Target Bioconjugation of Protein Through Chemical, Molecular Dynamics, and Artificial Intelligence Approaches
Abstract
1. Introduction
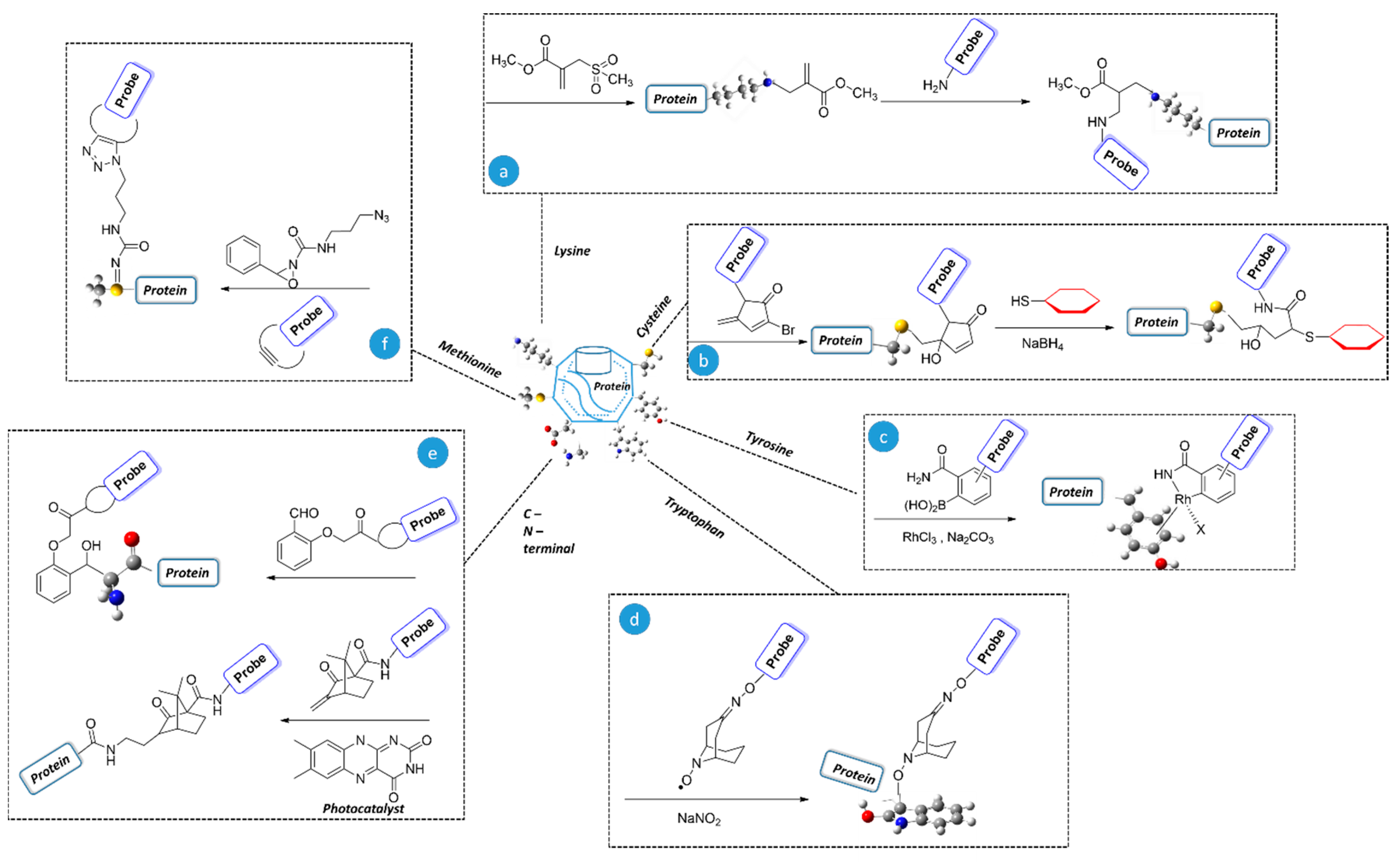
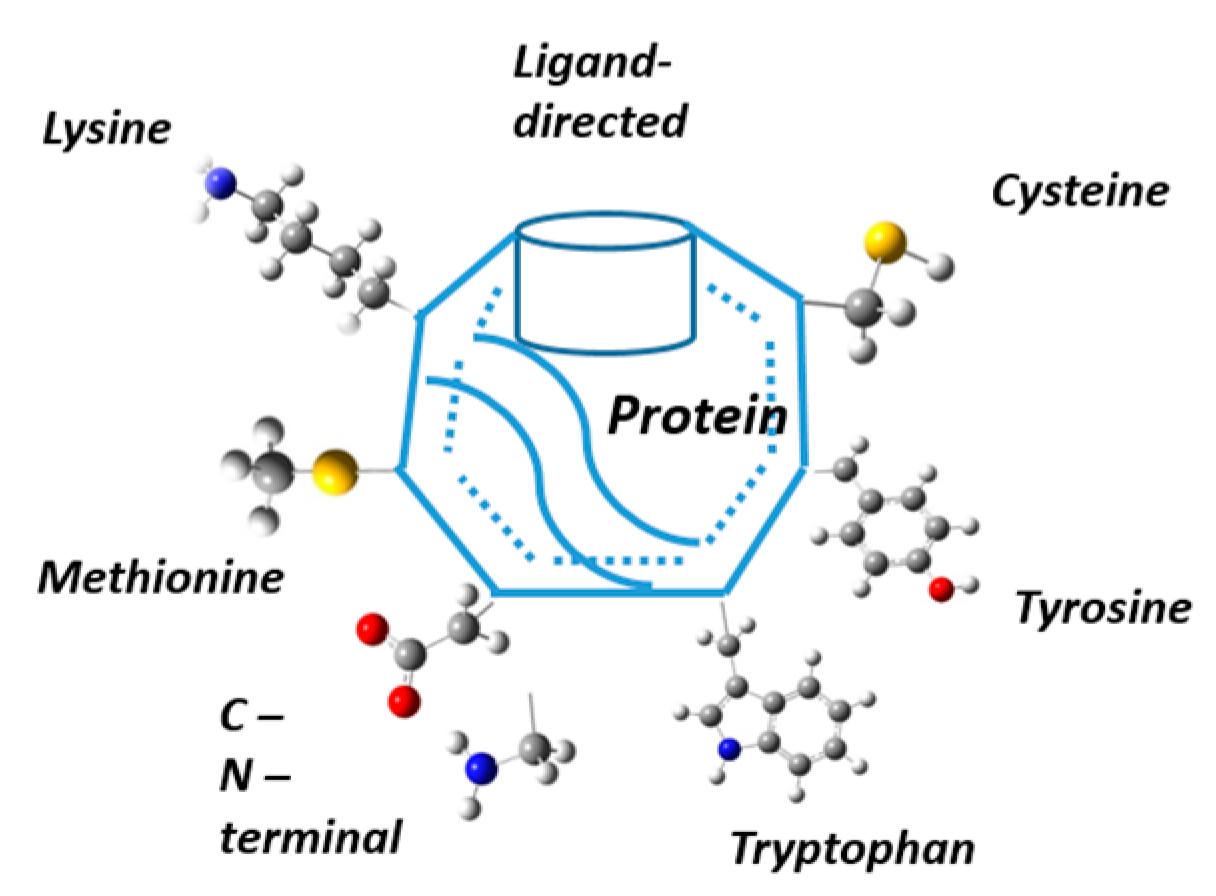
2. Modification of Naturally Occurring Amino Acids Through Chemo and Regioselective Approaches
3. Protein-Selective Proximity-Driven Chemical Modification
4. Traceless Affinity Labeling
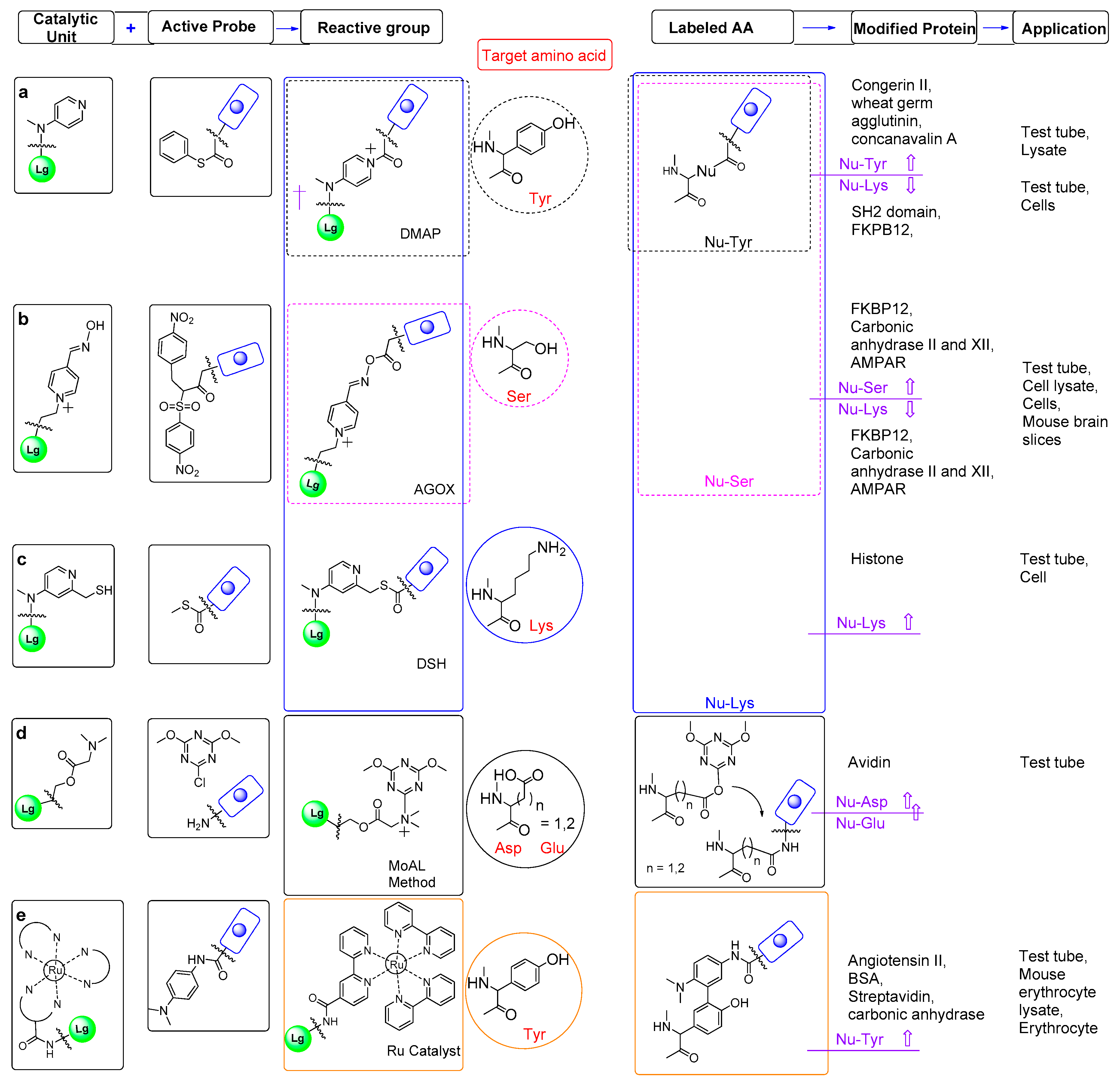
5. Affinity-Guided Catalyst
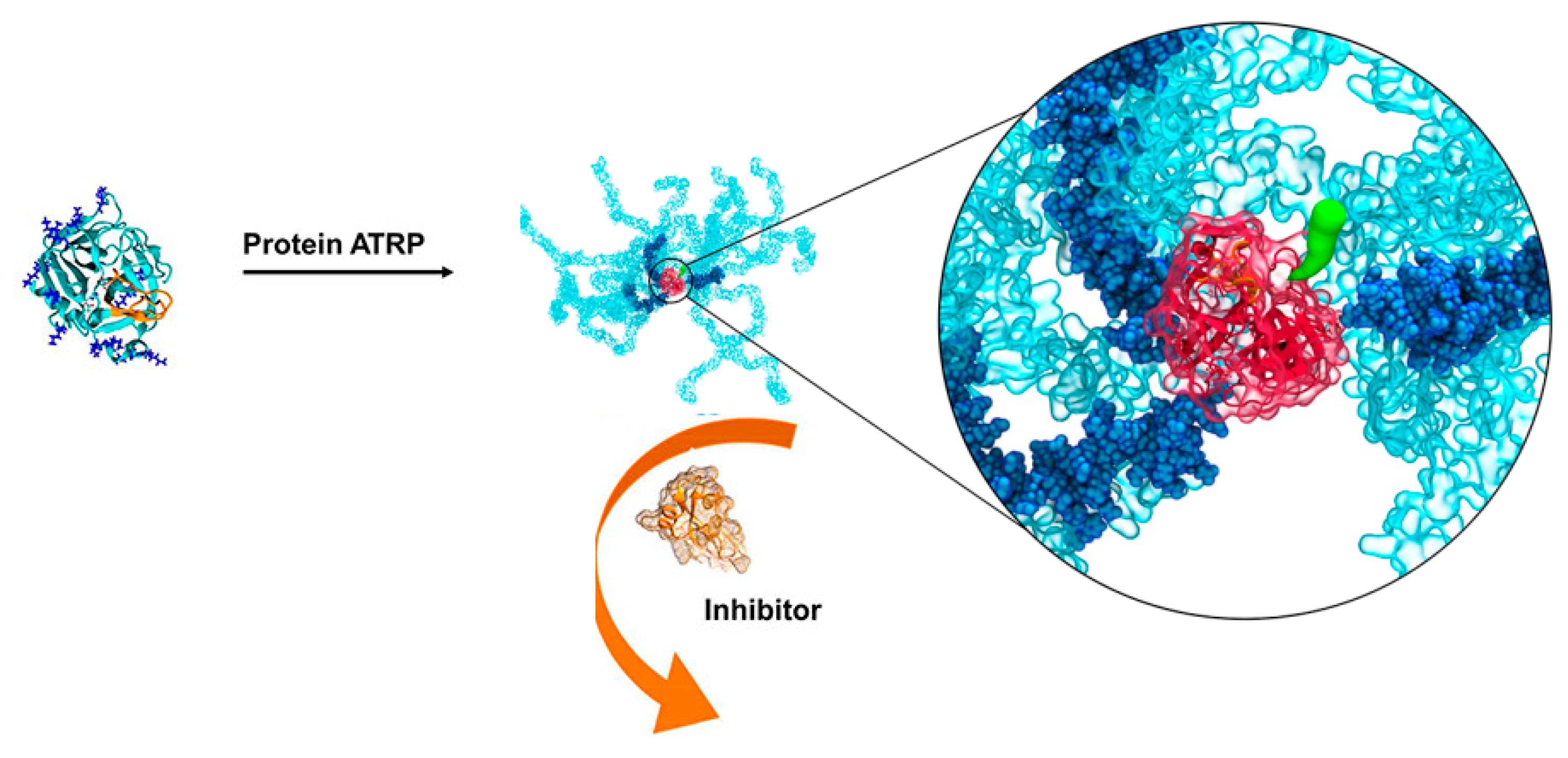
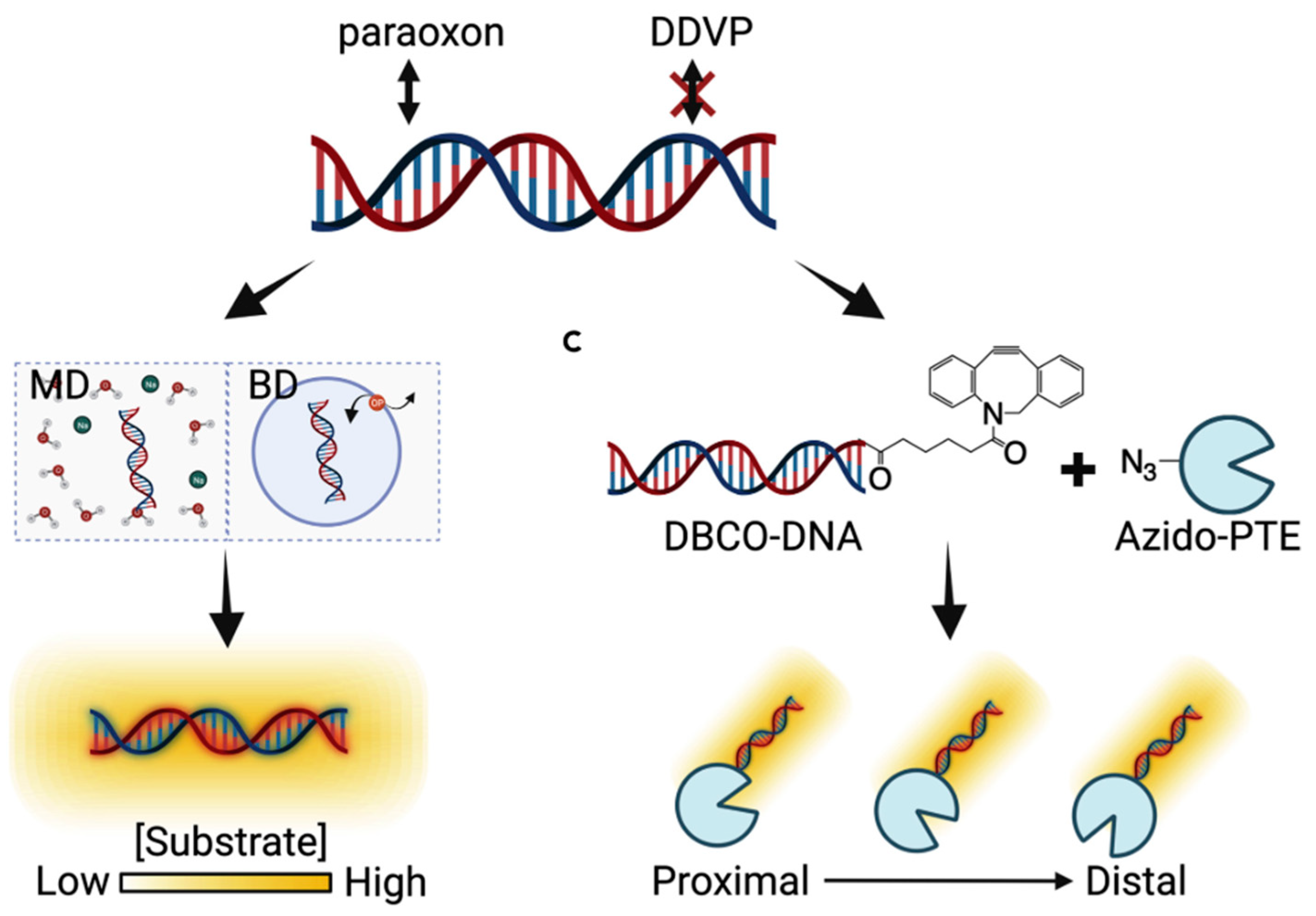
6. Molecular Dynamics-Guided Labeling
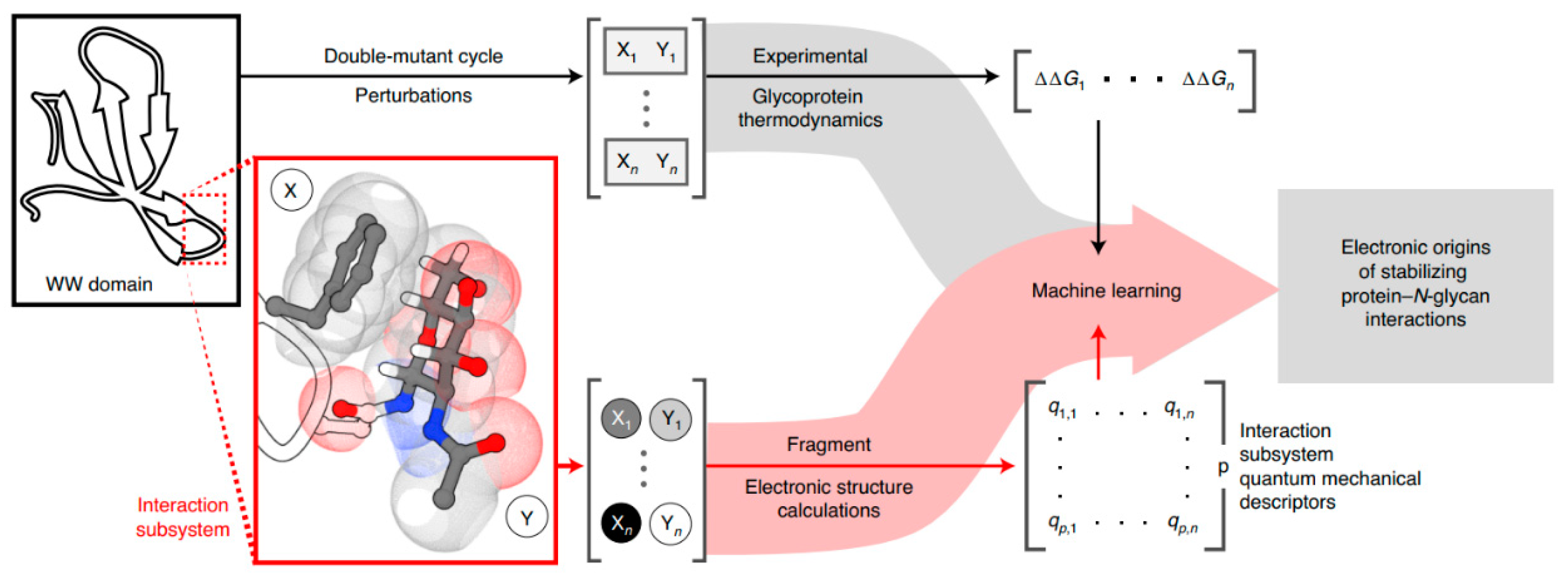
7. Artificial Intelligence in Protein Labeling
8. Biomedical Applications
9. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gagner, J.E.; Kim, W.; Chaikof, E.L. Designing protein-based biomaterials for medical applications. Acta Biomater. 2014, 10, 1542–1557. [Google Scholar] [CrossRef] [PubMed]
- Shadish, J.A.; DeForest, C.A. Site-Selective Protein Modification: From Functionalized Proteins to Functional Biomaterials. Matter 2020, 2, 50–77. [Google Scholar] [CrossRef]
- Krall, N.; da Cruz, F.P.; Boutureira, O.; Bernardes, G.J.L. Site-selective protein-modification chemistry for basic biology and drug development. Nat. Chem. 2016, 8, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Reddy, N.C.; Kumar, M.; Molla, R.; Rai, V. Chemical methods for modification of proteins. Org. Biomol. Chem. 2020, 18, 4669–4691. [Google Scholar] [CrossRef] [PubMed]
- Rawale, D.G.; Thakur, K.; Adusumalli, S.R.; Rai, V. Chemical Methods for Selective Labeling of Proteins. Eur. J. Org. Chem. 2019, 2019, 6749–6763. [Google Scholar] [CrossRef]
- de Graaf, A.J.; Kooijman, M.; Hennink, W.E.; Mastrobattista, E. Nonnatural Amino Acids for Site-Specific Protein Conjugation. Bioconjugate Chem. 2009, 20, 1281–1295. [Google Scholar] [CrossRef]
- Spicer, C.D.; Davis, B.G. Selective chemical protein modification. Nat. Commun. 2014, 5, 4740. [Google Scholar] [CrossRef]
- Takaoka, Y.; Ojida, A.; Hamachi, I. Protein Organic Chemistry and Applications for Labeling and Engineering in Live-Cell Systems. Angew. Chem. Int. Ed. 2013, 52, 4088–4106. [Google Scholar] [CrossRef]
- Sletten, E.M.; Bertozzi, C.R. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem. Int. Ed. 2009, 48, 6974–6998. [Google Scholar] [CrossRef]
- Boutureira, O.; Bernardes, G.J. Advances in Chemical Protein Modification. Chem. Rev. 2015, 115, 2174–2195. [Google Scholar] [CrossRef]
- Rotem, D.; Jayasinghe, L.; Salichou, M.; Bayley, H. Protein Detection by Nanopores Equipped with Aptamers. J. Am. Chem. Soc. 2012, 134, 2781–2787. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angew. Chem. Int. Ed. 2005, 44, 7342–7372. [Google Scholar] [CrossRef]
- Ibraheem, A.; Campbell, R.E. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010, 14, 30–36. [Google Scholar] [CrossRef]
- Tamura, T.; Hamachi, I. Recent Progress in Design of Protein-Based Fluorescent Biosensors and Their Cellular Applications. ACS Chem. Biol. 2014, 9, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Tamura, T.; Hamachi, I. In Situ Construction of Protein-Based Semisynthetic Biosensors. ACS Sens. 2018, 3, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Rodríguez, A.; Davis, B.G. Chemical modification in the creation of novel biocatalysts. Curr. Opin. Chem. Biol. 2011, 15, 211–219. [Google Scholar] [CrossRef]
- Witus, L.S.; Francis, M.B. Using Synthetically Modified Proteins to Make New Materials. Accounts Chem. Res. 2011, 44, 774–783. [Google Scholar] [CrossRef]
- Liu, X.; Sun, J.; Gao, W. Site-selective protein modification with polymers for advanced biomedical applications. Biomaterials 2018, 178, 413–434. [Google Scholar] [CrossRef]
- Tamura, T.; Tsukiji, S.; Hamachi, I. Native FKBP12 Engineering by Ligand-Directed Tosyl Chemistry: Labeling Properties and Application to Photo-Cross-Linking of Protein Complexes in Vitro and in Living Cells. J. Am. Chem. Soc. 2012, 134, 2216–2226. [Google Scholar] [CrossRef]
- Tsai, Y.-H.; Essig, S.; James, J.R.; Lang, K.; Chin, J.W. Selective rapid and optically switchable regulation of protein function in live mammalian cells. Nat. Chem. 2015, 7, 554–561. [Google Scholar] [CrossRef]
