Metabolomic Changes in Plasma of Preterminal Stage of Rhesus Nonhuman Primates Exposed to Lethal Dose of Radiation
Abstract
:1. Introduction
2. Materials and Methods
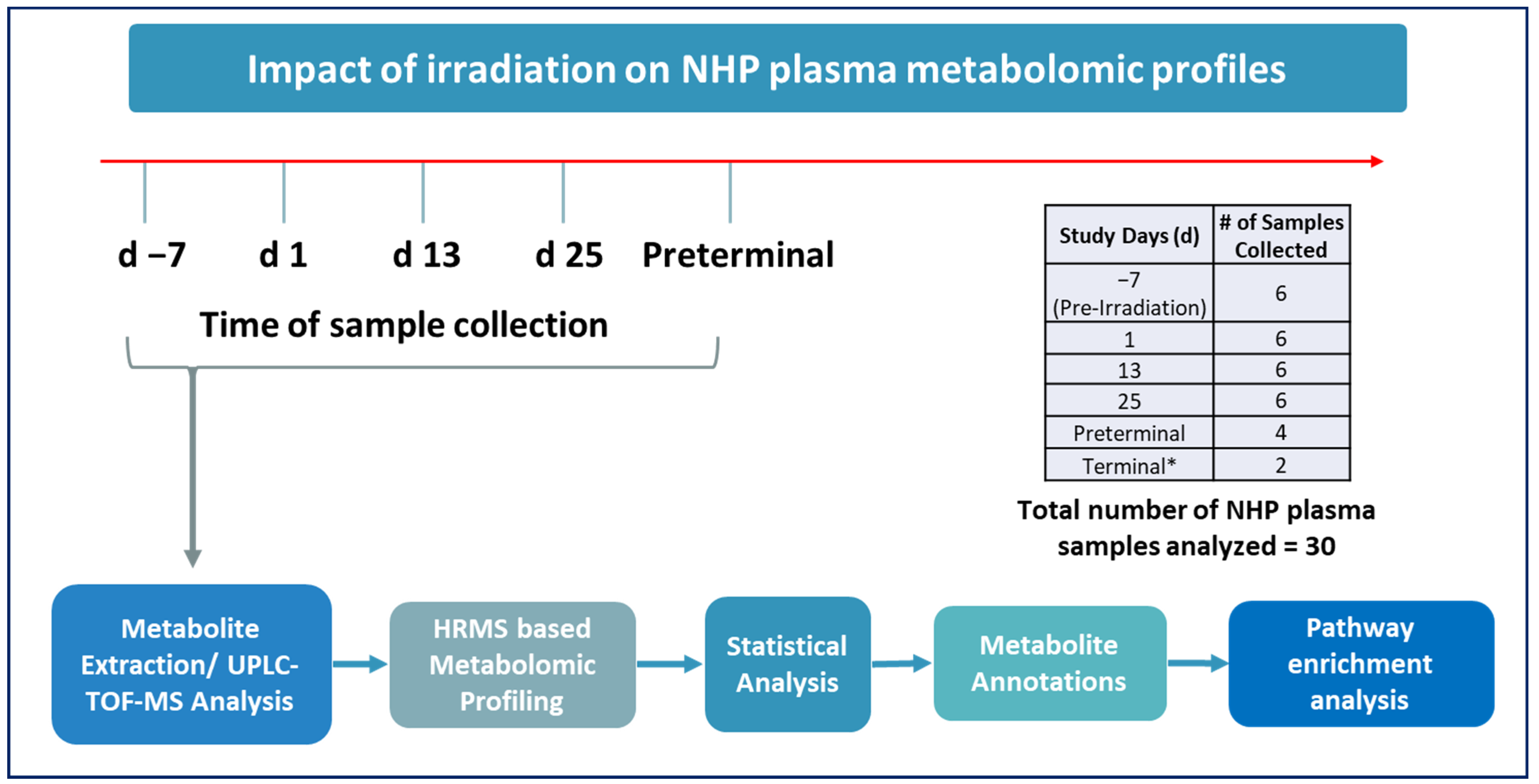
2.1. Experimental Design
2.2. Animals
2.3. Irradiation
2.4. Cage-Side Animal Observations
2.5. Blood Sample Collection
2.6. Euthanasia
