Targeted Metabolomics in High Performance Sports: Differences between the Resting Metabolic Profile of Endurance- and Strength-Trained Athletes in Comparison with Sedentary Subjects over the Course of a Training Year
Abstract
1. Introduction
2. Materials and Methods
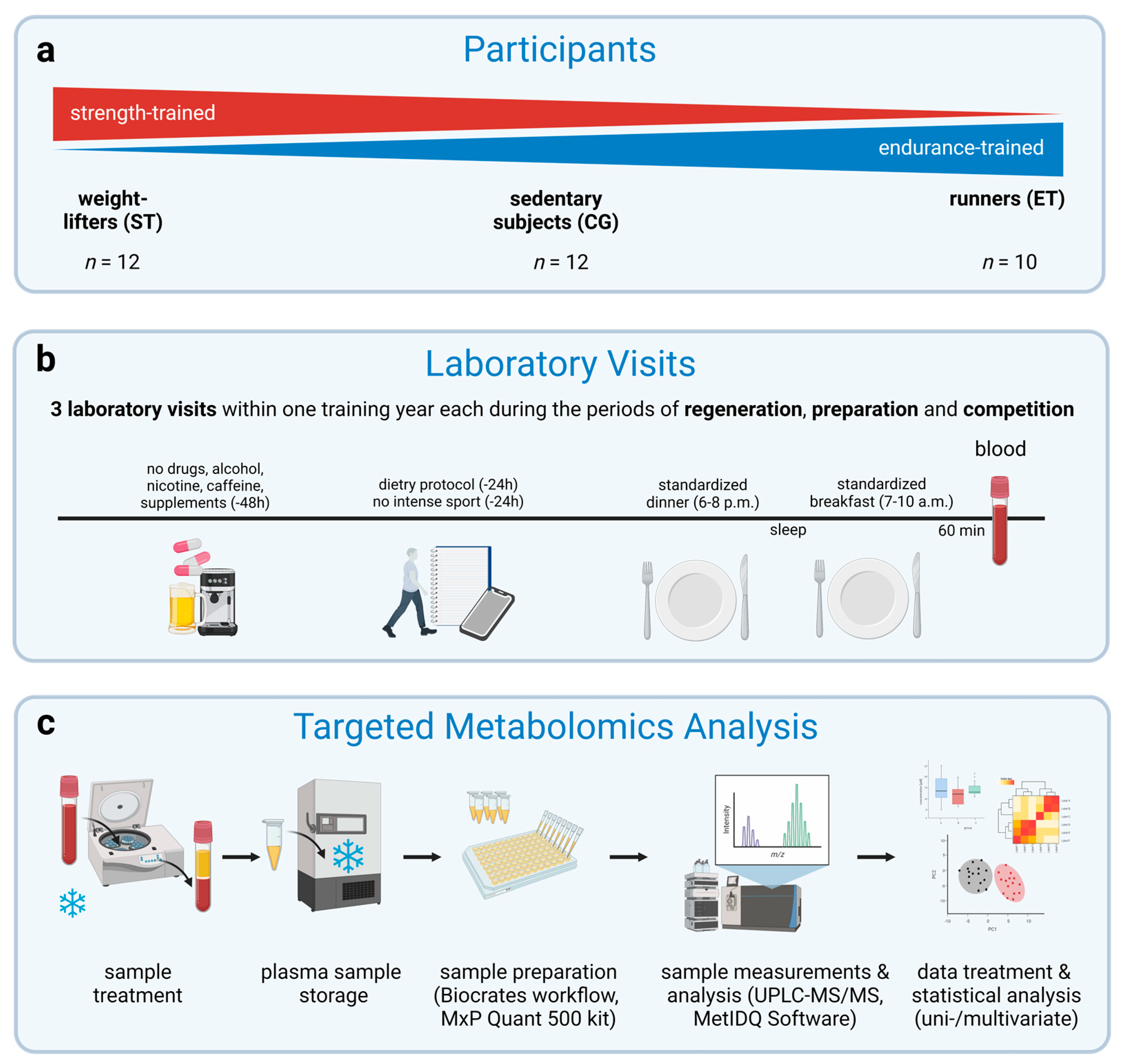
2.1. Participants
2.2. Experimental Design
2.3. Preliminary Testing
2.4. Standardization
2.5. Sample Handling
2.6. Targeted Metabolomics Analysis
2.7. Statistical Analysis
3. Results
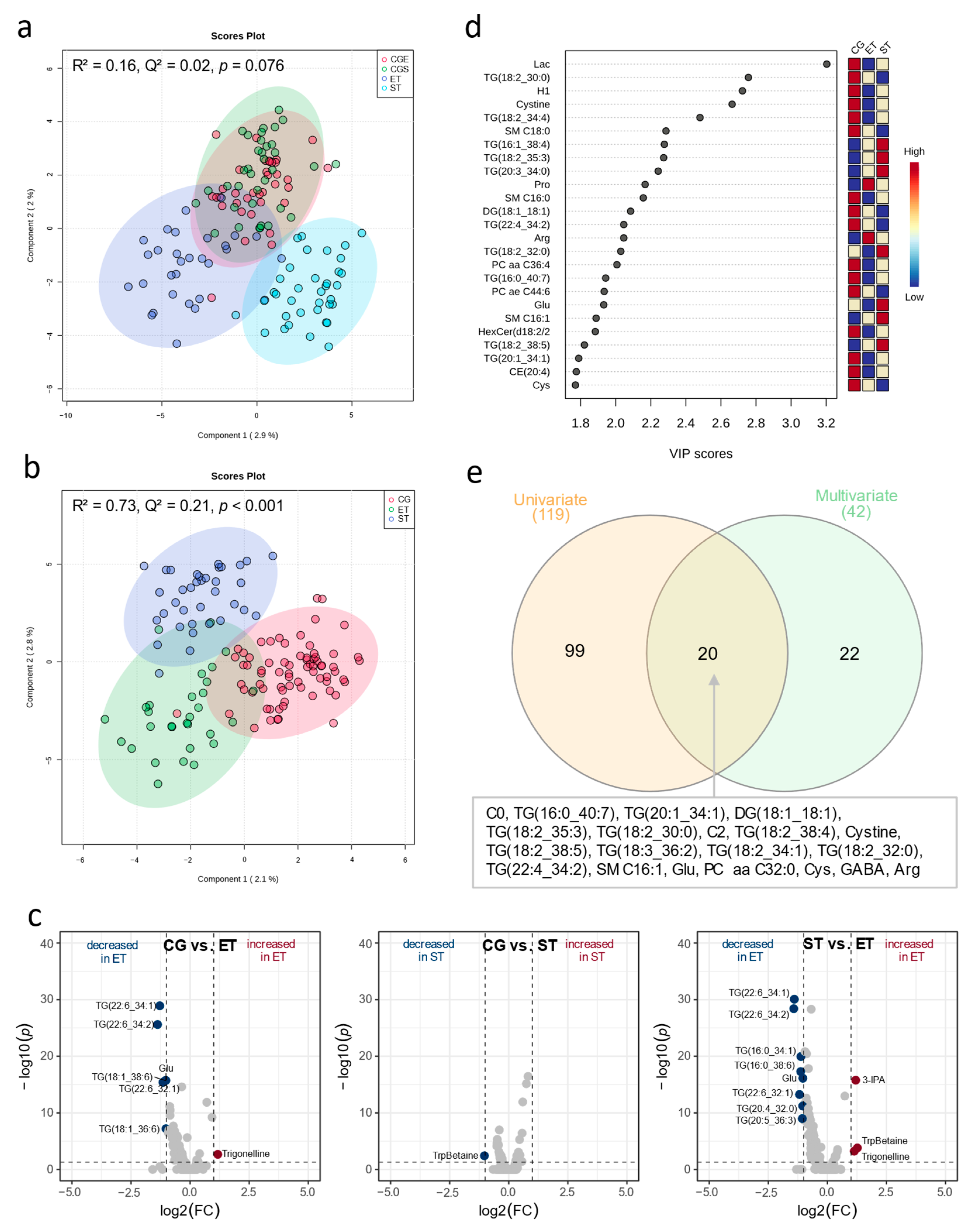
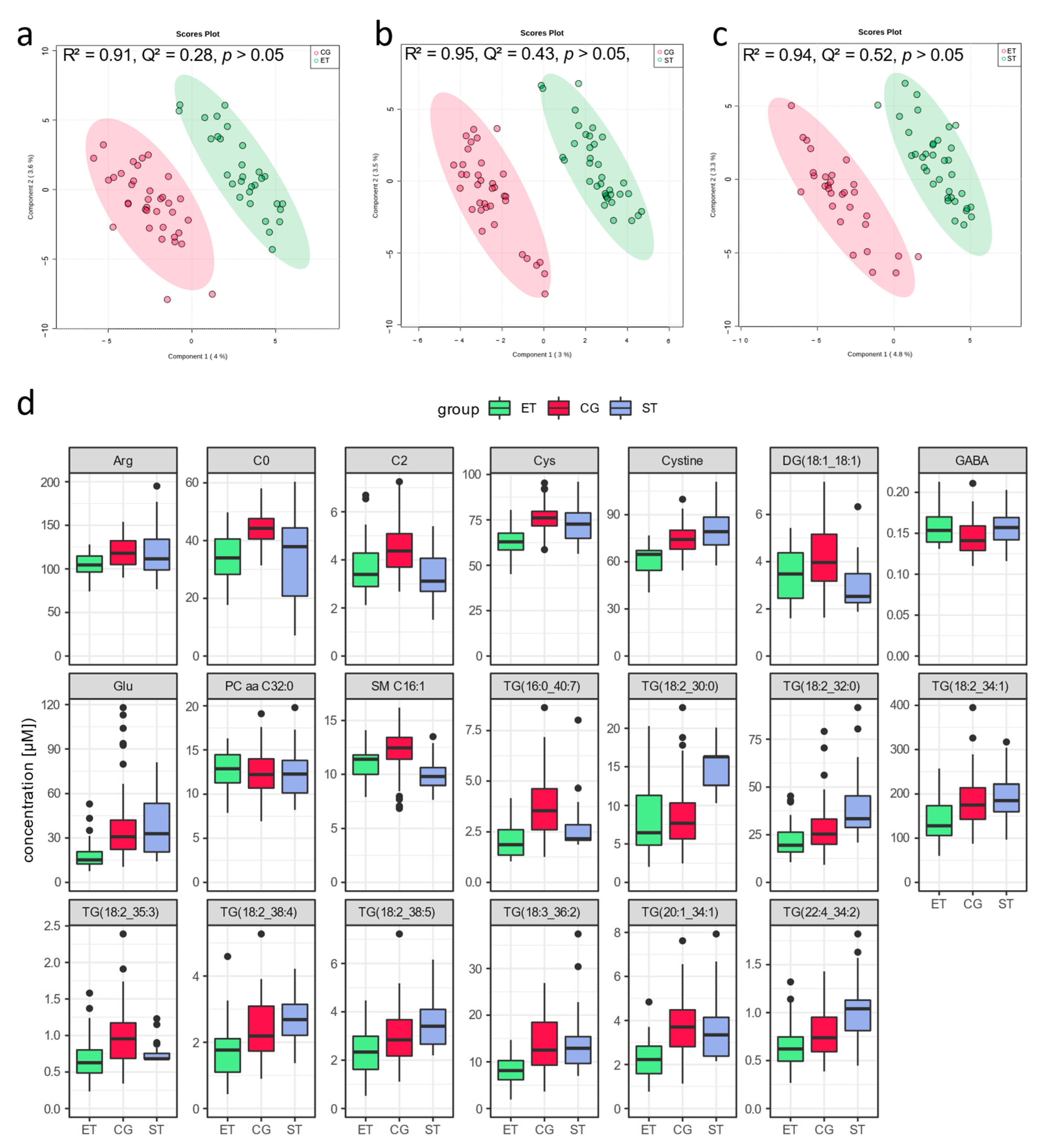
3.1. Resting Metabolic Profile
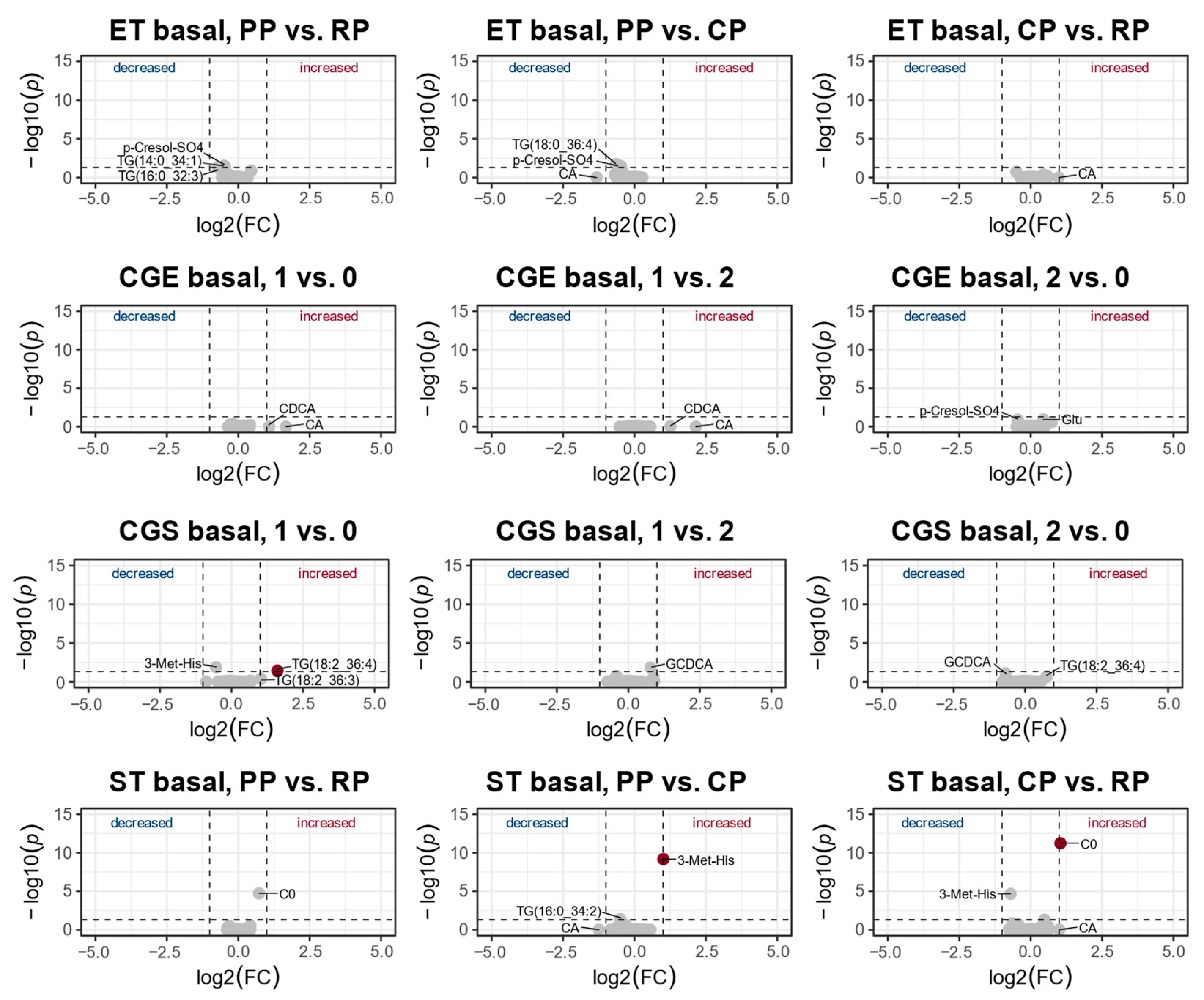
3.2. Changes in the Metabolic Profile over One Year of Training
4. Discussion
4.1. Resting Metabolic Profile
4.2. Changes of the Metabolic Profile over One Year of Training
4.3. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hughes, D.C.; Ellefsen, S.; Baar, K. Adaptations to Endurance and Strength Training. Cold Spring Harb. Perspect. Med. 2018, 8, a029769. [Google Scholar] [CrossRef] [PubMed]
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef]
- Mitchell, J.H.; Haskell, W.; Snell, P.; van Camp, S.P. Task Force 8: Classification of sports. J. Am. Coll. Cardiol. 2005, 45, 1364–1367. [Google Scholar] [CrossRef] [PubMed]
- Amann, M.; Calbet, J.A.L. Convective oxygen transport and fatigue. J. Appl. Physiol. 2008, 104, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, P.J.; Howarth, K.R.; Gibala, M.J.; Heigenhauser, G.J.F. Effects of 7 wk of endurance training on human skeletal muscle metabolism during submaximal exercise. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 2004, 97, 2148–2153. [Google Scholar] [CrossRef] [PubMed]
- Chesley, A.; Heigenhauser, G.J.; Spriet, L.L. Regulation of muscle glycogen phosphorylase activity following short-term endurance training. Am. J. Physiol. 1996, 270, E328–E335. [Google Scholar] [CrossRef]