- Dozier, J.K.; Distefano, M.D. Site-Specific PEGylation of Therapeutic Proteins. Int. J. Mol. Sci. 2015, 16, 25831–25864. [Google Scholar] [CrossRef]
- de Goeij, B.E.C.G.; Lambert, J.M. New developments for antibody-drug conjugate-based therapeutic approaches. Curr. Opin. Immunol. 2016, 40, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Chudasama, V.; Maruani, A.; Caddick, S. Recent advances in the construction of antibody–drug conjugates. Nat. Chem. 2016, 8, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef]
- Hoyt, E.A.; Cal, P.M.S.D.; Oliveira, B.L.; Bernardes, G.J.L. Contemporary approaches to site-selective protein modification. Nat. Rev. Chem. 2019, 3, 147–171. [Google Scholar] [CrossRef]
- Tamura, T.; Hamachi, I. Chemistry for Covalent Modification of Endogenous/Native Proteins: From Test Tubes to Complex Biological Systems. J. Am. Chem. Soc. 2019, 141, 2782–2799. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Hamachi, I. Recent Progress in Chemical Modification of Proteins. Anal. Sci. 2019, 35, 5–27. [Google Scholar] [CrossRef]
- Tamura, T.; Shigemitsu, H.; Hamachi, I. 4.12—In Situ Protein Labeling in Complex Environments. In Comprehensive Supramolecular Chemistry II; Atwood, J.L., Ed.; Elsevier: Oxford, UK, 2017; pp. 409–437. [Google Scholar]
- Hacker, S.M.; Backus, K.M.; Lazear, M.R.; Forli, S.; Correia, B.E.; Cravatt, B.F. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 2017, 9, 1181–1190. [Google Scholar] [CrossRef]
- Rosen, C.B.; Francis, M.B. Targeting the N terminus for site-selective protein modification. Nat. Chem. Biol. 2017, 13, 697–705. [Google Scholar] [CrossRef]
- Kozlowski, L.P. Proteome-pI: Proteome isoelectric point database. Nucleic Acids Res. 2016, 45, D1112–D1116. [Google Scholar] [CrossRef]
- Gunnoo, S.B.; Madder, A. Chemical Protein Modification through Cysteine. ChemBioChem 2016, 17, 529–553. [Google Scholar] [CrossRef]
- Koniev, O.; Wagner, A. Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chem. Soc. Rev. 2015, 44, 5495–5551. [Google Scholar] [CrossRef] [PubMed]
- deGruyter, J.N.; Malins, L.R.; Baran, P.S. Residue-Specific Peptide Modification: A Chemist’s Guide. Biochemistry 2017, 56, 3863–3873. [Google Scholar] [CrossRef] [PubMed]
- Nanna, A.R.; Li, X.; Walseng, E.; Pedzisa, L.; Goydel, R.S.; Hymel, D.; Burke, T.R.; Roush, W.R.; Rader, C. Harnessing a catalytic lysine residue for the one-step preparation of homogeneous antibody-drug conjugates. Nat. Commun. 2017, 8, 1112. [Google Scholar] [CrossRef]
- Cal, P.M.S.D.; Bernardes, G.J.L.; Gois, P.M.P. Cysteine-Selective Reactions for Antibody Conjugation. Angew. Chem. Int. Ed. 2014, 53, 10585–10587. [Google Scholar] [CrossRef] [PubMed]
- Chalker, J.M.; Bernardes, G.J.L.; Lin, Y.A.; Davis, B.G. Chemical Modification of Proteins at Cysteine: Opportunities in Chemistry and Biology. Chem. Asian J. 2009, 4, 630–640. [Google Scholar] [CrossRef]
- Bernardim, B.; Cal, P.M.; Matos, M.J.; Oliveira, B.L.; Martínez-Sáez, N.; Albuquerque, I.S.; Perkins, E.; Corzana, F.; Burtoloso, A.C.; Jiménez-Osés, G.; et al. Stoichiometric and irreversible cysteine-selective protein modification using carbonylacrylic reagents. Nat. Commun. 2016, 7, 13128. [Google Scholar] [CrossRef]
- Backus, K.M.; Correia, B.E.; Lum, K.M.; Forli, S.; Horning, B.D.; González-Páez, G.E.; Chatterjee, S.; Lanning, B.R.; Teijaro, J.R.; Olson, A.J.; et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 2016, 534, 570–574. [Google Scholar] [CrossRef]
- Matos, M.J.; Oliveira, B.L.; Martínez-Sáez, N.; Guerreiro, A.; Cal, P.M.S.D.; Bertoldo, J.; Maneiro, M.; Perkins, E.; Howard, J.; Deery, M.J.; et al. Chemo- and Regioselective Lysine Modification on Native Proteins. J. Am. Chem. Soc. 2018, 140, 4004–4017. [Google Scholar] [CrossRef]
- Jiang, H.; Chen, W.; Wang, J.; Zhang, R. Selective N-terminal modification of peptides and proteins: Recent progresses and applications. Chin. Chem. Lett. 2022, 33, 80–88. [Google Scholar] [CrossRef]
