Multi-Modality, Multi-Dimensional Characterization of Pediatric Non-Alcoholic Fatty Liver Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Experimental Measurements
2.3. Analysis
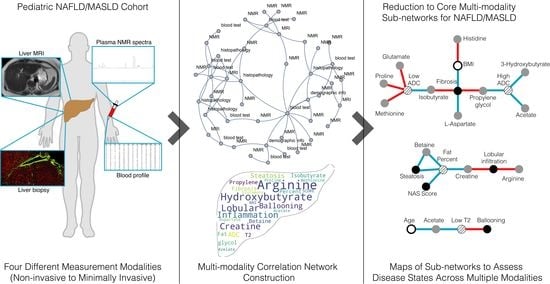
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADC | apparent diffusion coefficient |
| GLP-1 | glucagon-like peptide-1 |
| GLP-1 RA | glucagon-like peptide-1 receptor agonist |
| MAFLD | metabolic dysfunction-associated fatty liver disease |
| MASLD | metabolic dysfunction-associated steatotic liver disease |
| MCN | multimodal correlation network |
| MRI | magnetic resonance imaging |
| NAFLD | non-alcoholic fatty liver disease |
| NASH | non-alcoholic steatohepatitis |
| NMR | nuclear magnetic resonance |
| PCA | principal component analysis |
| ROS | reaction oxygen species |
References
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef] [PubMed]
- Nobili, V.; Alisi, A.; Valenti, L.; Miele, L.; Feldstein, A.E.; Alkhouri, N. NAFLD in children: New genes, new diagnostic modalities and new drugs. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J.; Van Natta, M.L.; Clark, J.; Neuschwander-Tetri, B.A.; Diehl, A.; Dasarathy, S.; Loomba, R.; Chalasani, N.; Kowdley, K.; Hameed, B.; et al. Prospective Study of Outcomes in Adults with Nonalcoholic Fatty Liver Disease. N. Engl. J. Med. 2021, 385, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Giardina, B.; Gevi, F.; Timperio, A.M.; Zolla, L. Clinical metabolomics: The next stage of clinical biochemistry. Blood Transfus. 2012, 10 (Suppl. S2), s19–s24. [Google Scholar] [PubMed]
- Nash, M.J.; Dobrinskikh, E.; Janssen, R.C.; Lovell, M.A.; Schady, D.A.; Levek, C.; Jones, K.L.; D’Alessandro, A.; Kievit, P.; Aagaard, K.M.; et al. Maternal Western diet is associated with distinct preclinical pediatric NAFLD phenotypes in juvenile nonhuman primate offspring. Hepatol. Commun. 2023, 7, e0014. [Google Scholar] [CrossRef]
- Diehl, A.M.; Day, C. Cause, Pathogenesis, and Treatment of Nonalcoholic Steatohepatitis. N. Engl. J. Med. 2017, 377, 2063–2072. [Google Scholar] [CrossRef]
- Mosca, A.; Panera, N.; Crudele, A.; Alisi, A. Noninvasive diagnostic tools for pediatric NAFLD: Where are we now? Expert Rev. Gastroenterol. Hepatol. 2020, 14, 1035–1046. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Makhlouf, H.R. Histology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis in adults and children. Clin. Liver Dis. 2016, 20, 293–312. [Google Scholar] [CrossRef]
- Manning, P.; Murphy, P.; Wang, K.; Hooker, J.; Wolfson, T.; Middleton, M.S.; Newton, K.P.; Behling, C.; Awai, H.I.; Durelle, J.; et al. Liver histology and diffusion-weighted MRI in children with nonalcoholic fatty liver disease: A MAGNET study. J. Magn. Reson. Imaging 2017, 46, 1149–1158. [Google Scholar] [CrossRef]
- Reeder, S.B.; Cruite, I.; Hamilton, G.; Sirlin, C.B. Quantitative Assessment of Liver Fat with Magnetic Resonance Imaging and Spectroscopy. J. Magn. Reson. Imaging 2011, 34, 729–749. [Google Scholar] [CrossRef]
- Ipsen, D.H.; Lykkesfeldt, J.; Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 2018, 75, 3313–3327. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, C.; Bullo, M.; Tinahones, F.J.; Martinez-Gonzalez, M.A.; Corella, D.; Fragkiadakis, G.A.; Lopez-Miranda, J.; Estruch, R.; Fito, M.; Jordi Salas-Salvado, J. Serum metabolites in non-alcoholic fatty- liver disease development or reversion; a targeted metabolomic approach within the PREDIMED trial. Nutr. Metab. 2017, 14, 58. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Noto, A.; Ibba, L.; Deidda, M.; Fanos, V.; Muntoni, S.; Leoni, V.P.; Atzori, L. Contribution of Metabolomics to the Understanding of NAFLD and NASH Syndromes: A Systematic Review. Metabolites 2021, 11, 694. [Google Scholar] [CrossRef]
