Metabolomics and Transcriptomics Revealed a Comprehensive Understanding of the Biochemical and Genetic Mechanisms Underlying the Color Variations in Chrysanthemums
Abstract
1. Introduction
2. Materials and Method
2.1. Materials
2.2. Equipment
2.3. Transcriptome Assays
2.4. PCA and OPLS-DA
2.5. Statistics Analysis
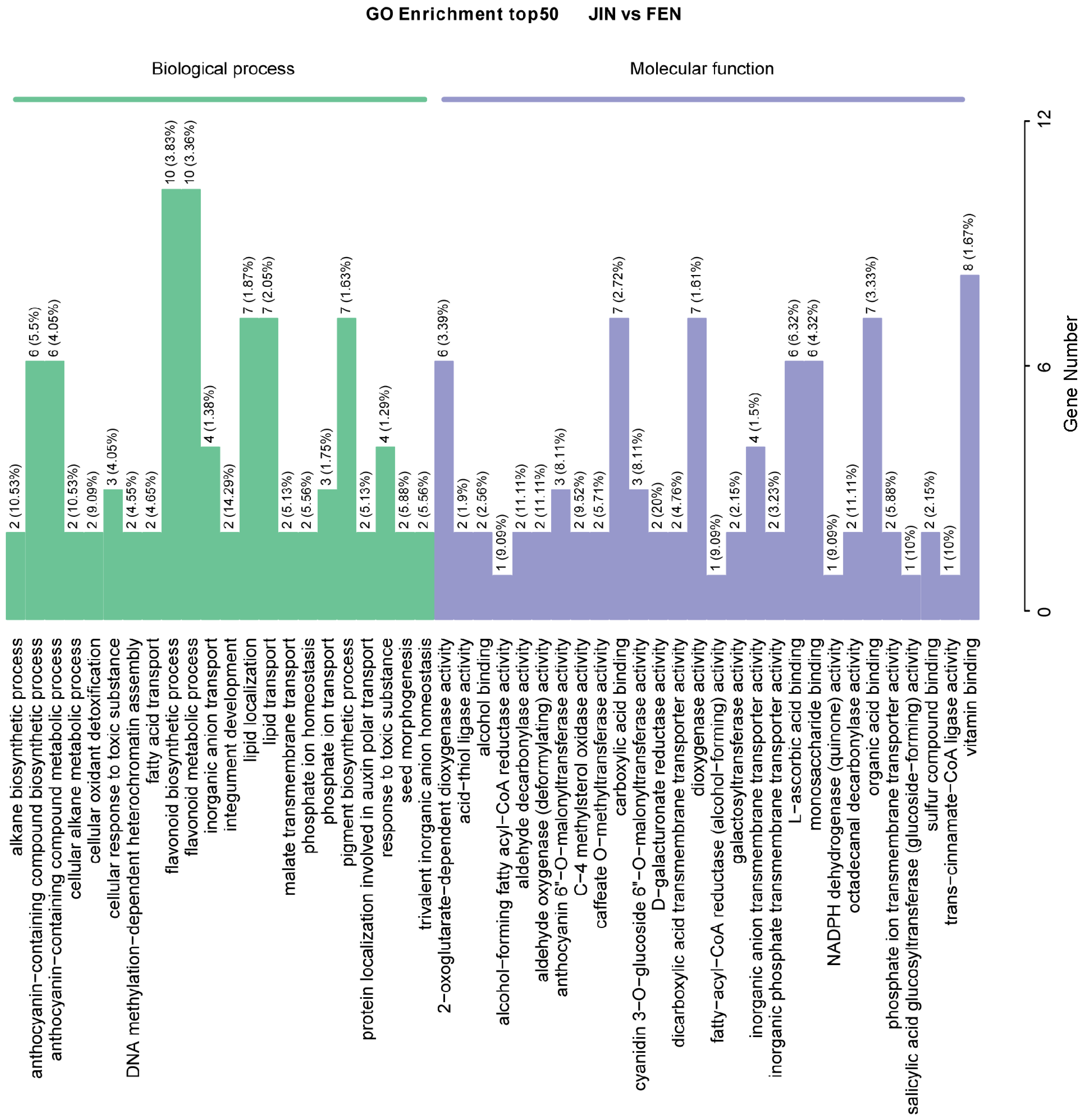
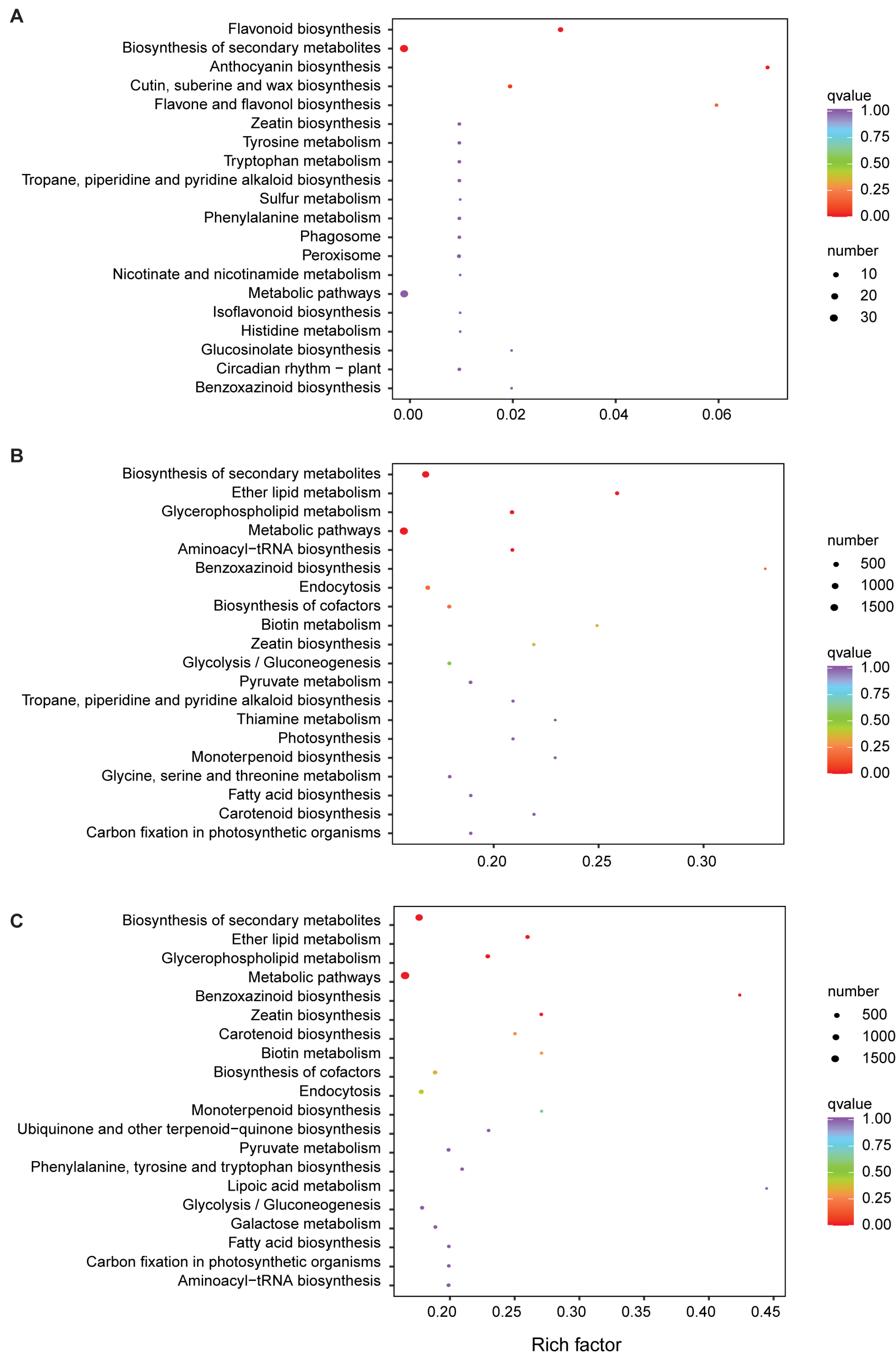
2.6. KEGG and GO
3. Results
3.1. UPLC-ESI-MS/MS-Based Quantitative Metabolomic Analysis
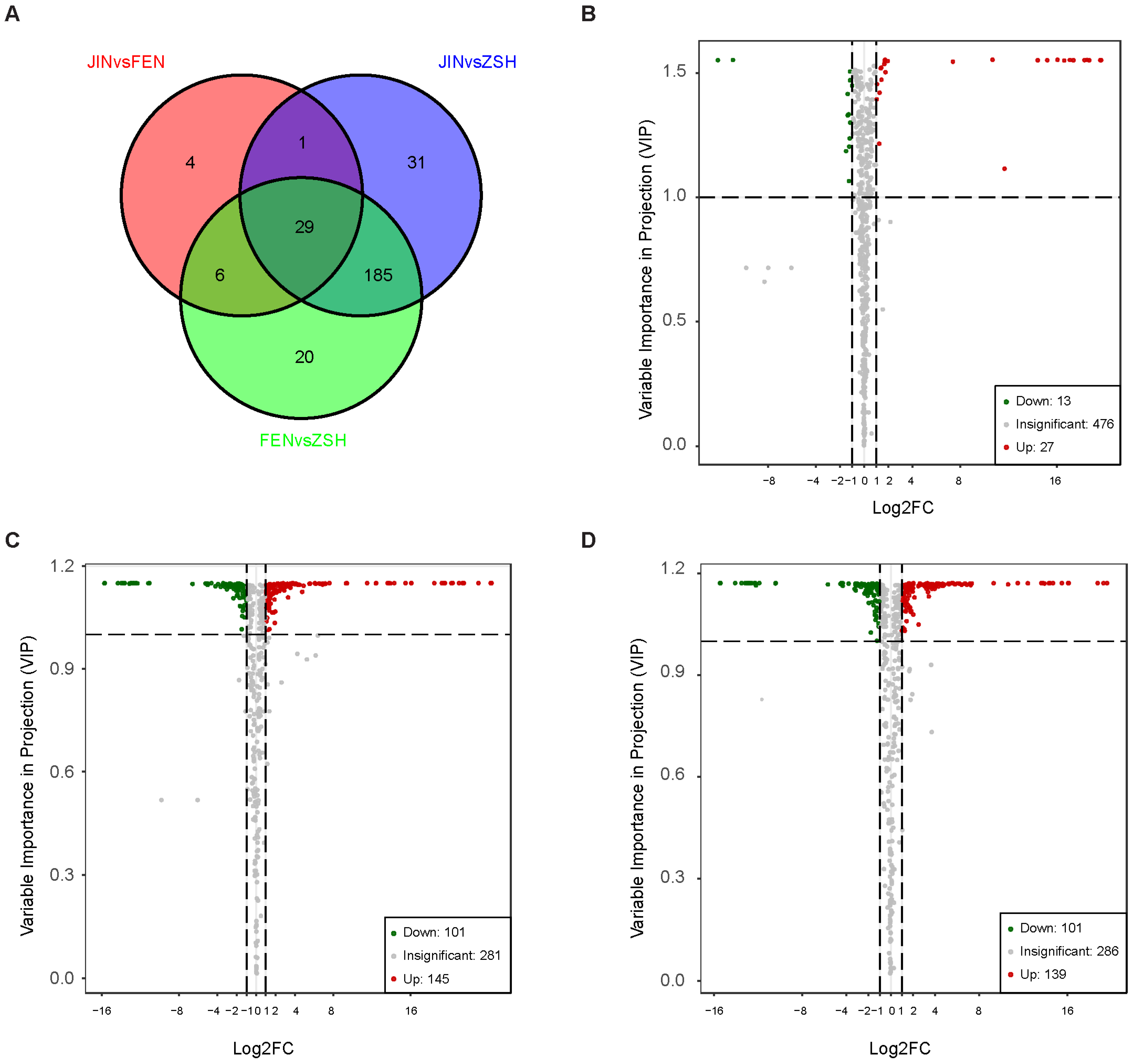
3.2. Analysis of Differentially Accumulated Metabolites (DAMs)
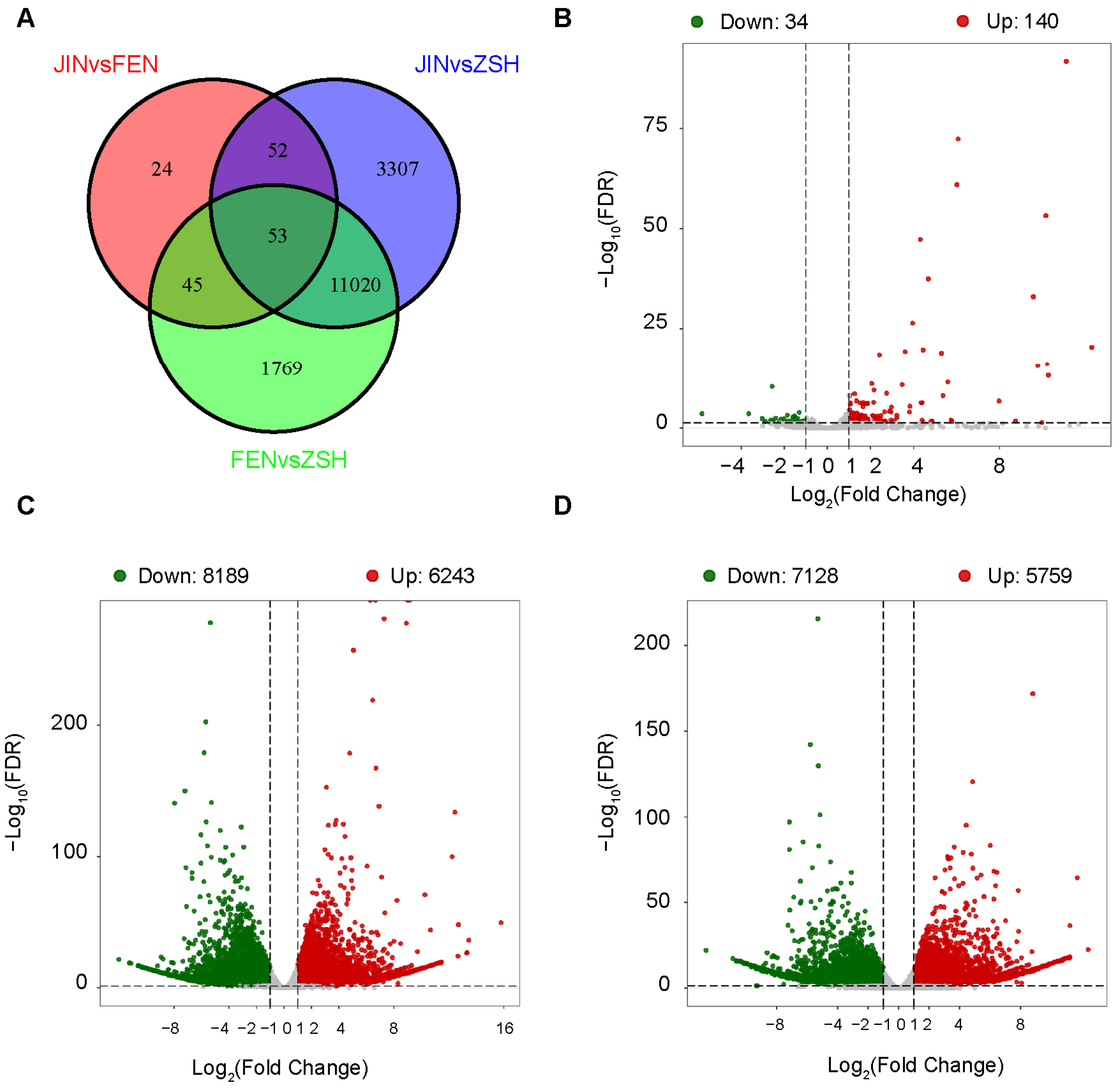
3.3. Analysis of Differentially Expressed Genes (DEGs)
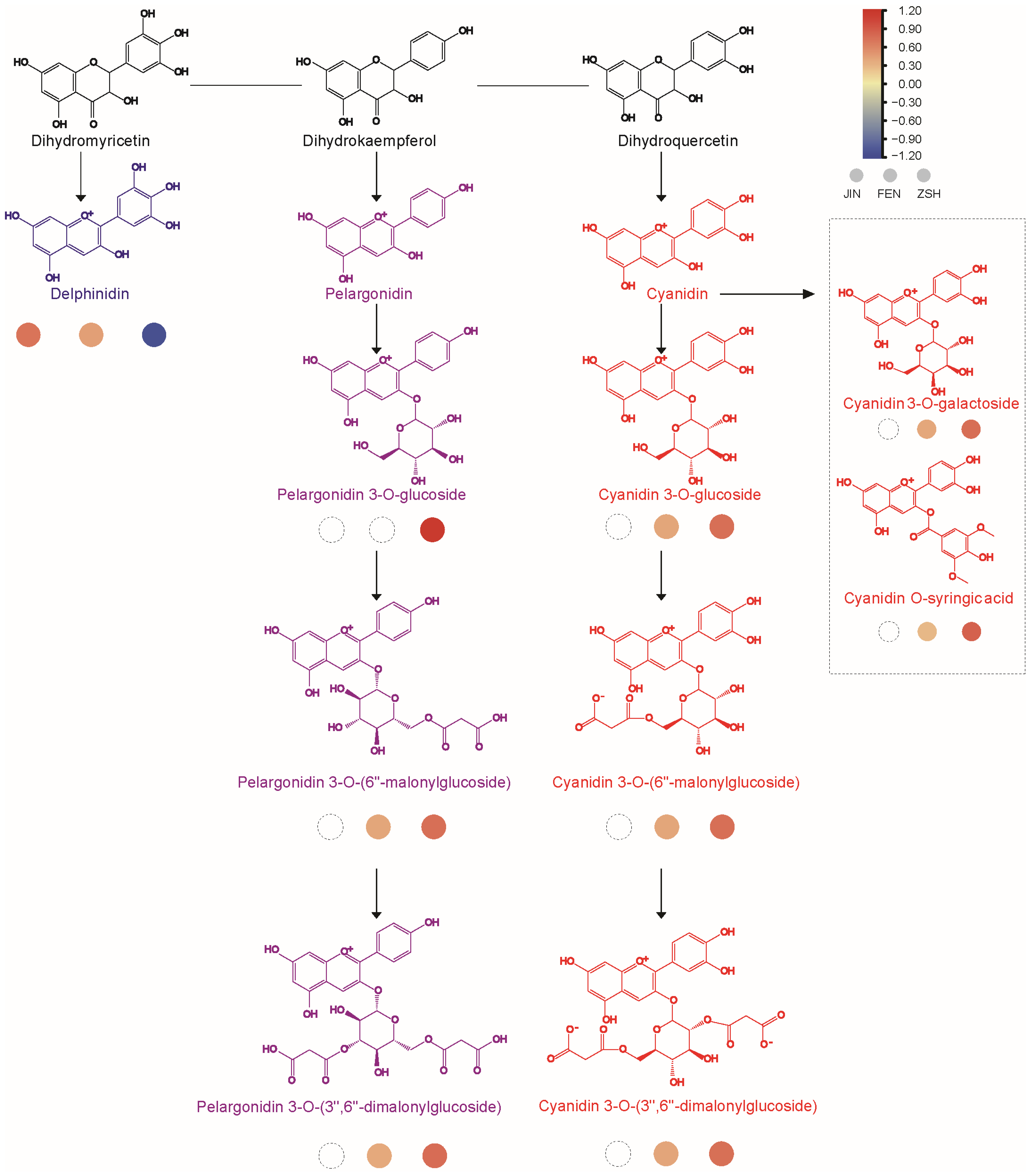
3.4. Anthocyanin Biosynthesis Pathway
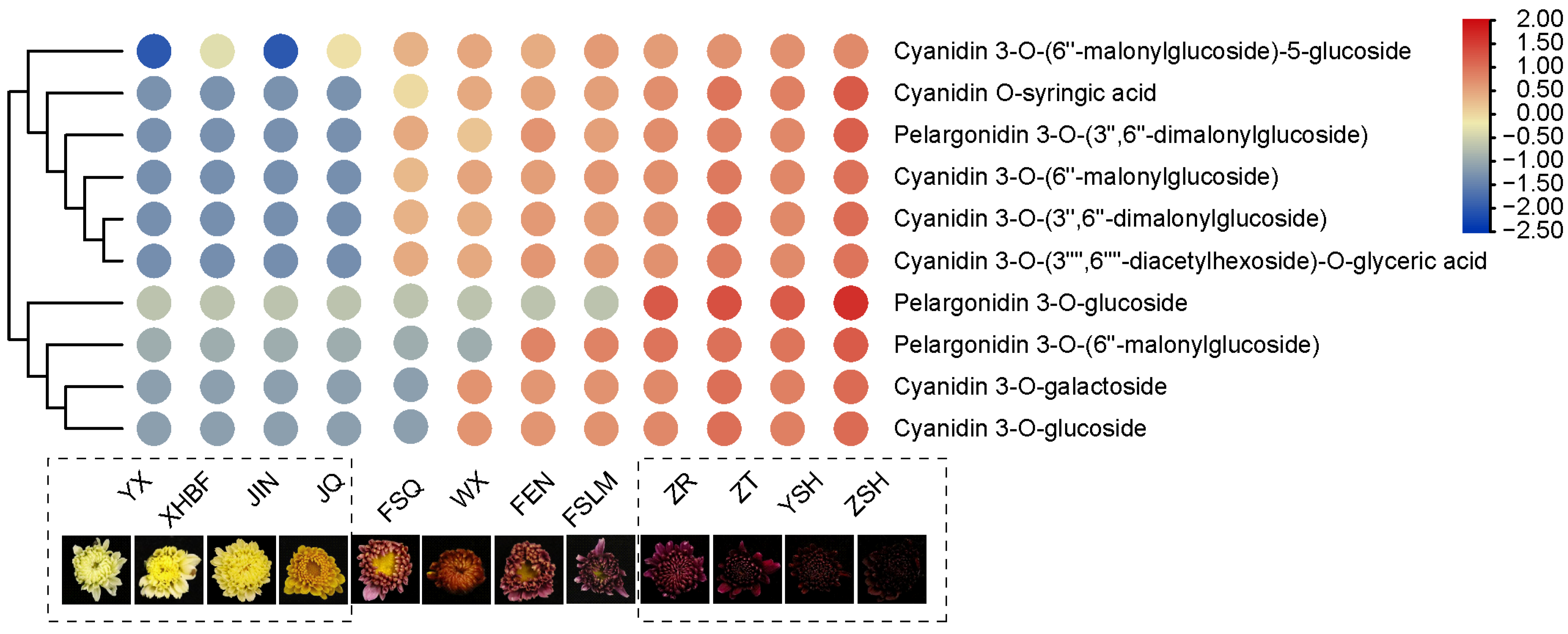
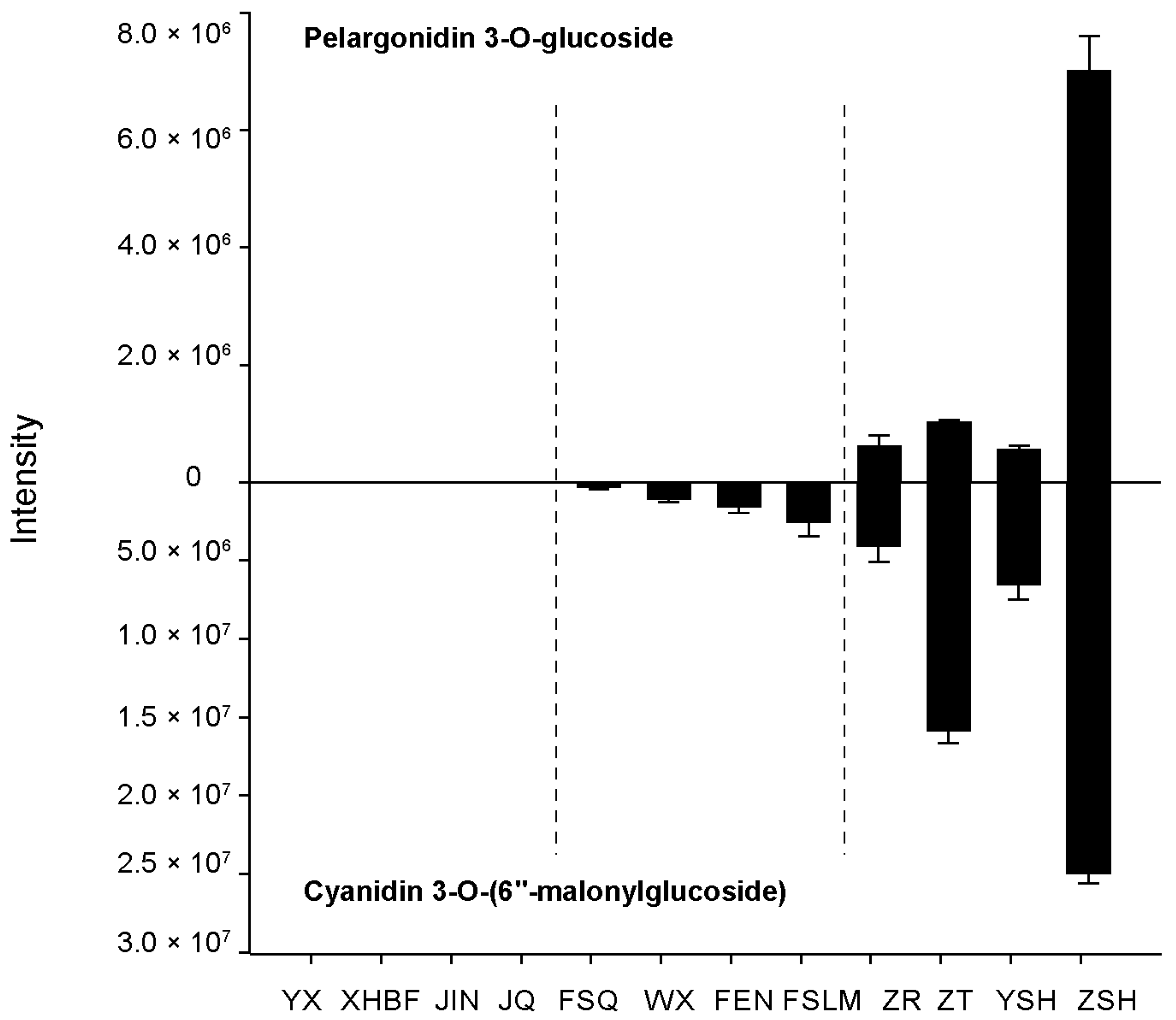
3.5. Verification of Metabolite Accumulation Patterns with Nine Different Color Cultivars and Identification of Two Metabolites as Potential Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liang, X.; Wu, H.; Su, W. A Rapid UPLC-PAD Fingerprint Analysis of Chrysanthemum Morifolium Ramat Combined with Chemometrics Methods. Food Anal. Methods 2014, 7, 197–204. [Google Scholar] [CrossRef]
