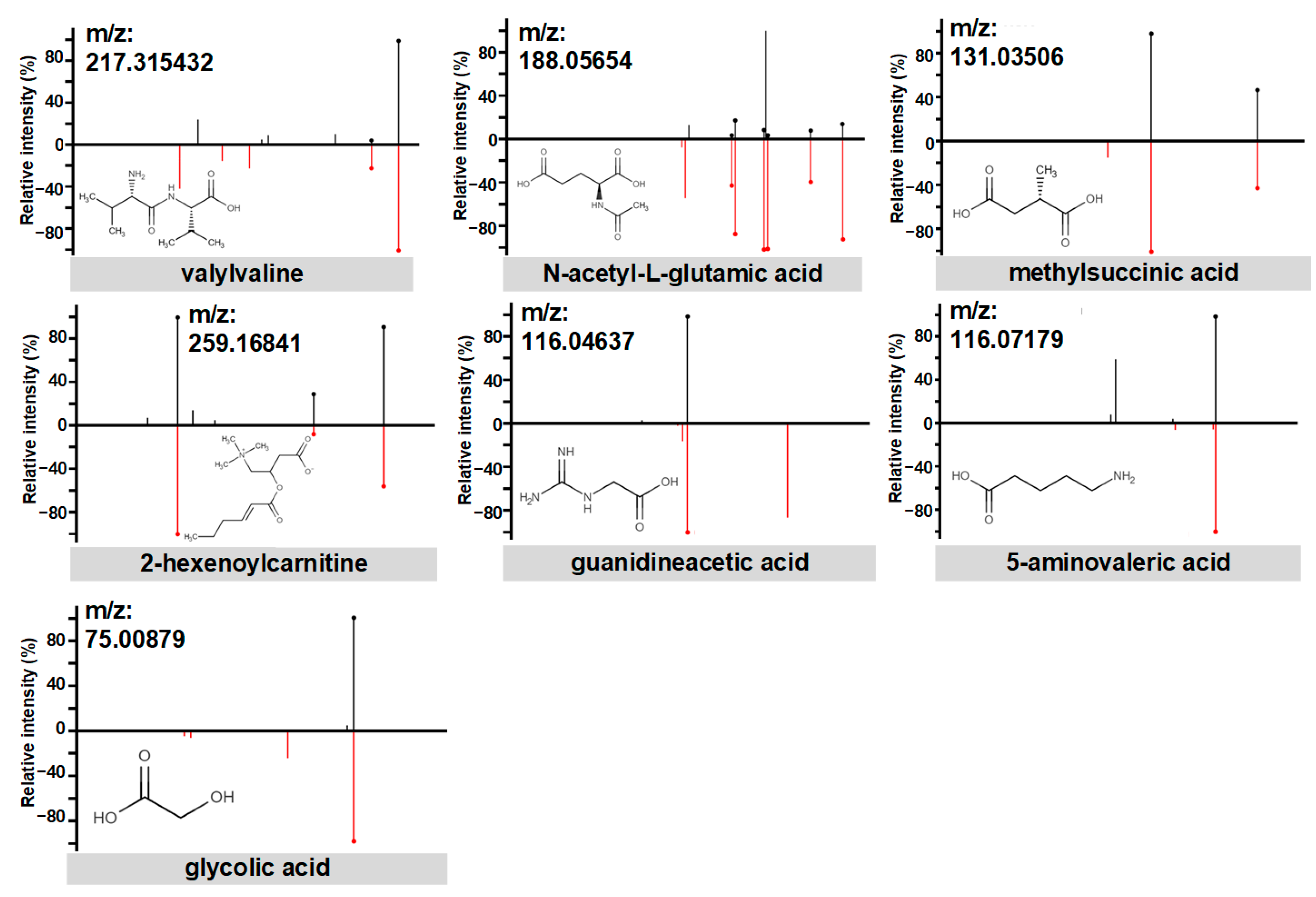
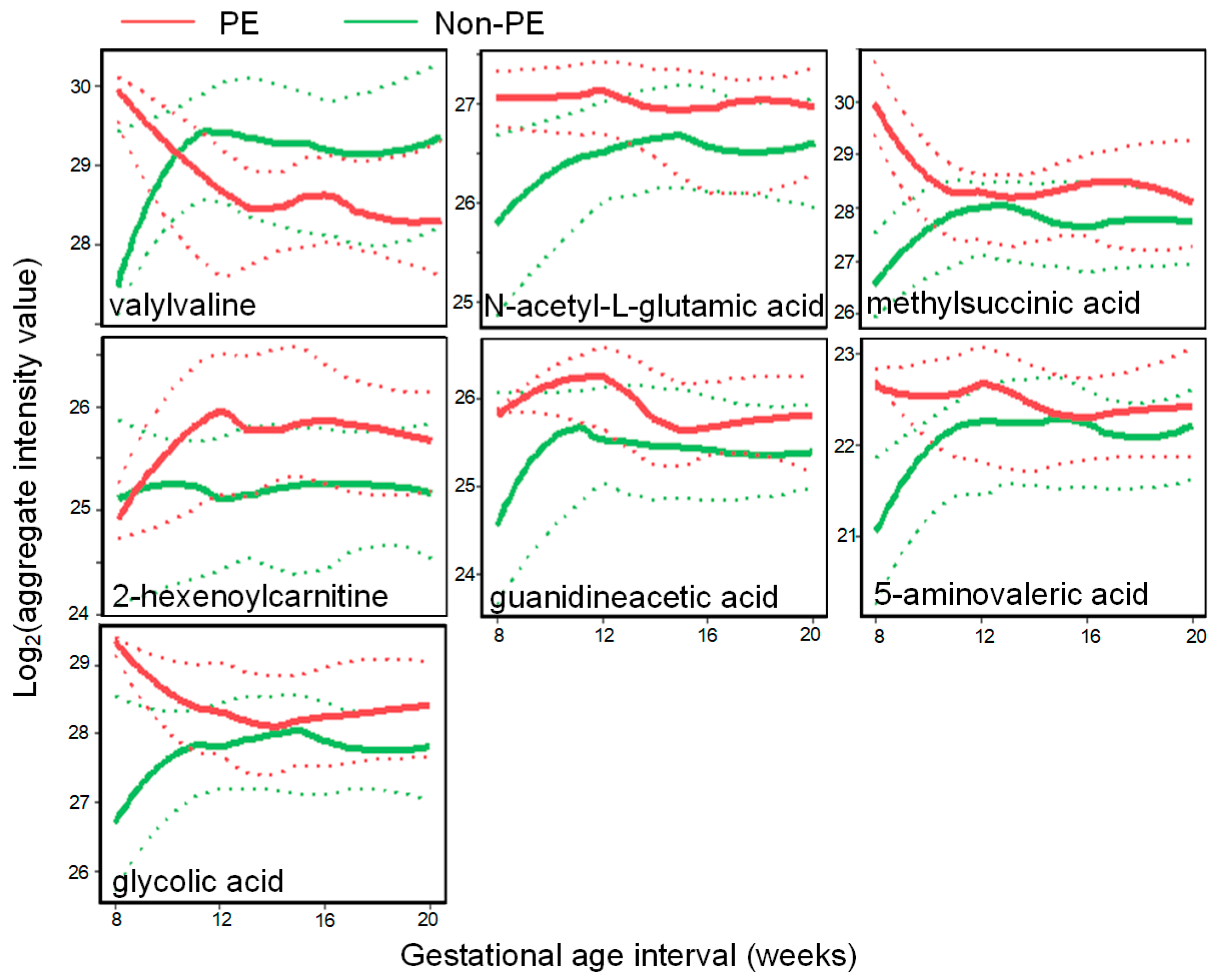
3.2. PE Predictive Metabolomics Biomarkers
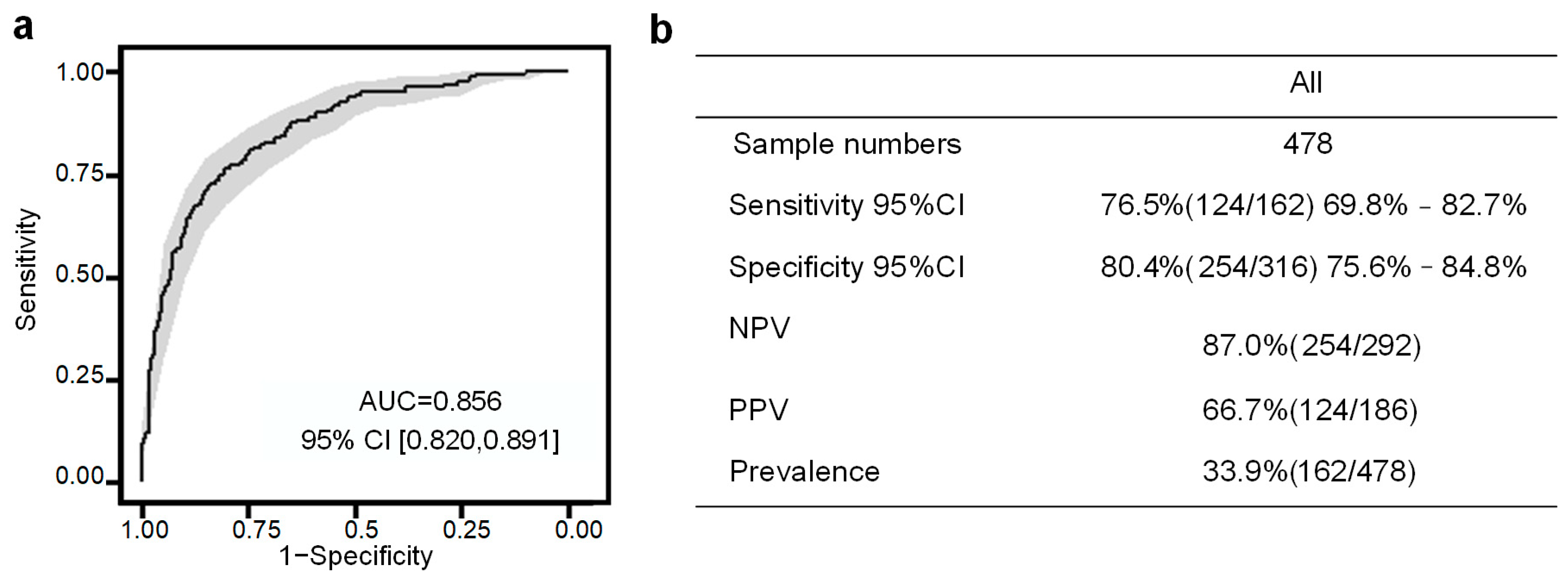
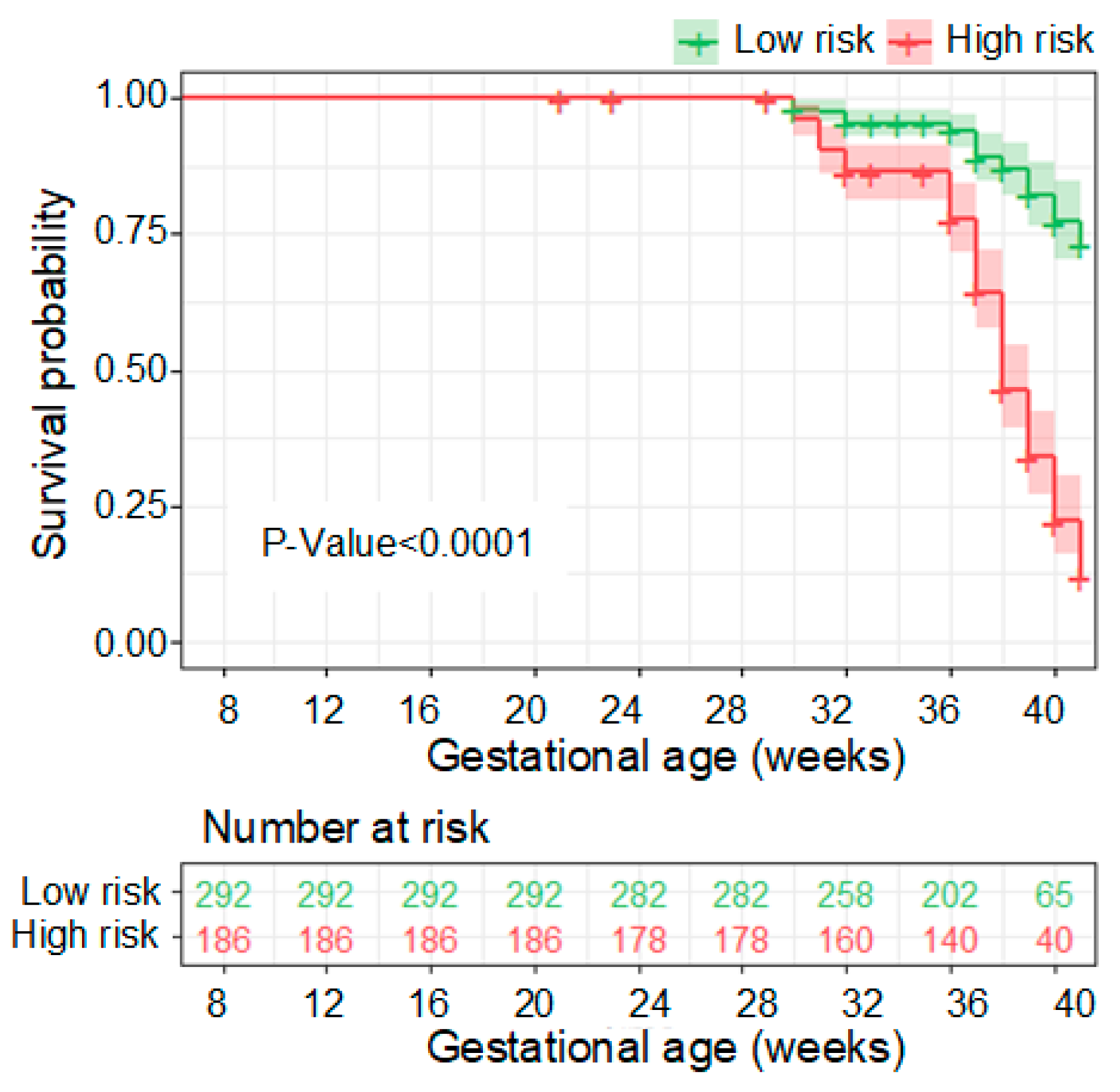
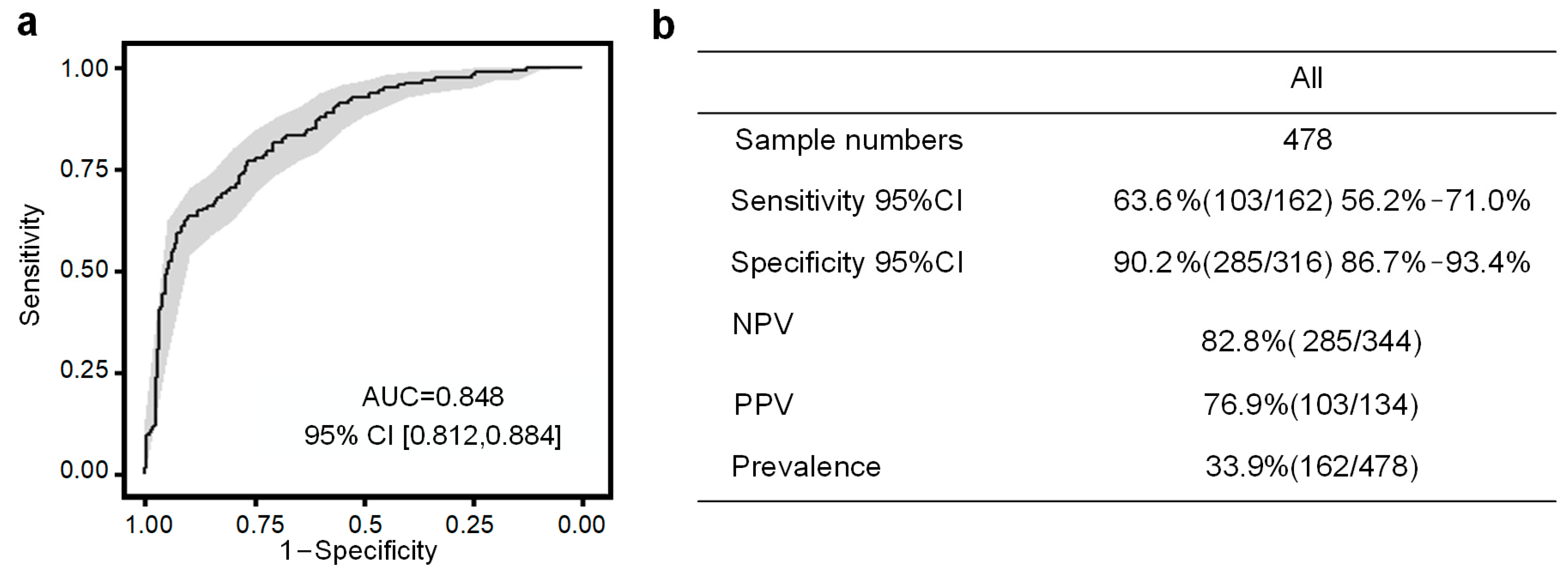
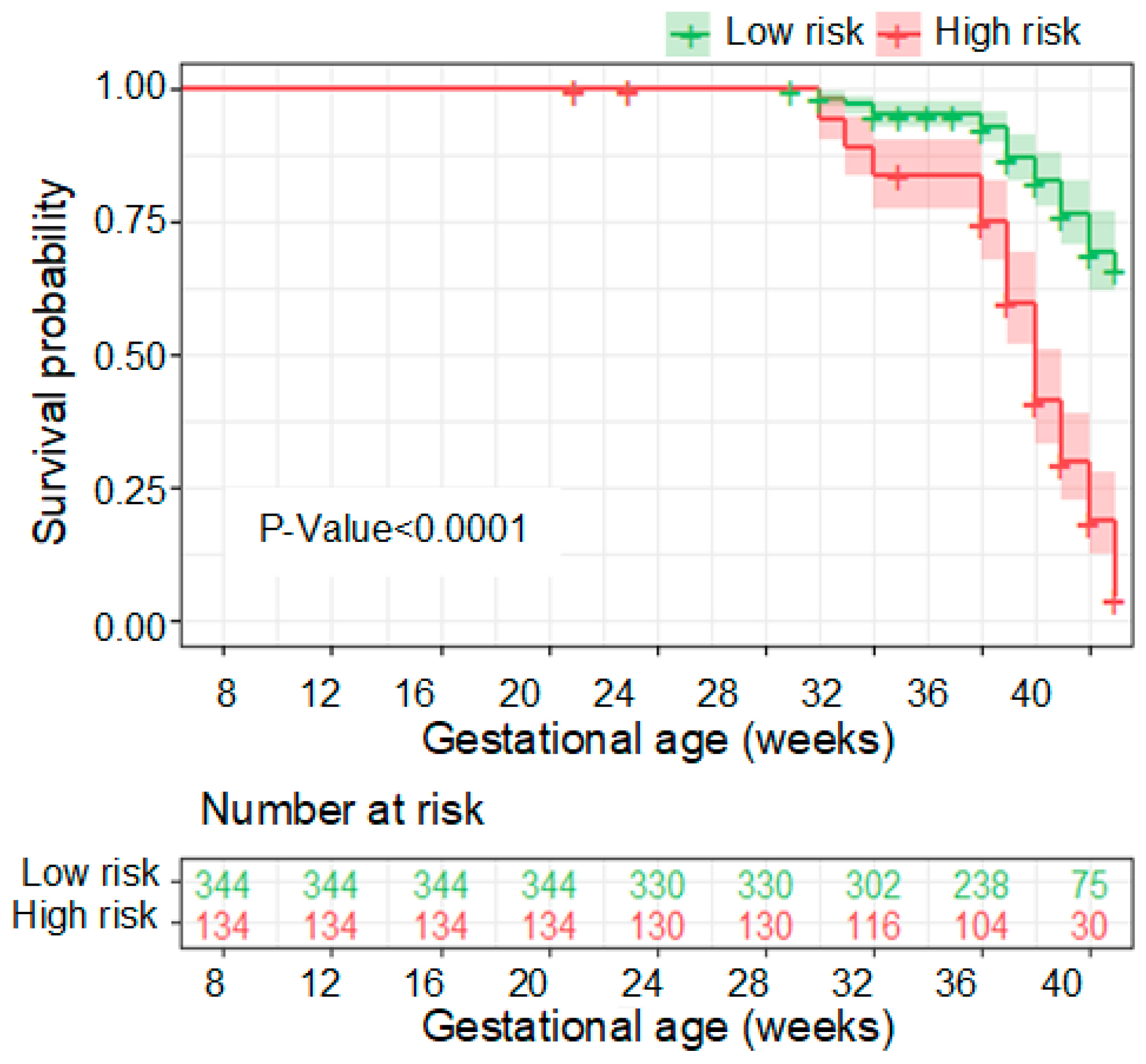
3.3. Performance of the PE Prediction Model
4. Discussion
4.1. Summary of Main Findings
4.2. Biological Implications of PE Biomarkers
4.3. Comparison with Prior Work and Limitations
4.4. Advantage of Urine Testing of PE Risk Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rich-Edwards, J.W.; Fraser, A.; Lawlor, D.A.; Catov, J.M. Pregnancy characteristics and women’s future cardiovascular health: An underused opportunity to improve women’s health? Epidemiol. Rev. 2014, 36, 57–70. [Google Scholar] [CrossRef]
- Duley, L. The global impact of preeclampsia and eclampsia. Semin. Perinatol. 2009, 33, 130–137. [Google Scholar] [CrossRef]
- Foisy, M.A.; Marchant, I.; Lessard, L.; Caron, L.; Bujold, E. Aspirin for the prevention of preeclampsia. J. Perinat. Med. 2022, 50, 1271–1272. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.; Shih, T.; Incerti, D.; Ton, T.G.N.; Lee, H.C.; Peneva, D.; Macones, G.A.; Sibai, B.M.; Jena, A.B. Short-term costs of preeclampsia to the United States health care system. Am. J. Obstet. Gynecol. 2017, 217, 237–248. [Google Scholar] [CrossRef]