- Loscalzo, J.; Barabasi, A.L. Systems biology and the future of medicine. In Wiley Interdisciplinary Reviews: Systems Biology and Medicine; John Wiley & Sons: New York, NY, USA, 2011; Volume 3, pp. 619–627. [Google Scholar]
- Silverman, E.K.; Schmidt, H.; Anastasiadou, E.; Altucci, L.; Angelini, M.; Badimon, L.; Balligand, J.L.; Benincasa, G.; Capasso, G.; Conte, F.; et al. Molecular networks in Network Medicine: Development and applications. In Wiley Interdisciplinary Reviews: Systems Biology and Medicine; John Wiley & Sons: New York, NY, USA, 2020; Volume 12, p. e1489. [Google Scholar]
- Jamshidi, N.; Miller, F.J.; Mandel, J.; Evans, T.; Kuo, M.D. Individualized therapy of HHT driven by network analysis of metabolomic profiles. BMC Syst. Biol. 2011, 5, 200. [Google Scholar] [CrossRef]
- Bydder, M.; Yokoo, T.; Hamilton, G.; Middleton, M.S.; Chavez, A.D.; Schwimmer, J.B.; Lavine, J.E.; Sirlin, C.B. Relaxation effects in the quantification of fat using gradient echo imaging. Magn. Reson. Imaging 2008, 26, 347–359. [Google Scholar] [CrossRef]
- Moran-Lev, H.; Cohen, S.; Webb, M.; Yerushalmy-Feler, A.; Amir, A.; Gal, D.L.; Lubetzky, R. Higher BMI predicts liver fibrosis among obese children and adolescents with NAFLD—An interventional pilot study. BMC Pediatr. 2021, 21, 385. [Google Scholar] [CrossRef]
- Nobili, V.; Alisi, A.; Newton, K.P.; Schwimmer, J.B. Comparison of the phenotype and approach to pediatric vs adult patients with nonalcoholic fatty liver disease. Gastroenterology 2016, 150, 1798–1810. [Google Scholar] [CrossRef]
- Parlati, L.; Regnier, M.; Guillou, H.; Postic, C. New targets for NAFLD. JHEP Rep. 2021, 3, 100346. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Kang, E.S.; Boutari, C.; Rhee, E.J.; Mantzoros, C.S. Current and emerging pharmacological options for the treatment of nonalcoholic steatohepatitis. Metabolism 2020, 111S, 154203. [Google Scholar] [CrossRef]
- Tacke, F.; Weiskirchen, R. Non-alcoholic fatty liver disease (NAFLD)/non-alcoholic steatohepatitis (NASH)-related liver fibrosis: Mechanisms, treatment and prevention. Ann. Transl. Med. 2021, 9, 729. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J.; Chalasani, N.; Kowdley, K.V.; McCullough, A.; Diehl, A.M.; Bass, N.M.; Neuschwander-Tetri, B.A.; Lavine, J.E.; Tonascia, J.; Unalp, A.; et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N. Engl. J. Med. 2010, 362, 1675–1685. [Google Scholar] [CrossRef] [PubMed]
- Shum, M.; Ngo, J.; Shirihai, O.S.; Liesa, M. Mitochondrial oxidative function in NAFLD: Friend or foe? Mol. Metab. 2021, 50, 101134. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism 2011, 60, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Männistö, V.T.; Simonen, M.; Hyysalo, J.; Soininen, P.; Kangas, A.J.; Kaminska, D.; Matte, A.K.; Venesmaa, S.; Käkelä, P.; Kärjä, V.; et al. Ketone body production is differentially altered in steatosis and non-alcoholic steatohepatitis in obese humans. Liver Int. 2015, 35, 1853–1861. [Google Scholar] [CrossRef]
- Tokushige, K.; Hashimoto, E.; Kodama, K.; Tobari, M.; Matsushita, N.; Kogiso, T.; Taniai, M.; Torii, N.; Shiratori, K.; Nishizaki, Y.; et al. Serum metabolomic profile and potential biomarkers for severity of fibrosis in nonalcoholic fatty liver disease. J. Gastroenterol. 2013, 48, 1392–1400. [Google Scholar] [CrossRef]
- Crespo, M.; Lappe, S.; Feldstein, A.E.; Alkhouri, N. Similarities and differences between pediatric and adult nonalcoholic fatty liver disease. Metabolism 2016, 65, 1161–1171. [Google Scholar] [CrossRef]
- Marques, P.; Francisco, V.; Martínez-Arenas, L.; Carvalho-Gomes, A.; Domingo, E.; Piqueras, L.; Berenguer, M.; Sanz, M.J. Overview of Cellular and Soluble Mediators in Systemic Inflammation Associated with Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2023, 24, 2313. [Google Scholar] [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wai-Sun Wong, V.; Dufour, J.F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef]