- Holloszy, J.O.; Coyle, E.F. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1984, 56, 831–838. [Google Scholar] [CrossRef]
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef]
- Seynnes, O.R.; de Boer, M.; Narici, M.V. Early skeletal muscle hypertrophy and architectural changes in response to high-intensity resistance training. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 2007, 102, 368–373. [Google Scholar] [CrossRef]
- Schranner, D.; Schönfelder, M.; Römisch-Margl, W.; Scherr, J.; Schlegel, J.; Zelger, O.; Riermeier, A.; Kaps, S.; Prehn, C.; Adamski, J.; et al. Physiological extremes of the human blood metabolome: A metabolomics analysis of highly glycolytic, oxidative, and anabolic athletes. Physiol. Rep. 2021, 9, e14885. [Google Scholar] [CrossRef]
- Sakaguchi, C.A.; Nieman, D.C.; Signini, E.F.; Abreu, R.M.; Catai, A.M. Metabolomics-Based Studies Assessing Exercise-Induced Alterations of the Human Metabolome: A Systematic Review. Metabolites 2019, 9, 164. [Google Scholar] [CrossRef] [PubMed]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sport. Med. Open 2018, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Mendes, P. Snapshots of systems. In Technological and Medical Implications of Metabolic Control Analysis; Springer: Berlin/Heidelberg, Germany, 2000; pp. 3–25. [Google Scholar]
- Schranner, D.; Kastenmüller, G.; Schönfelder, M.; Römisch-Margl, W.; Wackerhage, H. Metabolite Concentration Changes in Humans After a Bout of Exercise: A Systematic Review of Exercise Metabolomics Studies. Sports Med. Open 2020, 6, 273. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Gillitt, N.D.; Knab, A.M.; Shanely, R.A.; Pappan, K.L.; Jin, F.; Lila, M.A. Influence of a polyphenol-enriched protein powder on exercise-induced inflammation and oxidative stress in athletes: A randomized trial using a metabolomics approach. PLoS ONE 2013, 8, e72215. [Google Scholar] [CrossRef]