- Ohmiya, A. Molecular Mechanisms Underlying the Diverse Array of Petal Colors in Chrysanthemum Flowers. Breed. Sci. 2018, 68, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Li, M.; Li, Z.; Li, S.; Zhu, Y.; Hongxu; Wang, Z. Transcriptome Analysis of the Molecular Mechanism of Chrysanthemum Flower Color Change under Short-Day Photoperiods. Plant Physiol. Biochem. 2020, 146, 315–328. [Google Scholar] [CrossRef]
- Lv, S.; Xi, L.; Nie, J.; Wen, C.; Yuan, C.; Ma, N.; Zhao, L. Isolation and expression analysis of auxin efflux trans-porter CmPIN1 in Chrysanthemum (Chrysanthemum morifolium ‘Jinba’). J. China Agric. Univ. 2016, 21, 58–66. [Google Scholar]
- Hong, Y.; Bai, X.; Sun, W.; Jia, F.; Dai, S. The Numerical Classification of Chrysanthemum Flower Color Phe-notype. Acta Hortic. Sin. 2012, 39, 1330–1340. [Google Scholar]
- Kishimoto, S.; Maoka, T.; Nakayama, M.; Ohmiya, A. Carotenoid Composition in Petals of Chrysanthemum (Dendranthema Grandiflorum (Ramat.) Kitamura). Phytochemistry 2004, 65, 2781–2787. [Google Scholar] [CrossRef]
- Nakayama, M.; Koshioka, M.; Shibata, M.; Hiradate, S.; Sugie, H.; Yamaguchi, M. Identification of Cyanidin 3-O-(3″,6″-O-Dimalonyl-β-Glucopyranoside) as a Flower Pigment of Chrysanthemum (Dendranthema Grandiflorum). Biosci. Biotechnol. Biochem. 1997, 61, 1607–1608. [Google Scholar] [CrossRef]