2.7. Sample Preparation for LC-MS Analysis
2.8. Plasma Metabolomics Using UPLC QTOF Analysis
2.9. Statistical Analysis
3. Results
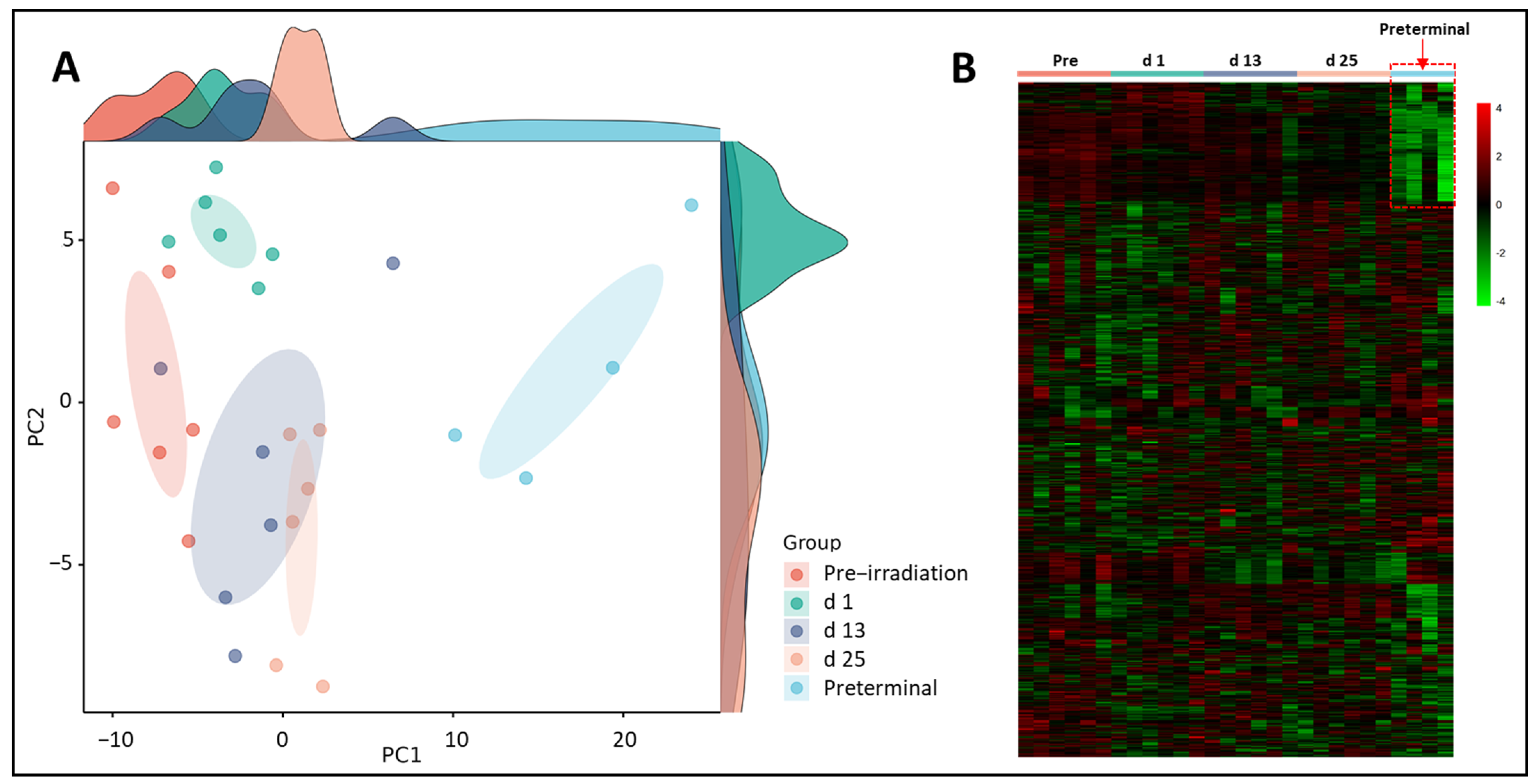
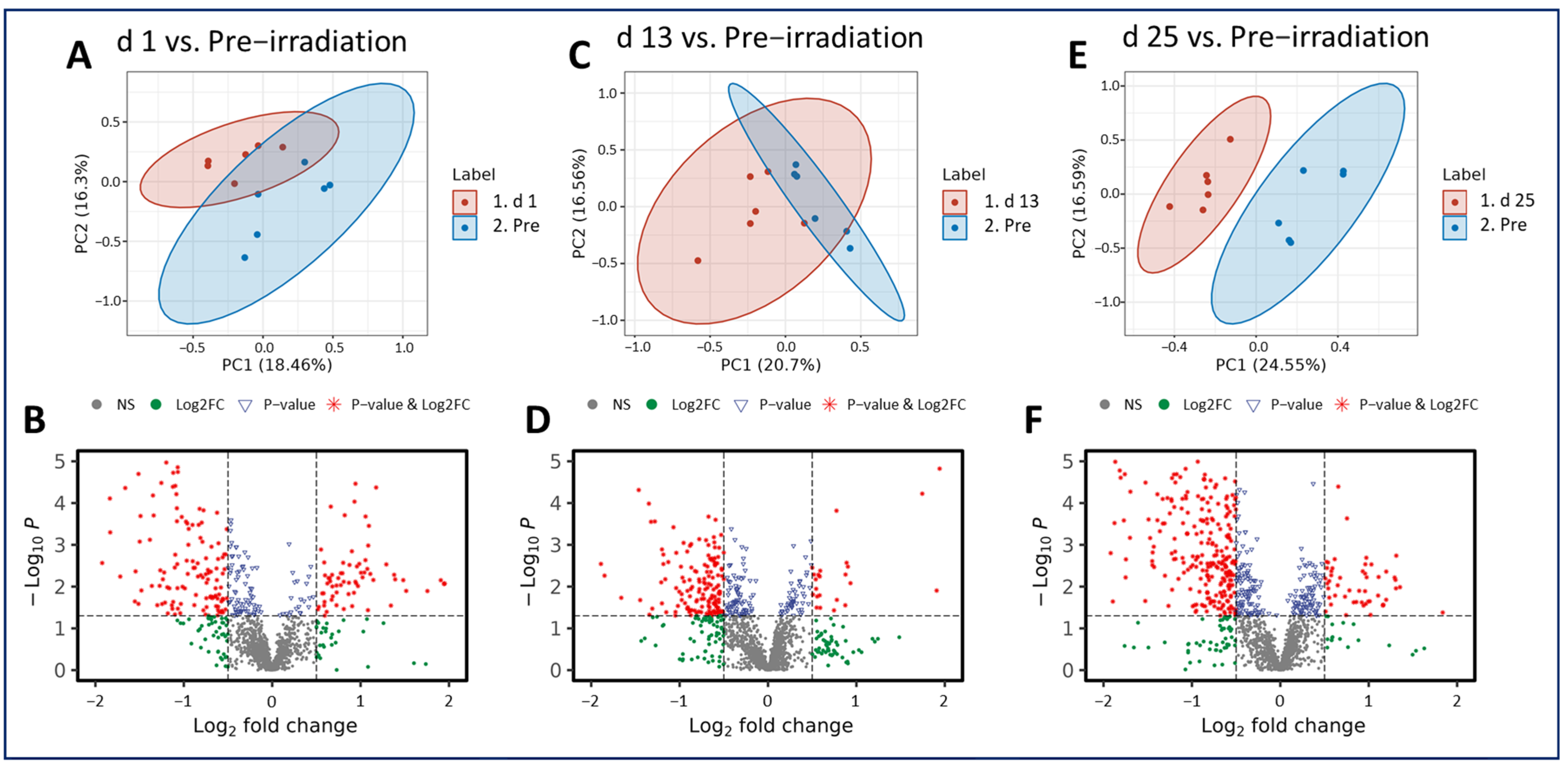
3.1. Putative Biochemical Changes Associated with Ionizing Radiation Exposure
3.2. Putative Biochemical Changes Associated with the Preterminal State after Ionizing Radiation Exposure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Health and Human Services. HHS Enhances Nation’s Health Preparedness for Radiological Threats. Available online: https://www.hhs.gov/about/news/2016/10/06/hhs-enhances-nation-s-health-preparedness-radiological-threats.html (accessed on 20 October 2023).