- Nieman, D.C.; Sha, W.; Pappan, K.L. IL-6 Linkage to Exercise-Induced Shifts in Lipid-Related Metabolites: A Metabolomics-Based Analysis. J. Proteome Res. 2017, 16, 970–977. [Google Scholar] [CrossRef]
- Howe, C.C.F.; Alshehri, A.; Muggeridge, D.; Mullen, A.B.; Boyd, M.; Spendiff, O.; Moir, H.J.; Watson, D.G. Untargeted Metabolomics Profiling of an 80.5 km Simulated Treadmill Ultramarathon. Metabolites 2018, 8, 14. [Google Scholar] [CrossRef]
- Khoramipour, K.; Gaeini, A.A.; Shirzad, E.; Gilany, K.; Chashniam, S.; Sandbakk, Ø. Metabolic load comparison between the quarters of a game in elite male basketball players using sport metabolomics. Eur. J. Sport. Sci. 2021, 21, 1022–1034. [Google Scholar] [CrossRef]
- Khoramipour, K.; Gaeini, A.A.; Shirzad, E.; Gilany, K.; Chamari, K.; Sandbakk, Ø. Using Metabolomics to Differentiate Player Positions in Elite Male Basketball Games: A Pilot Study. Front. Mol. Biosci. 2021, 8, 639786. [Google Scholar] [CrossRef]
- Paris, A.; Labrador, B.; Lejeune, F.-X.; Canlet, C.; Molina, J.; Guinot, M.; Mégret, A.; Rieu, M.; Thalabard, J.-C.; Le Bouc, Y. Metabolomic signatures in elite cyclists: Differential characterization of a seeming normal endocrine status regarding three serum hormones. Metabolomics 2021, 17, 67. [Google Scholar] [CrossRef]
- Lohman, T.G. Skinfolds and body density and their relation to body fatness: A review. Hum. Biol. 1981, 53, 181–225. [Google Scholar]
- Martin, C.K.; Correa, J.B.; Han, H.; Allen, H.R.; Rood, J.C.; Champagne, C.M.; Gunturk, B.K.; Bray, G.A. Validity of the Remote Food Photography Method (RFPM) for estimating energy and nutrient intake in near real-time. Obesity 2012, 20, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Mall, C.; Taylor, S.L.; Hitchcock, S.; Zhang, C.; Wettersten, H.I.; Jones, A.D.; Chapman, A.; Weiss, R.H. Mealtime, Temporal, and Daily Variability of the Human Urinary and Plasma Metabolomes in a Tightly Controlled Environment. PLoS ONE 2014, 9, e86223. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, S.; Bobeldijk, I.; Verheij, E.R.; Ramaker, R.; Kochhar, S.; Macdonald, I.A.; van Ommen, B.; Smilde, A.K. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Anal. Chem. 2006, 78, 567–574. [Google Scholar] [CrossRef]