- Antwi, E.; Amoakoh-Coleman, M.; Vieira, D.L.; Madhavaram, S.; Koram, K.A.; Grobbee, D.E.; Agyepong, I.A.; Klipstein-Grobusch, K. Systematic review of prediction models for gestational hypertension and preeclampsia. PLoS ONE 2020, 15, e0230955. [Google Scholar] [CrossRef] [PubMed]
- De Kat, A.C.; Hirst, J.; Woodward, M.; Kennedy, S.; Peters, S.A. Prediction models for preeclampsia: A systematic review. Pregnancy Hypertens. 2019, 16, 48–66. [Google Scholar] [CrossRef]
- Giannakou, K. Prediction of preeclampsia. Obstet. Med. 2021, 14, 220–224. [Google Scholar] [CrossRef]
- Eastwood, K.A.; Patterson, C.; Hunter, A.J.; McCance, D.R.; Young, I.S.; Holmes, V.A. Evaluation of the predictive value of placental vascularisation indices derived from 3-Dimensional power Doppler whole placental volume scanning for prediction of preeclampsia: A systematic review and meta-analysis. Placenta 2017, 51, 89–97. [Google Scholar] [CrossRef]
- Kleinrouweler, C.E.; Wiegerinck, M.M.; Ris-Stalpers, C.; Bossuyt, P.M.; van der Post, J.A.; von Dadelszen, P.; Mol, B.W.; Pajkrt, E.; EBM CONNECT Collaboration. Accuracy of circulating placental growth factor, vascular endothelial growth factor, soluble fms-like tyrosine kinase 1 and soluble endoglin in the prediction of preeclampsia: A systematic review and meta-analysis. BJOG 2012, 119, 778–787. [Google Scholar] [CrossRef]