- Lin, Y.A.; Chalker, J.M.; Floyd, N.; Bernardes, G.J.L.; Davis, B.G. Allyl Sulfides Are Privileged Substrates in Aqueous Cross-Metathesis: Application to Site-Selective Protein Modification. J. Am. Chem. Soc. 2008, 130, 9642–9643. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, G.J.L.; Chalker, J.M.; Errey, J.C.; Davis, B.G. Facile Conversion of Cysteine and Alkyl Cysteines to Dehydroalanine on Protein Surfaces: Versatile and Switchable Access to Functionalized Proteins. J. Am. Chem. Soc. 2008, 130, 5052–5053. [Google Scholar] [CrossRef] [PubMed]
- Dadová, J.; Galan, S.R.; Davis, B.G. Synthesis of modified proteins via functionalization of dehydroalanine. Curr. Opin. Chem. Biol. 2018, 46, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zang, C.; An, G.; Shang, M.; Cui, Z.; Chen, G.; Xi, Z.; Zhou, C. Cysteine-specific protein multi-functionalization and disulfide bridging using 3-bromo-5-methylene pyrrolones. Nat. Commun. 2020, 11, 1015. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, Q.L.; Song, Z.; Chan, A.S.; Xiong, X.F. Cleavable Cys labeling directed Lys site-selective stapling and single-site modification. Sci. China Chem. 2022, 65, 1356–1361. [Google Scholar] [CrossRef]
- Wang, L.; Gruzdys, V.; Pang, N.; Meng, F.; Sun, X.-L. Primary arylamine-based tyrosine-targeted protein modification. RSC Adv. 2014, 4, 39446–39452. [Google Scholar] [CrossRef]
- Sato, S.; Nakamura, K.; Nakamura, H. Tyrosine-Specific Chemical Modification with in Situ Hemin-Activated Luminol Derivatives. ACS Chem. Biol. 2015, 10, 2633–2640. [Google Scholar] [CrossRef]
- Joshi, N.S.; Whitaker, L.R.; Francis, M.B. A Three-Component Mannich-Type Reaction for Selective Tyrosine Bioconjugation. J. Am. Chem. Soc. 2004, 126, 15942–15943. [Google Scholar] [CrossRef]
- Tilley, S.D.; Francis, M.B. Tyrosine-Selective Protein Alkylation Using π-Allylpalladium Complexes. J. Am. Chem. Soc. 2006, 128, 1080–1081. [Google Scholar] [CrossRef]
- Seki, Y.; Ishiyama, T.; Sasaki, D.; Abe, J.; Sohma, Y.; Oisaki, K.; Kanai, M. Transition Metal-Free Tryptophan-Selective Bioconjugation of Proteins. J. Am. Chem. Soc. 2016, 138, 10798–10801. [Google Scholar] [CrossRef]
- Hansen, M.B.; Hubálek, F.; Skrydstrup, T.; Hoeg-Jensen, T. Chemo- and Regioselective Ethynylation of Tryptophan-Containing Peptides and Proteins. Chem. A Eur. J. 2016, 22, 1572–1576. [Google Scholar] [CrossRef] [PubMed]
- Antos, J.M.; McFarland, J.M.; Iavarone, A.T.; Francis, M.B. Chemoselective Tryptophan Labeling with Rhodium Carbenoids at Mild pH. J. Am. Chem. Soc. 2009, 131, 6301–6308. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yang, X.; Jia, S.; Weeks, A.M.; Hornsby, M.; Lee, P.S.; Nichiporuk, R.V.; Iavarone, A.T.; Wells, J.A.; Toste, F.D.; et al. Redox-based reagents for chemoselective methionine bioconjugation. Science 2017, 355, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Ohata, J.; Miller, M.K.; Mountain, C.M.; Vohidov, F.; Ball, Z.T. A Three-Component Organometallic Tyrosine Bioconjugation. Angew. Chem. Int. Ed. 2018, 57, 2827–2830. [Google Scholar] [CrossRef]
- Maruyama, K.; Ishiyama, T.; Seki, Y.; Sakai, K.; Togo, T.; Oisaki, K.; Kanai, M. Protein Modification at Tyrosine with Iminoxyl Radicals. J. Am. Chem. Soc. 2021, 143, 19844–19855. [Google Scholar] [CrossRef]
- Imiołek, M.; Karunanithy, G.; Ng, W.-L.; Baldwin, A.J.; Gouverneur, V.E.; Davis, B.G. Selective Radical Trifluoromethylation of Native Residues in Proteins. J. Am. Chem. Soc. 2018, 140, 1568–1571. [Google Scholar] [CrossRef]
- Taylor, M.T.; Nelson, J.E.; Suero, M.G.; Gaunt, M.J. A protein functionalization platform based on selective reactions at methionine residues. Nature 2018, 562, 563–568. [Google Scholar] [CrossRef]
- MacDonald, I.J.; Munch, H.K.; Moore, T.; Francis, M.B. One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nat. Chem. Biol. 2015, 11, 326–331. [Google Scholar] [CrossRef]
- Chen, D.; Disotuar, M.M.; Xiong, X.; Wang, Y.; Chou, D.H.-C. Selective N-terminal functionalization of native peptides and proteins. Chem. Sci. 2017, 8, 2717–2722. [Google Scholar] [CrossRef]
- McGrath, N.A.; Andersen, K.A.; Davis, A.K.F.; Lomax, J.E.; Raines, R.T. Diazo compounds for the bioreversible esterification of proteins. Chem. Sci. 2015, 6, 752–755. [Google Scholar] [CrossRef]