- Gosden, C.; Gardener, D. Weapons of mass destruction–threats and responses. BMJ 2005, 331, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Gale, R.P.; Armitage, J.O.; Hashmi, S.K. Emergency response to radiological and nuclear accidents and incidents. Br. J. Haematol. 2021, 192, 968–972. [Google Scholar] [CrossRef] [PubMed]
- Gale, R.P.; Baranov, A. If the unlikely becomes likely: Medical response to nuclear accidents. Bull. At. Sci. 2011, 67, 10–18. [Google Scholar] [CrossRef]
- Gusev, I.A.; Guskova, A.K.; Mettler, F.A. Medical Management of Radiation Accidents; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Cohen, J. Biodefense: 10 years after reinventing Project BioShield. Science 2011, 333, 1216–1218. [Google Scholar] [CrossRef] [PubMed]
- Russell, P.K. Project BioShield: What it is, why it is needed, and its accomplishments so far. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2007, 45 (Suppl. S1), S68–S72. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services. Project BioShield Annual Report, January 2014–December 2014. Available online: https://www.medicalcountermeasures.gov/media/36816/pbs-report-2014.pdf (accessed on 29 February 2023).
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist, 7th ed.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2012. [Google Scholar]
- DiCarlo, A.L.; Maher, C.; Hick, J.L.; Hanfling, D.; Dainiak, N.; Chao, N.; Bader, J.L.; Coleman, C.N.; Weinstock, D.M. Radiation injury after a nuclear detonation: Medical consequences and the need for scarce resources allocation. Disaster Med. Public Health Prep. 2011, 5 (Suppl. S1), S32–S44. [Google Scholar] [CrossRef]
- Waselenko, J.K.; MacVittie, T.J.; Blakely, W.F.; Pesik, N.; Wiley, A.L.; Dickerson, W.E.; Tsu, H.; Confer, D.L.; Coleman, C.N.; Seed, T.; et al. Medical management of the acute radiation syndrome: Recommendations of the Strategic National Stockpile Radiation Working Group. Ann. Intern. Med. 2004, 140, 1037–1051. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Guidance Document: Product Development under the Animal Rule. Available online: http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm399217.pdf (accessed on 20 October 2023).
- Singh, V.K.; Newman, V.L.; Romaine, P.L.; Hauer-Jensen, M.; Pollard, H.B. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev. Mol. Diagn. 2016, 16, 65–81. [Google Scholar] [CrossRef]
- Coy, S.L.; Cheema, A.K.; Tyburski, J.B.; Laiakis, E.C.; Collins, S.P.; Fornace, A., Jr. Radiation metabolomics and its potential in biodosimetry. Int. J. Radiat. Biol. 2011, 87, 802–823. [Google Scholar] [CrossRef]
- Singh, V.K.; Seed, T.M.; Cheema, A.K. Metabolomics-based predictive biomarkers of radiation injury and countermeasure efficacy: Current status and future perspectives. Expert Rev. Mol. Diagn. 2021, 21, 641–654. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Fornace, A.J., Jr.; Laiakis, E.C. Metabolomic applications in radiation biodosimetry: Exploring radiation effects through small molecules. Int. J. Radiat. Biol. 2017, 93, 1151–1176. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.D.; Goans, R.E.; Blain, P.G.; Thomas, S.H. Diagnosis and treatment of polonium poisoning. Clin. Toxicol. 2009, 47, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.; Fell, T.; Leggett, R.; Lloyd, D.; Puncher, M.; Youngman, M. The polonium-210 poisoning of Mr Alexander Litvinenko. J. Radiol. Prot. Off. J. Soc. Radiol. Prot. 2017, 37, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Nathwani, A.C.; Down, J.F.; Goldstone, J.; Yassin, J.; Dargan, P.I.; Virchis, A.; Gent, N.; Lloyd, D.; Harrison, J.D. Polonium-210 poisoning: A first-hand account. Lancet 2016, 388, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- International Atomic Energy Agency. The Radiological Accident in Goiânia. Available online: http://www-pub.iaea.org/MTCD/Publications/PDF/Pub815_web.pdf (accessed on 2 October 2023).