- Velauthar, L.; Plana, M.N.; Kalidindi, M.; Zamora, J.; Thilaganathan, B.; Illanes, S.E.; Khan, K.S.; Aquilina, J.; Thangaratinam, S. First-trimester uterine artery Doppler and adverse pregnancy outcome: A meta-analysis involving 55,974 women. Ultrasound Obstet. Gynecol. 2014, 43, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.; Reddy, M.; Nolan, R.; Camunas-Soler, J.; Khodursky, A.; Scheller, N.M.; Cantonwine, D.E.; Engelbrechtsen, L.; Mi, J.D.; Dutta, A.; et al. RNA profiles reveal signatures of future health and disease in pregnancy. Nature 2022, 601, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Akolekar, R.; Syngelaki, A.; Poon, L.; Wright, D.; Nicolaides, K.H. Competing risks model in early screening for preeclampsia by biophysical and biochemical markers. Fetal Diagn. Ther. 2013, 33, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Anderson, U.D.; Gram, M.; Akerstrom, B.; Hansson, S.R. First trimester prediction of preeclampsia. Curr. Hypertens. Rep. 2015, 17, 584. [Google Scholar] [CrossRef] [PubMed]
- Kolialexi, A.; Mavreli, D.; Tounta, G.; Mavrou, A.; Papantoniou, N. Urine proteomic studies in preeclampsia. Proteom. Clin. Appl. 2015, 9, 501–506. [Google Scholar] [CrossRef]
- Guo, H.X.; Zhu, Y.B.; Wu, C.P.; Zhong, M.; Hu, S.W. Potential urine biomarkers for gestational hypertension and preeclampsia. Mol. Med. Rep. 2019, 19, 2463–2470. [Google Scholar] [CrossRef]