- Ohmiya, A.; Sasaki, K.; Nashima, K.; Oda-Yamamizo, C.; Hirashima, M.; Sumitomo, K. Transcriptome Analysis in Petals and Leaves of Chrysanthemums with Different Chlorophyll Levels. BMC Plant Biol. 2017, 17, 202. [Google Scholar] [CrossRef]
- Park, C.; Chae, S.; Park, S.-Y.; Kim, J.; Kim, Y.; Chung, S.; Arasu, M.; Al-Dhabi, N.; Park, S. Anthocyanin and Carotenoid Contents in Different Cultivars of Chrysanthemum (Dendranthema Grandiflorum Ramat.) Flower. Molecules 2015, 20, 11090–11102. [Google Scholar] [CrossRef] [PubMed]
- Feild, T.S.; Lee, D.W.; Holbrook, N.M. Why Leaves Turn Red in Autumn. The Role of Anthocyanins in Senescing Leaves of Red-Osier Dogwood. Plant Physiol. 2001, 127, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Miller, R.; Owens, S.J.; Rørslett, B. Plants and Colour: Flowers and Pollination. Opt. Laser Technol. 2011, 43, 282–294. [Google Scholar] [CrossRef]
- Steyn, W.J.; Wand, S.J.E.; Holcroft, D.M.; Jacobs, G. Anthocyanins in Vegetative Tissues: A Proposed Unified Function in Photoprotection. New Phytol. 2002, 155, 349–361. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J. Recent Advances on the Development and Regulation of Flower Color in Ornamental Plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The Origin and Evolution of Plant Flavonoid Metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent Advances on the Regulation of Anthocyanin Synthesis in Reproductive Organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Jaakola, L. New Insights into the Regulation of Anthocyanin Biosynthesis in Fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Cain, C.C.; Saslowsky, D.E.; Walker, R.A.; Shirley*, B.W. Expression of Chalcone Synthase and Chalcone Isomerase Proteins in Arabidopsis Seedlings. Plant Mol. Biol. 1997, 35, 377–381. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A Colorful Model for the Regulation and Evolution of Biochemical Pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Irmisch, S.; Ruebsam, H.; Jancsik, S.; Man Saint Yuen, M.; Madilao, L.L.; Bohlmann, J. Flavonol Biosynthesis Genes and Their Use in Engineering the Plant Antidiabetic Metabolite Montbretin A. Plant Physiol. 2019, 180, 1277–1290. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An Apple MYB Transcription Factor, MdMYB3, Is Involved in Regulation of Anthocyanin Biosynthesis and Flower Development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, Y.; Oren-Shamir, M.; Ovadia, R.; De Jong, W.; Paran, I. The A Locus That Controls Anthocyanin Accumulation in Pepper Encodes a MYB Transcription Factor Homologous to Anthocyanin2 of Petunia. Theor. Appl. Genet. 2004, 109, 23–29. [Google Scholar] [CrossRef]