- Totaro, K.A.; Liao, X.; Bhattacharya, K.; Finneman, J.I.; Sperry, J.B.; Massa, M.A.; Thorn, J.; Ho, S.V.; Pentelute, B.L. Systematic Investigation of EDC/sNHS-Mediated Bioconjugation Reactions for Carboxylated Peptide Substrates. Bioconjugate Chem. 2016, 27, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Purushottam, L.; Adusumalli, S.R.; Singh, U.; Unnikrishnan, V.B.; Rawale, D.G.; Gujrati, M.; Mishra, R.K.; Rai, V. Single-site glycine-specific labeling of proteins. Nat. Commun. 2019, 10, 2539. [Google Scholar] [CrossRef] [PubMed]
- Bloom, S.; Liu, C.; Kölmel, D.K.; Qiao, J.X.; Zhang, Y.; Poss, M.A.; Ewing, W.R.; MacMillan, D.W.C. Decarboxylative alkylation for site-selective bioconjugation of native proteins via oxidation potentials. Nat. Chem. 2018, 10, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Hao, M.; Liu, J.; Chen, Y.; Wufuer, G.; Zhu, J.; Zhang, X.; Zheng, T.; Fang, M.; Zhang, S.; et al. Design of a recombinant asparaginyl ligase for site-specific modification using efficient recognition and nucleophile motifs. Commun. Chem. 2024, 7, 87. [Google Scholar] [CrossRef]
- Ma, Q.; He, B.; Tang, G.; Xie, R.; Zheng, P. Enzymatic Protein Immobilization on Amino-Functionalized Nanoparticles. Molecules 2023, 28, 379. [Google Scholar] [CrossRef]
- Amaike, K.; Tamura, T.; Hamachi, I. Recognition-driven chemical labeling of endogenous proteins in multi-molecular crowding in live cells. Chem. Commun. 2017, 53, 11972–11983. [Google Scholar] [CrossRef]
- Hayashi, T.; Hamachi, I. Traceless Affinity Labeling of Endogenous Proteins for Functional Analysis in Living Cells. Accounts Chem. Res. 2012, 45, 1460–1469. [Google Scholar] [CrossRef]
- Tsukiji, S.; Hamachi, I. Ligand-directed tosyl chemistry for in situ native protein labeling and engineering in living systems: From basic properties to applications. Curr. Opin. Chem. Biol. 2014, 21, 136–143. [Google Scholar] [CrossRef]
- Chowdhry, V.; Westheimer, F.H. Photoaffinity Labeling of Biological Systems. Annu. Rev. Biochem. 1979, 48, 293–325. [Google Scholar] [CrossRef]
- Chen, G.; Heim, A.; Riether, D.; Yee, D.; Milgrom, Y.; Gawinowicz, M.A.; Sames, D. Reactivity of Functional Groups on the Protein Surface: Development of Epoxide Probes for Protein Labeling. J. Am. Chem. Soc. 2003, 125, 8130–8133. [Google Scholar] [CrossRef]
- Hamachi, I.; Nagase, T.; Shinkai, S. A General Semisynthetic Method for Fluorescent Saccharide-Biosensors Based on a Lectin. J. Am. Chem. Soc. 2000, 122, 12065–12066. [Google Scholar] [CrossRef]
- Nagase, T.; Shinkai, S.; Hamachi, I. Post-photoaffinity labeling modification using aldehyde chemistry to produce a fluorescent lectin toward saccharide-biosensors. Chem. Commun. 2001, 3, 229–230. [Google Scholar] [CrossRef]
- Nagase, T.; Nakata, E.; Shinkai, S.; Hamachi, I. Construction of Artificial Signal Transducers on a Lectin Surface by Post-Photoaffinity-Labeling Modification for Fluorescent Saccharide Biosensors. Chem. A Eur. J. 2003, 9, 3660–3669. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, Y.; Tsutsumi, H.; Kasagi, N.; Nakata, E.; Hamachi, I. One-Pot and Sequential Organic Chemistry on an Enzyme Surface to Tether a Fluorescent Probe at the Proximity of the Active Site with Restoring Enzyme Activity. J. Am. Chem. Soc. 2006, 128, 3273–3280. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, H.; Miyagawa, M.; Koshi, Y.; Takaoka, Y.; Tsukiji, S.; Hamachi, I. Affinity-Labeling-Based Introduction of a Reactive Handle for Natural Protein Modification. Chem. Asian J. 2008, 3, 1134–1139. [Google Scholar] [CrossRef] [PubMed]
- Tsukiji, S.; Miyagawa, M.; Takaoka, Y.; Tamura, T.; Hamachi, I. Ligand-directed tosyl chemistry for protein labeling in vivo. Nat. Chem. Biol. 2009, 5, 341–343. [Google Scholar] [CrossRef]
- Tsukiji, S.; Wang, H.; Miyagawa, M.; Tamura, T.; Takaoka, Y.; Hamachi, I. Quenched Ligand-Directed Tosylate Reagents for One-Step Construction of Turn-On Fluorescent Biosensors. J. Am. Chem. Soc. 2009, 131, 9046–9054. [Google Scholar] [CrossRef]
- Takaoka, Y.; Sun, Y.; Tsukiji, S.; Hamachi, I. Mechanisms of chemical protein19F-labeling and NMR-based biosensor construction in vitro and in cells using self-assembling ligand-directed tosylate compounds. Chem. Sci. 2011, 2, 511–520. [Google Scholar] [CrossRef]