- Buhimschi, I.A.; Zhao, G.; Funai, E.F.; Harris, N.; Sasson, I.E.; Bernstein, I.M.; Saade, G.R.; Buhimschi, C.S. Proteomic profiling of urine identifies specific fragments of SERPINA1 and albumin as biomarkers of preeclampsia. Am. J. Obstet. Gynecol. 2008, 199, 551.e1–551.e16. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, Y.; Jin, X.; Zhang, L.; Zhou, Y.; Niu, J.; Chen, J.; Gu, Y. Urinary proteomics analysis for renal injury in hypertensive disorders of pregnancy with iTRAQ labeling and LC-MS/MS. Proteom. Clin. Appl. 2011, 5, 300–310. [Google Scholar] [CrossRef]
- Kononikhin, A.S.; Starodubtseva, N.L.; Bugrova, A.E.; Shirokova, V.A.; Chagovets, V.V.; Indeykina, M.I.; Popov, I.A.; Kostyukevich, Y.I.; Vavina, O.V.; Muminova, K.T.; et al. An untargeted approach for the analysis of the urine peptidome of women with preeclampsia. J. Proteom. 2016, 149, 38–43. [Google Scholar] [CrossRef]
- Ghaemi, M.S.; Tarca, A.L.; Romero, R.; Stanley, N.; Fallahzadeh, R.; Tanada, A.; Culos, A.; Ando, K.; Han, X.; Blumenfeld, Y.J.; et al. Proteomic signatures predict preeclampsia in individual cohorts but not across cohorts—Implications for clinical biomarker studies. J. Matern. Fetal Neonatal Med. 2022, 35, 5621–5628. [Google Scholar] [CrossRef]
- Zhou, X.; Niu, J.M.; Ji, W.J.; Zhang, Z.; Wang, P.P.; Ling, X.F.; Li, Y.M. Precision test for precision medicine: Opportunities, challenges and perspectives regarding preeclampsia as an intervention window for future cardiovascular disease. Am. J. Transl. Res. 2016, 8, 1920–1934. [Google Scholar]