- Wei, Z.-Z.; Hu, K.-D.; Zhao, D.-L.; Tang, J.; Huang, Z.-Q.; Jin, P.; Li, Y.-H.; Han, Z.; Hu, L.-Y.; Yao, G.-F.; et al. MYB44 Competitively Inhibits the Formation of the MYB340-BHLH2-NAC56 Complex to Regulate Anthocyanin Biosynthesis in Purple-Fleshed Sweet Potato. BMC Plant Biol. 2020, 20, 258. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D. Cetyltrimethyl Ammonium Bromide (CTAB) DNA Miniprep for Plant DNA Isolation. Cold Spring Harb. Protoc. 2009, pdb.prot5177. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for Linking Genomes to Life and the Environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Tang, M.; Xue, W.; Li, X.; Wang, L.; Wang, M.; Wang, W.; Yin, X.; Chen, B.; Qu, X.; Li, J.; et al. Mitotically Heritable Epigenetic Modifications of CmMYB6 Control Anthocyanin Biosynthesis in Chrysanthemum. New Phytol. 2022, 236, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Cruz, L.; Basílio, N.; Mateus, N.; De Freitas, V.; Pina, F. Natural and Synthetic Flavylium-Based Dyes: The Chemistry Behind the Color. Chem. Rev. 2022, 122, 1416–1481. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhu, J.; Li, B.; Li, S.; Yang, Y.; Wang, Q.; Xu, W.; Wang, L. Functional Identification of Anthocyanin Glucosyltransferase Genes: A Ps3GT Catalyzes Pelargonidin to Pelargonidin 3-O-Glucoside Painting the Vivid Red Flower Color of Paeonia. Planta 2023, 257, 65. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Q.; Fu, N.; Song, M.; Han, X.; Yang, Q.; Zhang, Y.; Tong, Z.; Zhang, J. Cyanidin-3-O-Glucoside Contributes to Leaf Color Change by Regulating Two BHLH Transcription Factors in Phoebe Bournei. Int. J. Mol. Sci. 2023, 24, 3829. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum Biotechnology: Quo Vadis? Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Harnly, J.M. Identification of the Phenolic Components of Chrysanthemum Flower (Chrysanthemum Morifolium Ramat). Food Chem. 2010, 120, 319–326. [Google Scholar] [CrossRef]
- Su, J.; Jiang, J.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.; Chen, F. Current Achievements and Future Prospects in the Genetic Breeding of Chrysanthemum: A Review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple Functional Roles of Anthocyanins in Plant-Environment Interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Putri, S.P.; Yamamoto, S.; Tsugawa, H.; Fukusaki, E. Current Metabolomics: Technological Advances. J. Biosci. Bioeng. 2013, 116, 9–16. [Google Scholar] [CrossRef]
- Shi, Y.; Guo, Y.; Wang, Y.; Li, M.; Li, K.; Liu, X.; Fang, C.; Luo, J. Metabolomic Analysis Reveals Nutritional Diversity among Three Staple Crops and Three Fruits. Foods 2022, 11, 550. [Google Scholar] [CrossRef]