- Da Silva, F.C.; Hunt, J.G.; Ramalho, A.T.; Crispim, V.R. Dose reconstruction of a Brazilian industrial gamma radiography partial-body overexposure case. J. Radiol. Prot. Off. J. Soc. Radiol. Prot. 2005, 25, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Olabisi, A.O. Nonhuman primates as models for the discovery and development of radiation countermeasures. Expert Opin. Drug Discov. 2017, 12, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Schule, S.; Gluzman-Poltorak, Z.; Vainstein, V.; Basile, L.A.; Haimerl, M.; Stroszczynski, C.; Majewski, M.; Schwanke, D.; Port, M.; Abend, M.; et al. Gene Expression Changes in a Prefinal Health Stage of Lethally Irradiated Male and Female Rhesus Macaques. Radiat. Res. 2023, 199, 17–24. [Google Scholar] [CrossRef] [PubMed]
- National Research Council of the National Academy of Sciences. Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Singh, V.K.; Fatanmi, O.O.; Wise, S.Y.; Carpenter, A.D.; Olsen, C.H. Determination of lethality curve for cobalt-60 gamma-radiation source in rhesus macaques using subject-based supportive care. Radiat. Res. 2022, 198, 599–614. [Google Scholar] [CrossRef]
- Li, Y.; Singh, J.; Varghese, R.; Zhang, Y.; Fatanmi, O.O.; Cheema, A.K.; Singh, V.K. Transcriptome of rhesus macaque (Macaca mulatta) exposed to total-body irradiation. Sci. Rep. 2021, 11, 6295. [Google Scholar] [CrossRef]
- Li, Y.; Girgis, M.; Wise, S.Y.; Fatanmi, O.O.; Seed, T.M.; Maniar, M.; Cheema, A.K.; Singh, V.K. Analysis of the metabolomic profile in serum of irradiated nonhuman primates treated with Ex-Rad, a radiation countermeasure. Sci. Rep. 2021, 11, 11449. [Google Scholar] [CrossRef]
- Phipps, A.J.; Bergmann, J.N.; Albrecht, M.T.; Singh, V.K.; Homer, M.J. Model for evaluating antimicrobial therapy to prevent life-threatening bacterial infections following exposure to a medically significant radiation dose. Antimicrob. Agents Chemother. 2022, 66, e0054622. [Google Scholar] [CrossRef] [PubMed]
- Cheema, A.K.; Li, Y.; Moulton, J.; Girgis, M.; Wise, S.Y.; Carpenter, A.; Fatanmi, O.O.; Singh, V.K. Identification of novel biomarkers for acute radiation injury using multiomics approach and nonhuman primate model. Int. J. Radiat. Oncol. Biol. Phys. 2022, 114, 310–320. [Google Scholar] [CrossRef] [PubMed]
- American Veterinary Medical Association. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition. Available online: https://www.avma.org/sites/default/files/2020-01/2020-Euthanasia-Final-1-17-20.pdf (accessed on 24 December 2023).