- Tamura, T.; Kioi, Y.; Miki, T.; Tsukiji, S.; Hamachi, I. Fluorophore Labeling of Native FKBP12 by Ligand-Directed Tosyl Chemistry Allows Detection of Its Molecular Interactions in Vitro and in Living Cells. J. Am. Chem. Soc. 2013, 135, 6782–6785. [Google Scholar] [CrossRef]
- Yamaura, K.; Kuwata, K.; Tamura, T.; Kioi, Y.; Takaoka, Y.; Kiyonaka, S.; Hamachi, I. Live cell off-target identification of lapatinib using ligand-directed tosyl chemistry. Chem. Commun. 2014, 50, 14097–14100. [Google Scholar] [CrossRef]
- Takaoka, Y.; Kioi, Y.; Morito, A.; Otani, J.; Arita, K.; Ashihara, E.; Ariyoshi, M.; Tochio, H.; Shirakawa, M.; Hamachi, I. Quantitative comparison of protein dynamics in live cells and in vitro by in-cell 19F-NMR. Chem. Commun. 2013, 49, 2801–2803. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, S.-H.; Yasui, R.; Miki, T.; Ojida, A.; Hamachi, I. Ligand-Directed Acyl Imidazole Chemistry for Labeling of Membrane-Bound Proteins on Live Cells. J. Am. Chem. Soc. 2012, 134, 3961–3964. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ke, J.; Zhou, X.E.; Yi, W.; Brunzelle, J.S.; Li, J.; Yong, E.-L.; Xu, H.E.; Melcher, K. Structural basis for molecular recognition of folic acid by folate receptors. Nature 2013, 500, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Wakayama, S.; Kiyonaka, S.; Arai, I.; Kakegawa, W.; Matsuda, S.; Ibata, K.; Nemoto, Y.L.; Kusumi, A.; Yuzaki, M.; Hamachi, I. Chemical labelling for visualizing native AMPA receptors in live neurons. Nat. Commun. 2017, 8, 14850. [Google Scholar] [CrossRef]
- Miki, T.; Fujishima, S.-H.; Komatsu, K.; Kuwata, K.; Kiyonaka, S.; Hamachi, I. LDAI-Based Chemical Labeling of Intact Membrane Proteins and Its Pulse-Chase Analysis under Live Cell Conditions. Chem. Biol. 2014, 21, 1013–1022. [Google Scholar] [CrossRef]
- Matsuo, K.; Kioi, Y.; Yasui, R.; Takaoka, Y.; Miki, T.; Fujishima, S.-H.; Hamachi, I. One-step construction of caged carbonic anhydrase I using a ligand-directed acyl imidazole-based protein labeling method. Chem. Sci. 2013, 4, 2573–2580. [Google Scholar] [CrossRef]
- Yamaura, K.; Kiyonaka, S.; Numata, T.; Inoue, R.; Hamachi, I. Discovery of allosteric modulators for GABAA receptors by ligand-directed chemistry. Nat. Chem. Biol. 2016, 12, 822–830. [Google Scholar] [CrossRef]
- Takaoka, Y.; Nishikawa, Y.; Hashimoto, Y.; Sasaki, K.; Hamachi, I. Ligand-directed dibromophenyl benzoate chemistry for rapid and selective acylation of intracellular natural proteins. Chem. Sci. 2015, 6, 3217–3224. [Google Scholar] [CrossRef]
- Matsuo, K.; Nishikawa, Y.; Masuda, M.; Hamachi, I. Live-Cell Protein Sulfonylation Based on Proximity-driven N-Sulfonyl Pyridone Chemistry. Angew. Chem. Int. Ed. 2018, 57, 659–662. [Google Scholar] [CrossRef]
- Tamura, T.; Ueda, T.; Goto, T.; Tsukidate, T.; Shapira, Y.; Nishikawa, Y.; Fujisawa, A.; Hamachi, I. Rapid labelling and covalent inhibition of intracellular native proteins using ligand-directed N-acyl-N-alkyl sulfonamide. Nat. Commun. 2018, 9, 1870. [Google Scholar] [CrossRef]
- Lang, K.; Chin, J.W. Cellular Incorporation of Unnatural Amino Acids and Bioorthogonal Labeling of Proteins. Chem. Rev. 2014, 114, 4764–4806. [Google Scholar] [CrossRef] [PubMed]
- Lanning, B.R.; Whitby, L.R.; Dix, M.M.; Douhan, J.; Gilbert, A.M.; Hett, E.C.; Johnson, T.O.; Joslyn, C.; Kath, J.C.; Niessen, S. A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nat. Chem. Biol. 2014, 10, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Dalton, S.E.; Dittus, L.; Thomas, D.A.; Convery, M.A.; Nunes, J.; Bush, J.T.; Evans, J.P.; Werner, T.; Bantscheff, M.; Murphy, J.A.; et al. Selectively Targeting the Kinome-Conserved Lysine of PI3Kδ as a General Approach to Covalent Kinase Inhibition. J. Am. Chem. Soc. 2018, 140, 932–939. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Tamura, T.; Hamachi, I. Development of a Cell-Based Ligand-Screening System for Identifying Hsp90 Inhibitors. Biochemistry 2020, 59, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Asanuma, M.; Nakanishi, S.; Saito, Y.; Okazaki, M.; Dodo, K.; Sodeoka, M. Turn-ON fluorescent affinity labeling using a small bifunctional O-nitrobenzoxadiazole unit. Chem. Sci. 2014, 5, 1021–1029. [Google Scholar] [CrossRef]