- Roberts, J.M.; August, P.A.; Bakris, G.; Barton, J.R.; Bernstein, I.M.; Druzin, M.; Gaiser, R.R.; Granger, J.P.; Jeyabalan, A.; Johnson, D.D.; et al. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet. Gynecol. 2013, 122, 1122–1131. [Google Scholar] [CrossRef]
- Sylvester, K.G.; Hao, S.; Li, Z.; Han, Z.; Tian, L.; Ladella, S.; Wong, R.J.; Shaw, G.M.; Stevenson, D.K.; Cohen, H.J.; et al. Gestational dating by urine metabolic profile at high resolution weekly sampling timepoints: Discovery and validation. Front. Mol. Med. 2022, 2, 6. [Google Scholar] [CrossRef]
- Kim, T.; Tang, O.; Vernon, S.T.; Kott, K.A.; Koay, Y.C.; Park, J.; James, D.E.; Grieve, S.M.; Speed, T.P.; Yang, P.; et al. A hierarchical approach to removal of unwanted variation for large-scale metabolomics data. Nat. Commun. 2021, 12, 4992. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Heinemann, J. Cluster analysis of untargeted metabolomic experiments. Methods Mol. Biol. 2019, 1859, 275–285. [Google Scholar] [CrossRef]
- Viant, M.R.; Kurland, I.J.; Jones, M.R.; Dunn, W.B. How close are we to complete annotation of metabolomes? Curr. Opin. Chem. Biol. 2017, 36, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Syrjanen, S.; Piironen, P.; Markkanen, H. Free amino-acid content of wax-stimulated human whole saliva as related to periodontal disease. Arch. Oral Biol. 1987, 32, 607–610. [Google Scholar] [CrossRef]