- Singh, V.K.; Kulkarni, S.; Fatanmi, O.O.; Wise, S.Y.; Newman, V.L.; Romaine, P.L.; Hendrickson, H.; Gulani, J.; Ghosh, S.P.; Kumar, K.S.; et al. Radioprotective efficacy of gamma-tocotrienol in nonhuman primates. Radiat. Res. 2016, 185, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Girgis, M.; Jayatilake, M.; Serebrenik, A.A.; Cheema, A.K.; Kaytor, M.D.; Singh, V.K. Pharmacokinetic and metabolomic studies with a BIO 300 oral powder formulation in nonhuman primates. Sci. Rep. 2022, 12, 13475. [Google Scholar] [CrossRef] [PubMed]
- Cheema, A.K.; Li, Y.; Singh, J.; Johnson, R.; Girgis, M.; Wise, S.Y.; Fatanmi, O.O.; Kaytor, M.D.; Singh, V.K. Microbiome study in irradiated mice treated with BIO 300, a promising radiation countermeasure. Anim. Microbiome 2021, 3, 71. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.S.; Uppal, M.; Randhawa, S.; Cheema, M.S.; Aghdam, N.; Usala, R.L.; Ghosh, S.P.; Cheema, A.K.; Dritschilo, A. Radiation metabolomics: Current status and future directions. Front. Oncol. 2016, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Crook, A.; De Lima Leite, A.; Payne, T.; Bhinderwala, F.; Woods, J.; Singh, V.K.; Powers, R. Radiation exposure induces cross-species temporal metabolic changes that are mitigated in mice by amifostine. Sci. Rep. 2021, 11, 14004. [Google Scholar] [CrossRef]
- Cheema, A.K.; Li, Y.; Girgis, M.; Jayatilake, M.; Fatanmi, O.O.; Wise, S.Y.; Seed, T.M.; Singh, V.K. Alterations in tissue metabolite profiles with amifostine-prophylaxed mice exposed to gamma radiation. Metabolites 2020, 10, 211. [Google Scholar] [CrossRef]
- Cheema, A.K.; Li, Y.; Girgis, M.; Jayatilake, M.; Simas, M.; Wise, S.Y.; Olabisi, A.O.; Seed, T.M.; Singh, V.K. Metabolomic studies in tissues of mice treated with amifostine and exposed to gamma-radiation. Sci. Rep. 2019, 9, 15701. [Google Scholar] [CrossRef]
- Cheema, A.K.; Hinzman, C.P.; Mehta, K.Y.; Hanlon, B.K.; Garcia, M.; Fatanmi, O.O.; Singh, V.K. Plasma derived exosomal biomarkers of exposure to ionizing radiation in nonhuman primates. Int. J. Mol. Sci. 2018, 19, 3427. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Garcia, M.; Fornace, A.J., Jr.; Singh, V.K. Nonhuman primates with acute radiation syndrome: Results from a global serum metabolomics study after 7.2 Gy total-body irradiation. Radiat. Res. 2018, 190, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Singh, V.K.; Fornace, A.J. Lipidomic signatures of nonhuman primates with radiation-induced hematopoietic syndrome. Sci. Rep. 2017, 7, 9777. [Google Scholar] [CrossRef] [PubMed]
- Cheema, A.K.; Mehta, K.Y.; Rajagopal, M.U.; Wise, S.Y.; Fatanmi, O.O.; Singh, V.K. Metabolomic studies of tissue injury in nonhuman primates exposed to gamma-radiation. Int. J. Mol. Sci. 2019, 20, 3360. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.D.; Li, Y.; Fatanmi, O.O.; Wise, S.Y.; Petrus, S.A.; Janocha, B.L.; Cheema, A.K.; Singh, V.K. Metabolomic profiles in tissues of nonhuman primates exposed to total- or partial-body radiation. Radiat. Res. 2023, in press.