- Hong, S.; Wang, J.; Wang, Q.; Zhang, G.; Zhao, Y.; Ma, Q.; Wu, Z.; Ma, J.; Gu, C. Decoding the Formation of Diverse Petal Colors of Lagerstroemia Indica by Integrating the Data from Transcriptome and Metabolome. Front. Plant Sci. 2022, 13, 970023. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Q.; Xie, X.; Cai, Y.; Li, J.; Feng, Y.; Zhang, Y. Integrated Metabolomics and Transcriptomics Analyses Reveal the Molecular Mechanisms Underlying the Accumulation of Anthocyanins and Other Flavonoids in Cowpea Pod ( Vigna Unguiculata L.). J. Agric. Food Chem. 2020, 68, 9260–9275. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, K.; Zhou, X.; Fang, C. Integrative Analysis of Metabolome and Transcriptome Reveals the Role of Strigolactones in Wounding-Induced Rice Metabolic Re-Programming. Metabolites 2022, 12, 789. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guo, Z.; Wang, K.; Wang, R.; Fang, C. Comparative Metabolomic Analysis Reveals the Role of OsHPL1 in the Cold-Induced Metabolic Changes in Rice. Plants 2023, 12, 2032. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, D.; Zhuang, F.; Wang, J.; Gao, R.; Zhang, Q.; Wang, X.; Zhang, G.; Fang, M.; Zhang, Y.; Li, Y.; et al. Metabolomics and Transcriptomics Revealed a Comprehensive Understanding of the Biochemical and Genetic Mechanisms Underlying the Color Variations in Chrysanthemums. Metabolites 2023, 13, 742. https://doi.org/10.3390/metabo13060742
Wu D, Zhuang F, Wang J, Gao R, Zhang Q, Wang X, Zhang G, Fang M, Zhang Y, Li Y, et al. Metabolomics and Transcriptomics Revealed a Comprehensive Understanding of the Biochemical and Genetic Mechanisms Underlying the Color Variations in Chrysanthemums. Metabolites. 2023; 13(6):742. https://doi.org/10.3390/metabo13060742
Chicago/Turabian StyleWu, Di, Fengchao Zhuang, Jiarui Wang, Ruiqi Gao, Qiunan Zhang, Xiao Wang, Guochao Zhang, Minghui Fang, Yang Zhang, Yuhua Li, and et al. 2023. "Metabolomics and Transcriptomics Revealed a Comprehensive Understanding of the Biochemical and Genetic Mechanisms Underlying the Color Variations in Chrysanthemums" Metabolites 13, no. 6: 742. https://doi.org/10.3390/metabo13060742
APA StyleWu, D., Zhuang, F., Wang, J., Gao, R., Zhang, Q., Wang, X., Zhang, G., Fang, M., Zhang, Y., Li, Y., Guan, L., & Gao, Y. (2023). Metabolomics and Transcriptomics Revealed a Comprehensive Understanding of the Biochemical and Genetic Mechanisms Underlying the Color Variations in Chrysanthemums. Metabolites, 13(6), 742. https://doi.org/10.3390/metabo13060742