- Sass, J.O.; Mohr, V.; Olbrich, H.; Engelke, U.; Horvath, J.; Fliegauf, M.; Loges, N.T.; Schweitzer-Krantz, S.; Moebus, R.; Weiler, P.; et al. Mutations in ACY1, the gene encoding aminoacylase 1, cause a novel inborn error of metabolism. Am. J. Hum. Genet. 2006, 78, 401–409. [Google Scholar] [CrossRef]
- Nouioua, S.; Cheillan, D.; Zaouidi, S.; Salomons, G.S.; Amedjout, N.; Kessaci, F.; Boulahdour, N.; Hamadouche, T.; Tazir, M. Creatine deficiency syndrome. a treatable myopathy due to arginine-glycine amidinotransferase (AGAT) deficiency. Neuromuscul. Disord. 2013, 23, 670–674. [Google Scholar] [CrossRef]
- Cho, K.; Moon, J.S.; Kang, J.H.; Jang, H.B.; Lee, H.J.; Park, S.I.; Yu, K.S.; Cho, J.Y. Combined untargeted and targeted metabolomic profiling reveals urinary biomarkers for discriminating obese from normal-weight adolescents. Pediatr. Obes. 2017, 12, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Grosso, S.; Mostardini, R.; Farnetani, M.A.; Molinelli, M.; Berardi, R.; Dionisi-Vici, C.; Rizzo, C.; Morgese, G.; Balestri, P. Ethylmalonic encephalopathy: Further clinical and neuroradiological characterization. J. Neurol. 2002, 249, 1446–1450. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.; Wright, A.; Nicolaides, K.H. The competing risk approach for prediction of preeclampsia. Am. J. Obstet. Gynecol. 