- Lohse, J.; Swier, L.J.; Oudshoorn, R.C.; Médard, G.; Kuster, B.; Slotboom, D.-J.; Witte, M.D. Targeted Diazotransfer Reagents Enable Selective Modification of Proteins with Azides. Bioconjugate Chem. 2017, 28, 913–917. [Google Scholar] [CrossRef]
- Denda, M.; Morisaki, T.; Kohiki, T.; Yamamoto, J.; Sato, K.; Sagawa, I.; Inokuma, T.; Sato, Y.; Yamauchi, A.; Shigenaga, A. Labelling of endogenous target protein via N–S acyl transfer-mediated activation of N-sulfanylethylanilide. Org. Biomol. Chem. 2016, 14, 6244–6251. [Google Scholar] [CrossRef]
- Hughes, C.C.; Yang, Y.-L.; Liu, W.-T.; Dorrestein, P.C.; Clair, J.J.L.; Fenical, W. Marinopyrrole A Target Elucidation by Acyl Dye Transfer. J. Am. Chem. Soc. 2009, 131, 12094–12096. [Google Scholar] [CrossRef]
- Otsuki, S.; Nishimura, S.; Takabatake, H.; Nakajima, K.; Takasu, Y.; Yagura, T.; Sakai, Y.; Hattori, A.; Kakeya, H. Chemical tagging of a drug target using 5-sulfonyl tetrazole. Bioorganic Med. Chem. Lett. 2013, 23, 1608–1611. [Google Scholar] [CrossRef]
- Martín-Gago, P.; Fansa, E.K.; Winzker, M.; Murarka, S.; Janning, P.; Schultz-Fademrecht, C.; Baumann, M.; Wittinghofer, A.; Waldmann, H. Covalent Protein Labeling at Glutamic Acids. Cell Chem. Biol. 2017, 24, 589–597.e5. [Google Scholar] [CrossRef]
- Baggio, C.; Udompholkul, P.; Gambini, L.; Salem, A.F.; Jossart, J.; Perry, J.J.P.; Pellecchia, M. Aryl-fluorosulfate-based Lysine Covalent Pan-Inhibitors of Apoptosis Protein (IAP) Antagonists with Cellular Efficacy. J. Med. Chem. 2019, 62, 9188–9200. [Google Scholar] [CrossRef] [PubMed]
- Mortenson, D.E.; Brighty, G.J.; Plate, L.; Bare, G.; Chen, W.; Li, S.; Wang, H.; Cravatt, B.F.; Forli, S.; Powers, E.T.; et al. Inverse Drug Discovery” Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates. J. Am. Chem. Soc. 2018, 140, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Dong, J.; Plate, L.; Mortenson, D.E.; Brighty, G.J.; Li, S.; Liu, Y.; Galmozzi, A.; Lee, P.S.; Hulce, J.J.; et al. Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J. Am. Chem. Soc. 2016, 138, 7353–7364. [Google Scholar] [CrossRef] [PubMed]
- Koshi, Y.; Nakata, E.; Miyagawa, M.; Tsukiji, S.; Ogawa, T.; Hamachi, I. Target-Specific Chemical Acylation of Lectins by Ligand-Tethered DMAP Catalysts. J. Am. Chem. Soc. 2008, 130, 245–251. [Google Scholar] [CrossRef]
- Wang, H.; Koshi, Y.; Minato, D.; Nonaka, H.; Kiyonaka, S.; Mori, Y.; Tsukiji, S.; Hamachi, I. Chemical Cell-Surface Receptor Engineering Using Affinity-Guided, Multivalent Organocatalysts. J. Am. Chem. Soc. 2011, 133, 12220–12228. [Google Scholar] [CrossRef]
- Hayashi, T.; Yasueda, Y.; Tamura, T.; Takaoka, Y.; Hamachi, I. Analysis of Cell-Surface Receptor Dynamics through Covalent Labeling by Catalyst-Tethered Antibody. J. Am. Chem. Soc. 2015, 137, 5372–5380. [Google Scholar] [CrossRef]
- Hayashi, T.; Sun, Y.; Tamura, T.; Kuwata, K.; Song, Z.; Takaoka, Y.; Hamachi, I. Semisynthetic Lectin–4-Dimethylaminopyridine Conjugates for Labeling and Profiling Glycoproteins on Live Cell Surfaces. J. Am. Chem. Soc. 2013, 135, 12252–12258. [Google Scholar] [CrossRef]
- Takaoka, Y.; Uchinomiya, S.; Kobayashi, D.; Endo, M.; Hayashi, T.; Fukuyama, Y.; Hayasaka, H.; Miyasaka, M.; Ueda, T.; Shimada, I.; et al. Endogenous Membrane Receptor Labeling by Reactive Cytokines and Growth Factors to Chase Their Dynamics in Live Cells. Chem 2018, 4, 1451–1464. [Google Scholar] [CrossRef]
- Kunishima, M.; Nakanishi, S.; Nishida, J.; Tanaka, H.; Morisaki, D.; Hioki, K.; Nomoto, H. Convenient modular method for affinity labeling (MoAL method) based on a catalytic amidation. Chem. Commun. 2009, 37, 5597–5599. [Google Scholar] [CrossRef]
- Amamoto, Y.; Aoi, Y.; Nagashima, N.; Suto, H.; Yoshidome, D.; Arimura, Y.; Osakabe, A.; Kato, D.; Kurumizaka, H.; Kawashima, S.A.; et al. Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin. J. Am. Chem. Soc. 2017, 139, 7568–7576. [Google Scholar] [CrossRef]