- Do, K.T.; Wahl, S.; Raffler, J.; Molnos, S.; Laimighofer, M.; Adamski, J.; Suhre, K.; Strauch, K.; Peters, A.; Gieger, C.; et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics 2018, 14, 128. [Google Scholar] [CrossRef]
- Shah, J.S.; Rai, S.N.; DeFilippis, A.P.; Hill, B.G.; Bhatnagar, A.; Brock, G.N. Distribution based nearest neighbor imputation for truncated high dimensional data with applications to pre-clinical and clinical metabolomics studies. BMC Bioinform. 2017, 18, 114. [Google Scholar] [CrossRef]
- Van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis–A marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Weljie, A.M.; Bondareva, A.; Zang, P.; Jirik, F.R. 1H NMR metabolomics identification of markers of hypoxia-induced metabolic shifts in a breast cancer model system. J. Biomol. NMR 2011, 49, 185–193. [Google Scholar] [CrossRef]
- Zhong, F.; Liu, X.; Zhou, Q.; Hao, X.; Lu, Y.; Guo, S.; Wang, W.; Lin, D.; Chen, N. 1H NMR spectroscopy analysis of metabolites in the kidneys provides new insight into pathophysiological mechanisms: Applications for treatment with Cordyceps sinensis. Nephrol. Dial. Transplant. 2012, 27, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Opalka, J.R.; Gellerich, F.-N.; Zierz, S. Age and Sex Dependency of Carnitine Concentration in Human Serum and Skeletal Muscle. Clin. Chem. 2001, 47, 2150–2153. [Google Scholar] [CrossRef] [PubMed]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Zammit, V.A. Carnitine acyltransferases and their influence on CoA pools in health and disease. Mol. Asp. Med. 2004, 25, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Houten, S.M.; Wanders, R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 2010, 33, 469–477. [Google Scholar] [CrossRef]