2020, 223, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Guizani, M.; Valsamis, J.; Dutemeyer, V.; Kang, X.; Ceccotti, V.; Khalife, J.; Duiella, S.F.; Blavier, F.; Faraca, A.; Cos, T.; et al. First-Trimester combined multimarker prospective study for the detection of pregnancies at a high risk of developing preeclampsia using the fetal medicine foundation-algorithm. Fetal Diagn. Ther. 2018, 43, 266–273. [Google Scholar] [CrossRef]
- Kim, Y.R.; Jung, I.; Heo, S.J.; Chang, S.W.; Cho, H.Y. A preeclampsia risk prediction model based on maternal characteristics and serum markers in twin pregnancy. J. Matern. Fetal Neonatal Med. 2021, 34, 3623–3628. [Google Scholar] [CrossRef]
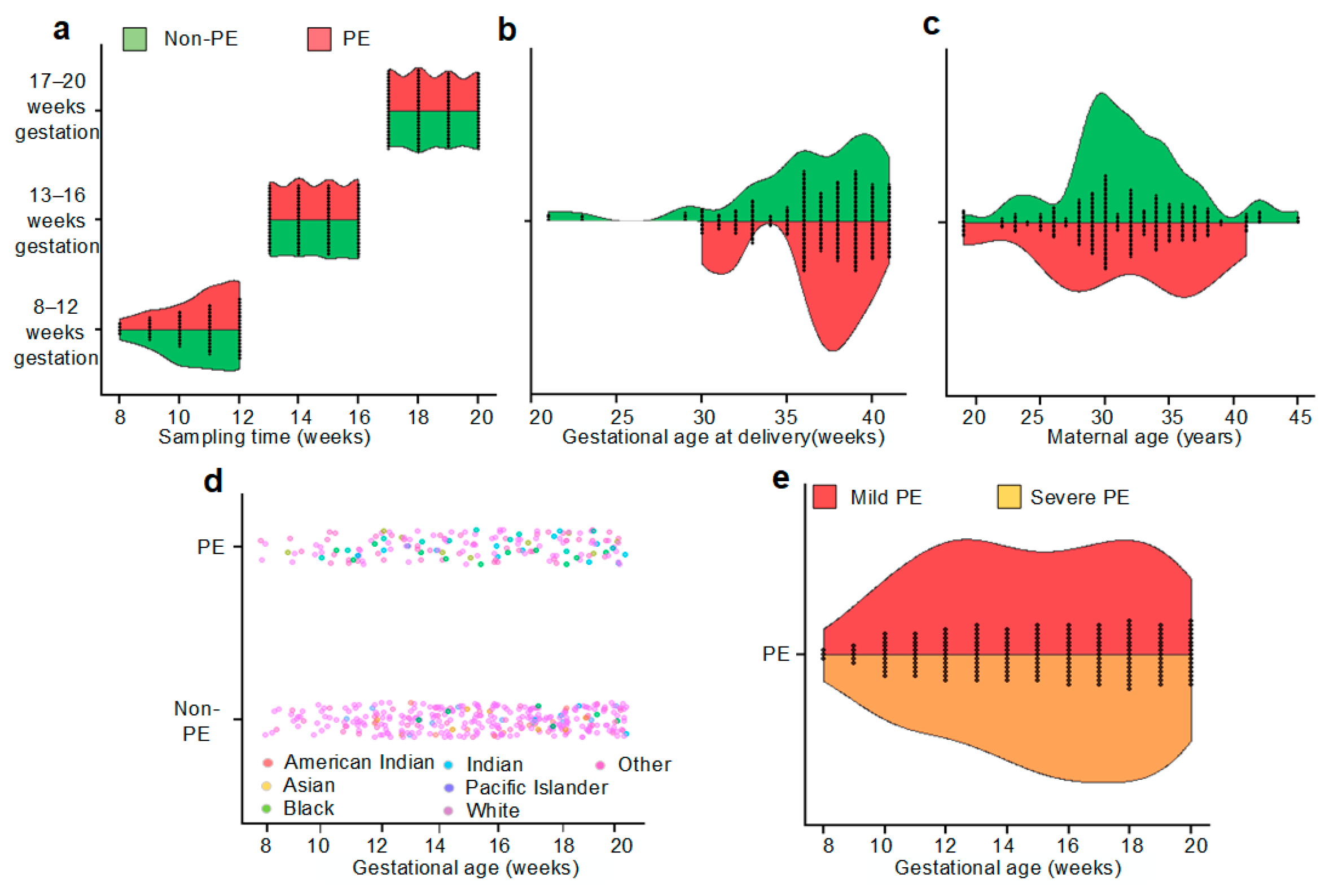
| PE (n = 20) | Non-PE (n = 40) | p-Value | |
|---|---|---|---|
| Number of samples | 162 | 316 | |
| Cohort, n (%) | <0.001 | ||
| Full-term with PE | 13 (65) | 0 (0) | |
| Pre-term with PE | 7 (35) | 0 (0) | |
| Pre-term without PE | 0 (0) | 21 (52.5) | |
| Full-term without PE | 0 (0) | 19 (47.5) | |
| Age, mean (IQR) | 35.1 (27.8, 36.2) | 32.1 (29, 35) | 0.84 |
| GA at delivery, mean (IQR) | 37 (36, 39) | 36 (33.8, 39) | 0.65 |
| Hypertensive disorder, n (%) | <0.001 | ||
| Mild PE | 10 (50) | 0 (0) | |
| Severe PE | 10 (50) | 0 (0) | |
| Non-PE | 0 (0) | 40 (100) | |
| Race, n (%) | 0.21 | ||
| American Indian | 0 (0) | 2 (5) | |
| Asian | 2 (10) | 1 (2.5) | |
| Black | 1 (5) | 1 (2.5) | |
| Indian | 2 (10) | 1 (2.5) | |
| Pacific Islander | 0 (0) | 1 (2.5) | |
| White | 9 (45) | 28 (70) | |
| Other | 6 (30) | 6 (15) | |