- Ishiguro, T.; Amamoto, Y.; Tanabe, K.; Liu, J.; Kajino, H.; Fujimura, A.; Aoi, Y.; Osakabe, A.; Horikoshi, N.; Kurumizaka, H.; et al. Synthetic Chromatin Acylation by an Artificial Catalyst System. Chem 2017, 2, 840–859. [Google Scholar] [CrossRef]
- Adamson, C.; Kajino, H.; Kawashima, S.A.; Yamatsugu, K.; Kanai, M. Live-Cell Protein Modification by Boronate-Assisted Hydroxamic Acid Catalysis. J. Am. Chem. Soc. 2021, 143, 14976–14980. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Song, Z.; Amaike, K.; Lee, S.; Yin, S.; Kiyonaka, S.; Hamachi, I. Affinity-Guided Oxime Chemistry for Selective Protein Acylation in Live Tissue Systems. J. Am. Chem. Soc. 2017, 139, 14181–14191. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Murakawa, S.; Higashiguchi, K.; Matsuda, K.; Tamura, T.; Hamachi, I. Lysine-Reactive N-Acyl-N-aryl Sulfonamide Warheads: Improved Reaction Properties and Application in the Covalent Inhibition of an Ibrutinib-Resistant BTK Mutant. J. Am. Chem. Soc. 2023, 145, 26202–26212. [Google Scholar] [CrossRef]
- Sato, S.; Nakamura, H. Ligand-Directed Selective Protein Modification Based on Local Single-Electron-Transfer Catalysis. Angew. Chem. Int. Ed. 2013, 52, 8681–8684. [Google Scholar] [CrossRef]
- Sato, S.; Morita, K.; Nakamura, H. Regulation of Target Protein Knockdown and Labeling Using Ligand-Directed Ru(bpy)3 Photocatalyst. Bioconjugate Chem. 2015, 26, 250–256. [Google Scholar] [CrossRef]
- Burridge, K.M.; Page, R.C.; Konkolewicz, D. Bioconjugates—From a specialized past to a diverse future. Polymer 2020, 211, 123062. [Google Scholar] [CrossRef]
- Veronese, F.M.; Mero, A. The Impact of PEGylation on Biological Therapies. BioDrugs 2008, 22, 315–329. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Götz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Scott, W.R.P.; Hünenberger, P.H.; Tironi, I.G.; Mark, A.E.; Billeter, S.R.; Fennen, J.; Torda, A.E.; Huber, T.; Krüger, P.; van Gunsteren, W.F. The GROMOS Biomolecular Simulation Program Package. J. Phys. Chem. A 1999, 103, 3596–3607. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.J.; Tirado-Rives, J.; Jorgensen, W.L. Improved Peptide and Protein Torsional Energetics with the OPLS-AA Force Field. J. Chem. Theory Comput. 2015, 11, 3499–3509. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Russell, A.J.; Carmali, S. Moving Protein PEGylation from an Art to a Data Science. Bioconjugate Chem. 2022, 33, 1643–1653. [Google Scholar] [CrossRef]
- Behera, S.; Balasubramanian, S. Molecular simulations explain the exceptional thermal stability, solvent tolerance and solubility of protein–polymer surfactant bioconjugates in ionic liquids. Phys. Chem. Chem. Phys. 2022, 24, 21904–21915. [Google Scholar] [CrossRef]
- Kaupbayeva, B.; Boye, S.; Munasinghe, A.; Murata, H.; Matyjaszewski, K.; Lederer, A.; Colina, C.M.; Russell, A.J. Molecular Dynamics-Guided Design of a Functional Protein–ATRP Conjugate That Eliminates Protein–Protein Interactions. Bioconjugate Chem. 2021, 32, 821–832. [Google Scholar] [CrossRef]
- Hong, X.; Cholko, T.; Chang, C.-E.A.; Wheeldon, I. Multiscale simulation-guided design of enzyme bioconjugates with enhanced catalysis. Chem Catal. 2022, 2, 2691–2703. [Google Scholar] [CrossRef]
- Tamasi, M.J.; Patel, R.A.; Borca, C.H.; Kosuri, S.; Mugnier, H.; Upadhya, R.; Murthy, N.S.; Webb, M.A.; Gormley, A.J. Machine Learning on a Robotic Platform for the Design of Polymer–Protein Hybrids. Adv. Mater. 2022, 34, 2201809. [Google Scholar] [CrossRef]
- Ardejani, M.S.; Noodleman, L.; Powers, E.T.; Kelly, J.W. Stereoelectronic effects in stabilizing protein–N-glycan interactions revealed by experiment and machine learning. Nat. Chem. 2021, 13, 480–487. [Google Scholar] [CrossRef]
- Bartlett, G.J.; Choudhary, A.; Raines, R.T.; Woolfson, D.N. n→π* interactions in proteins. Nat. Chem. Biol. 2010, 6, 615–620. [Google Scholar] [CrossRef]
- Li, J.; Chen, P.R. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 2016, 12, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Karpenko, I.A.; Hiblot, J.; Johnsson, K. Imaging and manipulating proteins in live cells through covalent labeling. Nat. Chem. Biol. 2015, 11, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Teicher, B.A.; Hassan, R. Antibody–drug conjugates for cancer therapy. Lancet Oncol. 2016, 17, e254–e262. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaïa, N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat. Rev. Drug Discov. 2017, 16, 315–337. [Google Scholar] [CrossRef]