- Karlic, H.; Lohninger, A. Supplementation of L-carnitine in athletes: Does it make sense? Nutrition 2004, 20, 709–715. [Google Scholar] [CrossRef]
- Phypers, B.; Pierce, J.T. Lactate physiology in health and disease. Contin. Educ. Anaesth. Crit. Care Pain 2006, 6, 128–132. [Google Scholar] [CrossRef]
- Pucino, V.; Bombardieri, M.; Pitzalis, C.; Mauro, C. Lactate at the crossroads of metabolism, inflammation, and autoimmunity. Eur. J. Immunol. 2017, 47, 14–21. [Google Scholar] [CrossRef]
- Baltazar, F.; Afonso, J.; Costa, M.; Granja, S. Lactate Beyond a Waste Metabolite: Metabolic Affairs and Signaling in Malignancy. Front. Oncol. 2020, 10, 231. [Google Scholar] [CrossRef]
- Broskey, N.T.; Zou, K.; Dohm, G.L.; Houmard, J.A. Plasma Lactate as a Marker for Metabolic Health. Exerc. Sport. Sci. Rev. 2020, 48, 119–124. [Google Scholar] [CrossRef]
- Kano, R.; Sato, K. A Competitive Sprinter’s Resting Blood Lactate Levels Fluctuate with a One-Year Training Cycle: Case Reports. J. Funct. Morphol. Kinesiol. 2021, 6, 95. [Google Scholar] [CrossRef]
- Roepstorff, C.; Vistisen, B.; Kiens, B. Intramuscular Triacylglycerol in Energy Metabolism during Exercise in Humans. Exerc. Sport. Sci. Rev. 2005, 33, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Perreault, L.; Newsom, S.A.; Strauss, A.; Kerege, A.; Kahn, D.E.; Harrison, K.A.; Snell-Bergeon, J.K.; Nemkov, T.; D’Alessandro, A.; Jackman, M.R.; et al. Intracellular localization of diacylglycerols and sphingolipids influences insulin sensitivity and mitochondrial function in human skeletal muscle. JCI Insight 2018, 3, e96805. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Salihovic, S.; Fall, T.; Hammar, U.; Ingelsson, E.; Ärnlöv, J.; Lind, L.; Sundström, J. Global Plasma Metabolomics to Identify Potential Biomarkers of Blood Pressure Progression. Arterioscler. Thromb. Vasc. Biol. 2020, 40, e227–e237. [Google Scholar] [CrossRef] [PubMed]