- Cheema, A.K.; Mehta, K.Y.; Santiago, P.T.; Fatanmi, O.O.; Kaytor, M.D.; Singh, V.K. Pharmacokinetic and metabolomic studies with BIO 300, a nanosuspension of genistein, in a nonhuman primate model. Int. J. Mol. Sci. 2019, 20, 1231. [Google Scholar] [CrossRef] [PubMed]
- Pannkuk, E.L.; Laiakis, E.C.; Fornace, A.J., Jr.; Fatanmi, O.O.; Singh, V.K. A metabolomic serum signature from nonhuman primates treated with a radiation countermeasure, gamma-tocotrienol, and exposed to ionizing radiation. Health Phys. 2018, 115, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Cheema, A.K.; Mehta, K.Y.; Fatanmi, O.O.; Wise, S.Y.; Hinzman, C.P.; Wolff, J.; Singh, V.K. A Metabolomic and lipidomic serum signature from nonhuman primates administered with a promising radiation countermeasure, gamma-tocotrienol. Int. J. Mol. Sci. 2018, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Maan, K.; Baghel, R.; Dhariwal, S.; Sharma, A.; Bakhshi, R.; Rana, P. Metabolomics and transcriptomics based multi-omics integration reveals radiation-induced altered pathway networking and underlying mechanism. NPJ Syst. Biol. Appl. 2023, 9, 42. [Google Scholar] [CrossRef]
- Upadhyay, M.; Rajagopal, M.; Gill, K.; Li, Y.; Bansal, S.; Sridharan, V.; Tyburski, J.B.; Boerma, M.; Cheema, A.K. Identification of plasma lipidome changes associated with low dose spacetType radiation exposure in a murine model. Metabolites 2020, 10, 252. [Google Scholar] [CrossRef]
- Pannkuk, E.L.; Laiakis, E.C.; Mak, T.D.; Astarita, G.; Authier, S.; Wong, K.; Fornace, A.J., Jr. A lipidomic and metabolomic serum signature from nonhuman primates exposed to ionizing radiation. Metabolomics 2016, 12, 80. [Google Scholar] [CrossRef]

| p-Value | ||||||
|---|---|---|---|---|---|---|
| Compound | Comparison | FC | Log2(FC) | WSR Test | MWU Test | CID Fragments |
| PC (20:2/18:0) | d 1 vs. pre-irradiation | 0.62122 | −0.68682 | <0.05 | 0.001 | 858.6165 798.5977 307.2622 283.2641 |
| d 13 vs. pre-irradiation | 0.87591 | −0.19114546 | 0.24 | 0.292 | ||
| d 25 vs. pre-irradiation | 0.79989 | −0.32212648 | <0.05 | 0.026 | ||
| PE (22:6/18:2) | d 1 vs. pre-irradiation | 0.59364 | −0.75234 | <0.05 | 0.005 | 786.5251 327.2357 283.2443 279.2334 |
| d 13 vs. pre-irradiation | 0.70821 | −0.49775088 | 0.24 | 0.061 | ||
| d 25 vs. pre-irradiation | 0.49562 | −1.01269369 | <0.05 | 0.003 | ||
| p-Value | ||||||
|---|---|---|---|---|---|---|
| Compound | Comparison | FC | Log2(FC) | WSR Test | MWU Test | CID Fragments |
| 5alpha-Pregnane-3,20-dione | d 1 vs. preterminal | 1.8672 | 0.900876466 | 0.01 | 0.002 | 315.2558 278.9958 266.9861 228.9908 315.2539 297.2443 278.9866 266.9792 |
| d 13 vs. preterminal | 1.5528 | 0.634872023 | 0.02 | 0.017 | ||
| d 25 vs. preterminal | 1.9819 | 0.986884171 | 0.01 | <0.001 | ||
| 17alpha-Hydroxypregnenolone | d 1 vs. preterminal | 9.4901 | 3.246423289 | 0.01 | <0.001 | 297.1469 282.1202 331.1852 315.1572 297.1487 282.1252 189.0946 135.0848 125.0597 |