- Eslam, M.; Alkhouri, N.; Vajro, P.; Baumann, U.; Weiss, R.; Socha, P.; Marcus, C.; Lee, W.S.; Kelly, D.; Porta, G.; et al. Defining paediatric metabolic (dysfunction)-associated fatty liver disease: An international expert consensus statement. Lancet Gastroenterol. Hepatol. 2021, 6, 864–873. [Google Scholar] [CrossRef]
- Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; Arab, J.P.; et al. A multi-society Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 2023, 101133. [Google Scholar] [CrossRef]
- Di Sessa, A.; Cirillo, G.; Guarino, S.; Marzuillo, P.; Miraglia Del Giudice, E. Pediatric non-alcoholic fatty liver disease: Current perspectives on diagnosis and management. Pediatr. Health Med. Ther. 2019, 10, 89–97. [Google Scholar] [CrossRef]
- Patel Chavez, C.; Cusi, K.; Kadiyala, S. The Emerging Role of Glucagon-like Peptide-1 Receptor Agonists for the Management of NAFLD. J. Clin. Endocrinol. Metab. 2022, 107, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Petracca, G.; Beatrice, G.; Csermely, A.; Lonardo, A.; Targher, G. Glucagon-Like Peptide-1 Receptor Agonists for Treatment of Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis: An Updated Meta-Analysis of Randomized Controlled Trials. Metabolites 2021, 11, 73. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Agren, R.; Kampf, C.; Asplund, A.; Uhlen, M.; Nielsen, J. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 3083. [Google Scholar] [CrossRef]
- Maldonado, E.M.; Fisher, C.P.; Mazzatti, D.J.; Barber, A.L.; Tindall, M.J.; Plant, N.J.; Kierzek, A.M.; Moore, J.B. Multi-scale, whole-system models of liver metabolic adaptation to fat and sugar in non-alcoholic fatty liver disease. NPJ Syst. Biol. Appl. 2018, 4, 33. [Google Scholar] [CrossRef]
- Berndt, N.; Bulik, S.; Wallach, I.; Wünsch, T.; König, M.; Stockmann, M.; Meierhofer, D.; Holzhütter, H.G. HEPATOKIN1 is a biochemistry-based model of liver metabolism for applications in medicine and pharmacology. Nat. Commun. 2018, 9, 2386. [Google Scholar] [CrossRef]
- Valkovic, L.; Gajdosik, M.; Traussnigg, S.; Wolf, P.; Chmelik, M.; Kienbacher, C.; Bogner, W.; Krebs, M.; Trauner, M.; Trattnig, S.; et al. Application of localized (3)(1)P MRS saturation transfer at 7 T for measurement of ATP metabolism in the liver: Reproducibility and initial clinical application in patients with non-alcoholic fatty liver disease. Eur. Radiol. 2014, 24, 1602–1609. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Fowler, K.J.; Ozturk, A.; Potu, C.K.; Louie, A.L.; Montes, V.; Henderson, W.C.; Wang, K.; Andre, M.P.; Samir, A.E.; et al. Liver fibrosis imaging: A clinical review of ultrasound and magnetic resonance elastography. J. Magn. Reson. Imaging 2020, 51, 25–42. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamshidi, N.; Feizi, A.; Sirlin, C.B.; Lavine, J.E.; Kuo, M.D. Multi-Modality, Multi-Dimensional Characterization of Pediatric Non-Alcoholic Fatty Liver Disease. Metabolites 2023, 13, 929. https://doi.org/10.3390/metabo13080929
Jamshidi N, Feizi A, Sirlin CB, Lavine JE, Kuo MD. Multi-Modality, Multi-Dimensional Characterization of Pediatric Non-Alcoholic Fatty Liver Disease. Metabolites. 2023; 13(8):929. https://doi.org/10.3390/metabo13080929
Chicago/Turabian StyleJamshidi, Neema, Alborz Feizi, Claude B. Sirlin, Joel E. Lavine, and Michael D. Kuo. 2023. "Multi-Modality, Multi-Dimensional Characterization of Pediatric Non-Alcoholic Fatty Liver Disease" Metabolites 13, no. 8: 929. https://doi.org/10.3390/metabo13080929
APA StyleJamshidi, N., Feizi, A., Sirlin, C. B., Lavine, J. E., & Kuo, M. D. (2023). Multi-Modality, Multi-Dimensional Characterization of Pediatric Non-Alcoholic Fatty Liver Disease. Metabolites, 13(8), 929. https://doi.org/10.3390/metabo13080929