- Perols, A.; Honarvar, H.; Strand, J.; Selvaraju, R.; Orlova, A.; Karlström, A.E.; Tolmachev, V. Influence of DOTA Chelator Position on Biodistribution and Targeting Properties of 111In-Labeled Synthetic Anti-HER2 Affibody Molecules. Bioconjug. Chem. 2012, 23, 1661–1670. [Google Scholar] [CrossRef]
- Junutula, J.R.; Raab, H.; Clark, S.; Bhakta, S.; Leipold, D.D.; Weir, S.; Chen, Y.; Simpson, M.; Tsai, S.P.; Dennis, M.S.; et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol. 2008, 26, 925–932. [Google Scholar] [CrossRef]
- Sachdeva, A.; Wang, K.; Elliott, T.; Chin, J.W. Concerted, Rapid, Quantitative, and Site-Specific Dual Labeling of Proteins. J. Am. Chem. Soc. 2014, 136, 7785–7788. [Google Scholar] [CrossRef]
- Lang, K.; Davis, L.; Torres-Kolbus, J.; Chou, C.; Deiters, A.; Chin, J.W. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 2012, 4, 298–304. [Google Scholar] [CrossRef]
- Zheng, S.; Zhang, G.; Li, J.; Chen, P.R. Monitoring Endocytic Trafficking of Anthrax Lethal Factor by Precise and Quantitative Protein Labeling. Angew. Chem. Int. Ed. 2014, 53, 6449–6453. [Google Scholar] [CrossRef]
- Zeglis, B.M.; Sevak, K.K.; Reiner, T.; Mohindra, P.; Carlin, S.D.; Zanzonico, P.; Weissleder, R.; Lewis, J.S. A Pretargeted PET Imaging Strategy Based on Bioorthogonal Diels–Alder Click Chemistry. J. Nucl. Med. 2013, 54, 1389–1396. [Google Scholar] [CrossRef]
- Chari, R.V.J.; Miller, M.L.; Widdison, W.C. Antibody–Drug Conjugates: An Emerging Concept in Cancer Therapy. Angew. Chem. Int. Ed. 2014, 53, 3796–3827. [Google Scholar] [CrossRef] [PubMed]
- Rezhdo, A.; Islam, M.; Huang, M.; Van Deventer, J.A. Future prospects for noncanonical amino acids in biological therapeutics. Curr. Opin. Biotechnol. 2019, 60, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Pasut, G.; Veronese, F.M. PEG conjugates in clinical development or use as anticancer agents: An overview. Adv. Drug Deliv. Rev. 2009, 61, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Gaertner, H.F.; Offord, R.E. Site-Specific Attachment of Functionalized Poly(ethylene glycol) to the Amino Terminus of Proteins. Bioconjugate Chem. 1996, 7, 38–44. [Google Scholar] [CrossRef]
- Gilmore, J.M.; Scheck, R.A.; Esser-Kahn, A.P.; Joshi, N.S.; Francis, M.B. N-Terminal Protein Modification through a Biomimetic Transamination Reaction. Angew. Chem. Int. Ed. 2006, 45, 5307–5311. [Google Scholar] [CrossRef]
- Toda, N.; Asano, S.; Barbas, C.F., III. Rapid, Stable, Chemoselective Labeling of Thiols with Julia–Kocieński-like Reagents: A Serum-Stable Alternative to Maleimide-Based Protein Conjugation. Angew. Chem. Int. Ed. 2013, 52, 12592–12596. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, S.J.; Yesmin, S.; Vittala, S.K.; Sepay, N.; Xia, F.; Ali, S.I.; Chang, W.-C.; Hung, Y.-C.; Ma, W.-L. Target Bioconjugation of Protein Through Chemical, Molecular Dynamics, and Artificial Intelligence Approaches. Metabolites 2024, 14, 668. https://doi.org/10.3390/metabo14120668
Abbas SJ, Yesmin S, Vittala SK, Sepay N, Xia F, Ali SI, Chang W-C, Hung Y-C, Ma W-L. Target Bioconjugation of Protein Through Chemical, Molecular Dynamics, and Artificial Intelligence Approaches. Metabolites. 2024; 14(12):668. https://doi.org/10.3390/metabo14120668
Chicago/Turabian StyleAbbas, Sk Jahir, Sabina Yesmin, Sandeepa K. Vittala, Nayim Sepay, Fangfang Xia, Sk Imran Ali, Wei-Chun Chang, Yao-Ching Hung, and Wen-Lung Ma. 2024. "Target Bioconjugation of Protein Through Chemical, Molecular Dynamics, and Artificial Intelligence Approaches" Metabolites 14, no. 12: 668. https://doi.org/10.3390/metabo14120668
APA StyleAbbas, S. J., Yesmin, S., Vittala, S. K., Sepay, N., Xia, F., Ali, S. I., Chang, W.-C., Hung, Y.-C., & Ma, W.-L. (2024). Target Bioconjugation of Protein Through Chemical, Molecular Dynamics, and Artificial Intelligence Approaches. Metabolites, 14(12), 668. https://doi.org/10.3390/metabo14120668