| Total number of previous pregnancies, mean (SD) | 1.1 (2.0) | 1.9 (2.2) | 0.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Sylvester, K.G.; Jin, B.; Wong, R.J.; Schilling, J.; Chou, C.J.; Han, Z.; Luo, R.Y.; Tian, L.; Ladella, S.; et al. Development of a Urine Metabolomics Biomarker-Based Prediction Model for Preeclampsia during Early Pregnancy. Metabolites 2023, 13, 715. https://doi.org/10.3390/metabo13060715
Zhang Y, Sylvester KG, Jin B, Wong RJ, Schilling J, Chou CJ, Han Z, Luo RY, Tian L, Ladella S, et al. Development of a Urine Metabolomics Biomarker-Based Prediction Model for Preeclampsia during Early Pregnancy. Metabolites. 2023; 13(6):715. https://doi.org/10.3390/metabo13060715
Chicago/Turabian StyleZhang, Yaqi, Karl G. Sylvester, Bo Jin, Ronald J. Wong, James Schilling, C. James Chou, Zhi Han, Ruben Y. Luo, Lu Tian, Subhashini Ladella, and et al. 2023. "Development of a Urine Metabolomics Biomarker-Based Prediction Model for Preeclampsia during Early Pregnancy" Metabolites 13, no. 6: 715. https://doi.org/10.3390/metabo13060715
APA StyleZhang, Y., Sylvester, K. G., Jin, B., Wong, R. J., Schilling, J., Chou, C. J., Han, Z., Luo, R. Y., Tian, L., Ladella, S., Mo, L., Marić, I., Blumenfeld, Y. J., Darmstadt, G. L., Shaw, G. M., Stevenson, D. K., Whitin, J. C., Cohen, H. J., McElhinney, D. B., & Ling, X. B. (2023). Development of a Urine Metabolomics Biomarker-Based Prediction Model for Preeclampsia during Early Pregnancy. Metabolites, 13(6), 715. https://doi.org/10.3390/metabo13060715