- Castro-Gómez, P.; Garcia-Serrano, A.; Visioli, F.; Fontecha, J. Relevance of dietary glycerophospholipids and sphingolipids to human health. Prostaglandins Leukot. Essent. Fat. Acids 2015, 101, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Tan-Chen, S.; Guitton, J.; Bourron, O.; Le Stunff, H.; Hajduch, E. Sphingolipid Metabolism and Signaling in Skeletal Muscle: From Physiology to Physiopathology. Front. Endocrinol. 2020, 11, 491. [Google Scholar] [CrossRef] [PubMed]
- Grebely, J.; Dore, G.J. An expanding role for primary care providers in the treatment of hepatitis C virus infection in the community. Hepatology 2011, 54, 2258–2260. [Google Scholar] [CrossRef] [PubMed]
- Bye, A.; Vettukattil, R.; Aspenes, S.T.; Giskeødegård, G.F.; Gribbestad, I.S.; Wisløff, U.; Bathen, T.F. Serum Levels of Choline-Containing Compounds Are Associated with Aerobic Fitness Level: The HUNT-Study. PLoS ONE 2012, 7, e42330. [Google Scholar] [CrossRef]
- Morris, C.; O’Grada, C.M.; Ryan, M.F.; Gibney, M.J.; Roche, H.M.; Gibney, E.R.; Brennan, L. Modulation of the lipidomic profile due to a lipid challenge and fitness level: A postprandial study. Lipids Health Dis. 2015, 14, 65. [Google Scholar] [CrossRef]
- Rennie, M.J.; Tipton, K.D. Protein and amino acid metabolism during and after exercise and the effects of nutrition. Annu. Rev. Nutr. 2000, 20, 457–483. [Google Scholar] [CrossRef]
- Morris, C.; Grada, C.O.; Ryan, M.; Roche, H.M.; de Vito, G.; Gibney, M.J.; Gibney, E.R.; Brennan, L. The relationship between aerobic fitness level and metabolic profiles in healthy adults. Mol. Nutr. Food Res. 2013, 57, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Lima, M.M.; Procopio, J.; Pithon-Curi, T.C.; Doi, S.Q.; Bazotte, R.B.; Curi, R. Glutamine and glutamate as vital metabolites. Braz. J. Med. Biol. Res. 2003, 36, 153–163. [Google Scholar] [CrossRef]