| d 13 vs. preterminal | 2.4951 | 1.319097638 | 0.01 | 0.012 | ||
| d 25 vs. preterminal | 3.7344 | 1.900876466 | 0.01 | 0.002 | ||
| Allopregnanolone | d 1 vs. preterminal | 1.715 | 0.778208576 | 0.01 | 0.004 | 319.2986 281.6630 272.6601 184.0767 148.5324 319.2920 281.6631 272.6624 184.0746 145.5324 |
| d 13 vs. preterminal | 1.4744 | 0.560127976 | 0.07 | 0.04 | ||
| d 25 vs. preterminal | 1.6643 | 0.734915511 | 0.04 | 0.015 | ||
| p-Value | ||||||
|---|---|---|---|---|---|---|
| Compound | Comparison | FC | Log2(FC) | WSR Test | MWU Test | CID Fragments |
| LysoPA (18:2) | d 1 vs. preterminal | 0.17627 | −2.50414114 | 0.01 | <0.001 | 433.2368 152.9961 |
| d 13 vs. preterminal | 0.25821 | −1.95338322 | 0.02 | 0.004 | ||
| d 25 vs. preterminal | 0.33539 | −1.57608842 | 0.01 | <0.001 | ||
| PE (22:6/18:2) | d 1 vs. preterminal | 0.56565 | −0.82201844 | 0.04 | 0.026 | 786.5251 327.2357 283.2443 279.2334 |
| d 13 vs. preterminal | 0.47415 | −1.07658456 | 0.02 | 0.025 | ||
| d 25 vs. preterminal | 0.67753 | −0.56164327 | 0.35 | 0.002 | ||
| Androsterone | d 1 vs. preterminal | 0.52337 | −0.93409687 | 0.01 | 0.001 | 291.1971 249.1860 233.1534 203.1080 |
| d 13 vs. preterminal | 0.65955 | −0.60044606 | 0.038 | 0.04 | ||
| d 25 vs. preterminal | 0.69581 | −0.52323 | 0.067 | 0.095 | ||
| Testosterone | d 1 vs. preterminal | 0.62021 | −0.68917131 | 0.114 | 0.018 | 281.6678 272.6580 162.9414 140.5447 104.1045 281.6606 272.6562 162.9465 148.5327 140.5424 104.1091 |
| d 13 vs. preterminal | 0.54735 | −0.86946444 | 0.019 | 0.01 | ||
| d 25 vs. preterminal | 0.64174 | −0.63993919 | 0.114 | 0.025 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carpenter, A.D.; Fatanmi, O.O.; Wise, S.Y.; Petrus, S.A.; Tyburski, J.B.; Cheema, A.K.; Singh, V.K. Metabolomic Changes in Plasma of Preterminal Stage of Rhesus Nonhuman Primates Exposed to Lethal Dose of Radiation. Metabolites 2024, 14, 18. https://doi.org/10.3390/metabo14010018
Carpenter AD, Fatanmi OO, Wise SY, Petrus SA, Tyburski JB, Cheema AK, Singh VK. Metabolomic Changes in Plasma of Preterminal Stage of Rhesus Nonhuman Primates Exposed to Lethal Dose of Radiation. Metabolites. 2024; 14(1):18. https://doi.org/10.3390/metabo14010018
Chicago/Turabian StyleCarpenter, Alana D., Oluseyi O. Fatanmi, Stephen Y. Wise, Sarah A. Petrus, John B. Tyburski, Amrita K. Cheema, and Vijay K. Singh. 2024. "Metabolomic Changes in Plasma of Preterminal Stage of Rhesus Nonhuman Primates Exposed to Lethal Dose of Radiation" Metabolites 14, no. 1: 18. https://doi.org/10.3390/metabo14010018
APA StyleCarpenter, A. D., Fatanmi, O. O., Wise, S. Y., Petrus, S. A., Tyburski, J. B., Cheema, A. K., & Singh, V. K. (2024). Metabolomic Changes in Plasma of Preterminal Stage of Rhesus Nonhuman Primates Exposed to Lethal Dose of Radiation. Metabolites, 14(1), 18. https://doi.org/10.3390/metabo14010018