- Smith, D.J.; Norris, S.R. Changes in glutamine and glutamate concentrations for tracking training tolerance. Med. Sci. Sport. Exerc. 2000, 32, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Mourtzakis, M.; Graham, T.E. Glutamate ingestion and its effects at rest and during exercise in humans. J. Appl. Physiol. 2002, 93, 1251–1259. [Google Scholar] [CrossRef]
- Cooper, A.J.L.; Jeitner, T.M. Central Role of Glutamate Metabolism in the Maintenance of Nitrogen Homeostasis in Normal and Hyperammonemic Brain. Biomolecules 2016, 6, 16. [Google Scholar] [CrossRef]
- Berridge, B.R.; van Vleet, J.F.; Herman, E. Chapter 46-Cardiac, Vascular, and Skeletal Muscle Systems. In Haschek and Rousseaux’s Handbook of Toxicologic Pathology (Third Edition); Haschek, W.M., Rousseaux, C.G., Wallig, M.A., Eds.; Academic Press: Boston, MA, USA, 2013; pp. 1567–1665. ISBN 978-0-12-415759-0. [Google Scholar]
- Kim, S.; Kim, J.; Yun, E.J.; Kim, K.H. Food metabolomics: From farm to human. Curr. Opin. Biotechnol. 2016, 37, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Malsagova, K.A.; Kopylov, A.T.; Stepanov, A.A.; Enikeev, D.V.; Potoldykova, N.V.; Balakin, E.I.; Pustovoyt, V.I.; Kaysheva, A.L. Molecular Profiling of Athletes Performing High-Intensity Exercises in Extreme Environments. Sports 2023, 11, 36. [Google Scholar] [CrossRef]
- Chinkes, D.L. Methods for measuring tissue protein breakdown rate in vivo. Curr. Opin. Clin. Nutr. Metab. Care 2005, 8, 534–537. [Google Scholar] [CrossRef]
- Raeder, C.; Wiewelhove, T.; Schneider, C.; Döweling, A.; Kellmann, M.; Meyer, T.; Pfeiffer, M.; Ferrauti, A. Effects of Active Recovery on Muscle Function Following High-Intensity Training Sessions in Elite Olympic Weightlifters; Ruhr-Universität Bochum: Bochum, Germany, 2017. [Google Scholar]
- Ra, S.-G.; Maeda, S.; Higashino, R.; Imai, T.; Miyakawa, S. Metabolomics of salivary fatigue markers in soccer players after consecutive games. Appl. Physiol. Nutr. Metab. 2014, 39, 1120–1126. [Google Scholar] [CrossRef]
- Madrid-Gambin, F.; Garcia-Aloy, M.; Vázquez-Fresno, R.; Vegas-Lozano, E.; de Villa Jubany, M.C.R.; Misawa, K.; Hase, T.; Shimotoyodome, A.; Andres-Lacueva, C. Impact of chlorogenic acids from coffee on urine metabolome in healthy human subjects. Food Res. Int. 2016, 89, 1064–1070. [Google Scholar] [CrossRef]
- Sato, S.; Parr, E.B.; Devlin, B.L.; Hawley, J.A.; Sassone-Corsi, P. Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Mol. Metab. 2018, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parstorfer, M.; Poschet, G.; Kronsteiner, D.; Brüning, K.; Friedmann-Bette, B. Targeted Metabolomics in High Performance Sports: Differences between the Resting Metabolic Profile of Endurance- and Strength-Trained Athletes in Comparison with Sedentary Subjects over the Course of a Training Year. Metabolites 2023, 13, 833. https://doi.org/10.3390/metabo13070833
Parstorfer M, Poschet G, Kronsteiner D, Brüning K, Friedmann-Bette B. Targeted Metabolomics in High Performance Sports: Differences between the Resting Metabolic Profile of Endurance- and Strength-Trained Athletes in Comparison with Sedentary Subjects over the Course of a Training Year. Metabolites. 2023; 13(7):833. https://doi.org/10.3390/metabo13070833
Chicago/Turabian StyleParstorfer, Mario, Gernot Poschet, Dorothea Kronsteiner, Kirsten Brüning, and Birgit Friedmann-Bette. 2023. "Targeted Metabolomics in High Performance Sports: Differences between the Resting Metabolic Profile of Endurance- and Strength-Trained Athletes in Comparison with Sedentary Subjects over the Course of a Training Year" Metabolites 13, no. 7: 833. https://doi.org/10.3390/metabo13070833
APA StyleParstorfer, M., Poschet, G., Kronsteiner, D., Brüning, K., & Friedmann-Bette, B. (2023). Targeted Metabolomics in High Performance Sports: Differences between the Resting Metabolic Profile of Endurance- and Strength-Trained Athletes in Comparison with Sedentary Subjects over the Course of a Training Year. Metabolites, 13(7), 833. https://doi.org/10.3390/metabo13070833




