Unravel the Supremacy of Klebsiella variicola over Native Microbial Strains for Aroma-Enhancing Compound Production in Reconstituted Tobacco Concentrate through Metagenomic Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experiment Materials, Strain, Fermentation Media and Conditions
2.2. Whole-Genome Shotgun Sequencing for Metagenomic Sequence Analysis
2.3. Gene Prediction
2.4. GC-MS Analysis of FRTLC
2.5. Determination of Routine Chemical Components
2.6. Evaluation of the Quality of Cigarettes
3. Results
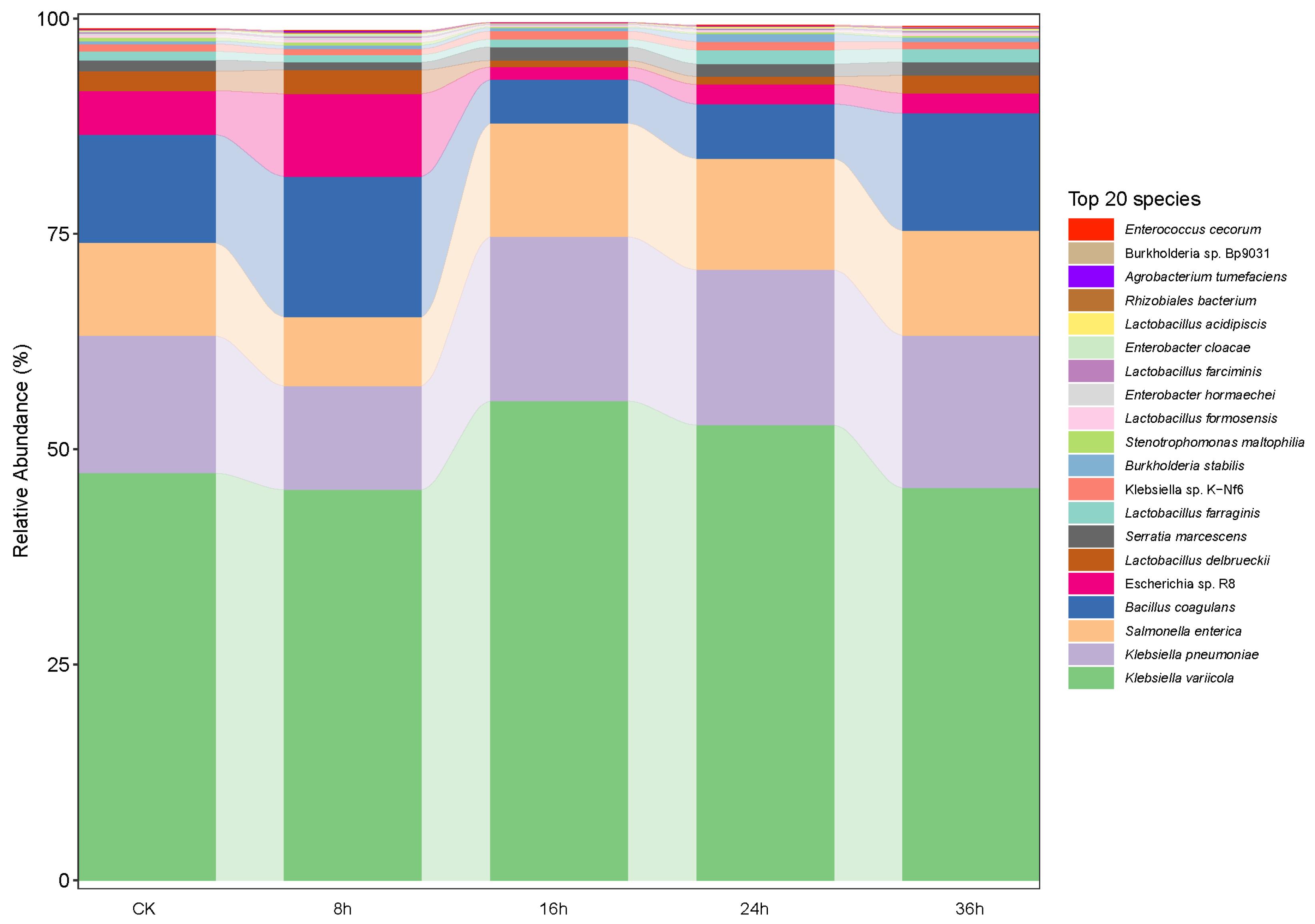
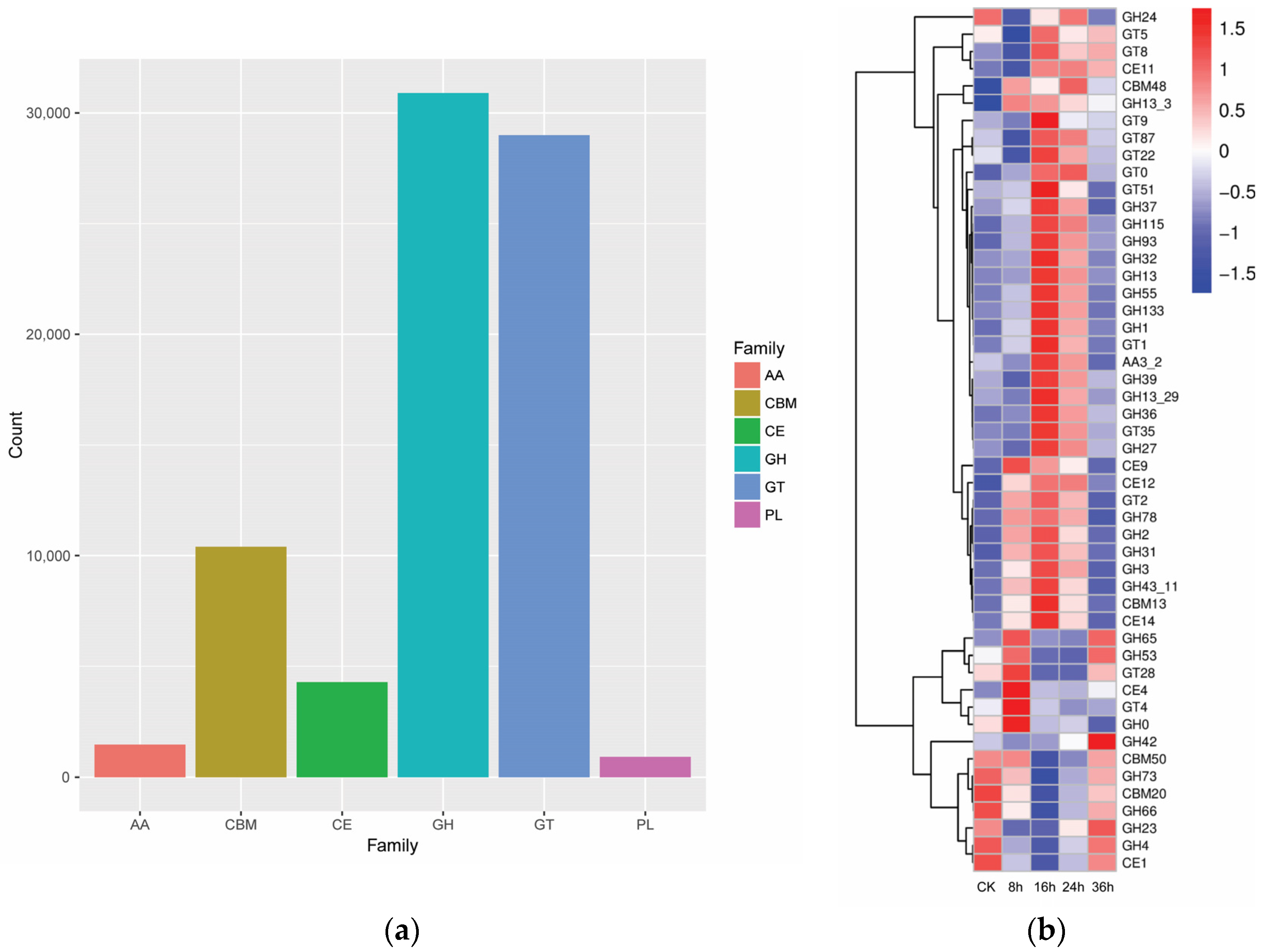
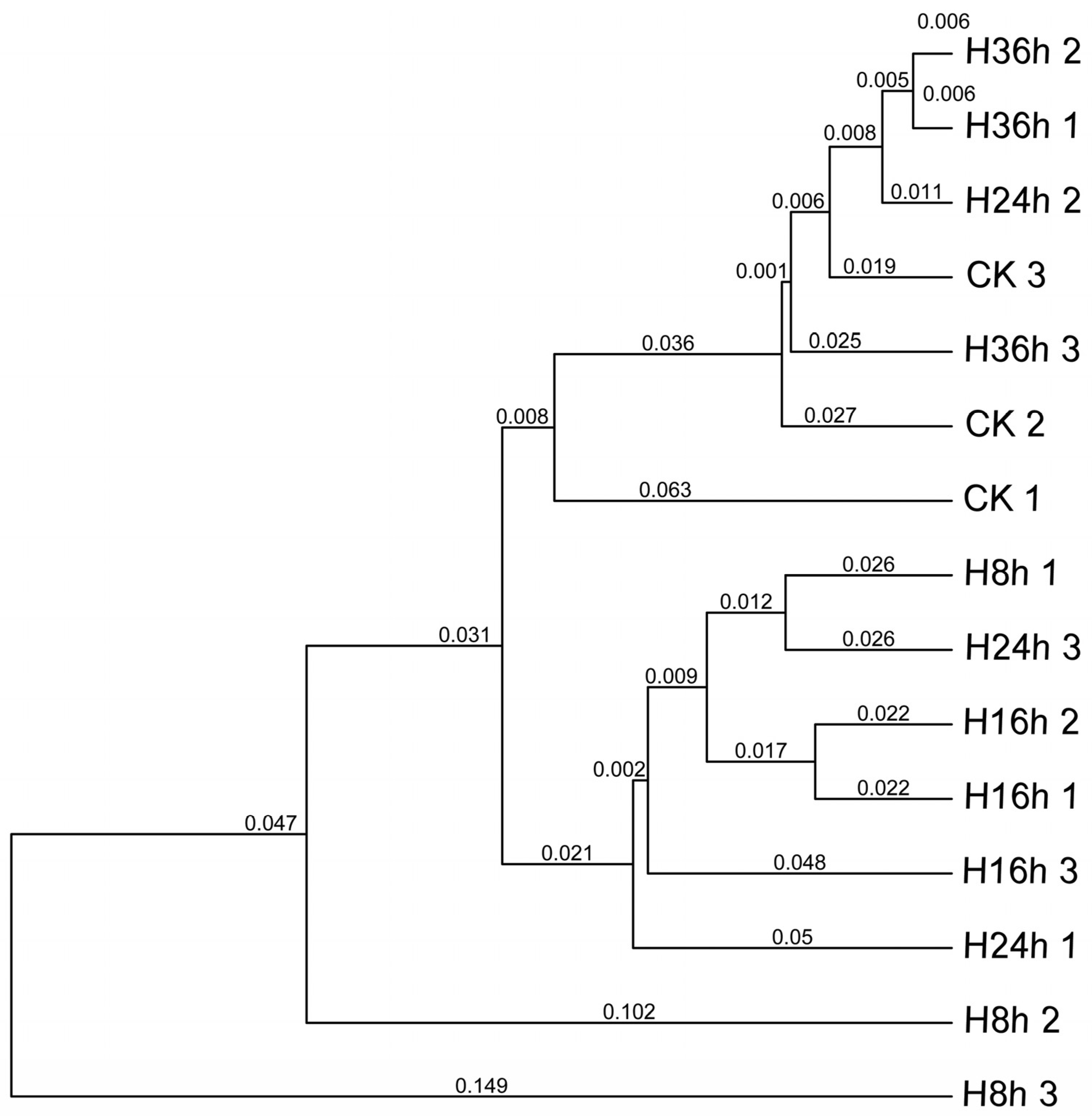
3.1. Dynamics of Microbial Community and Functional Genes in FRTLC
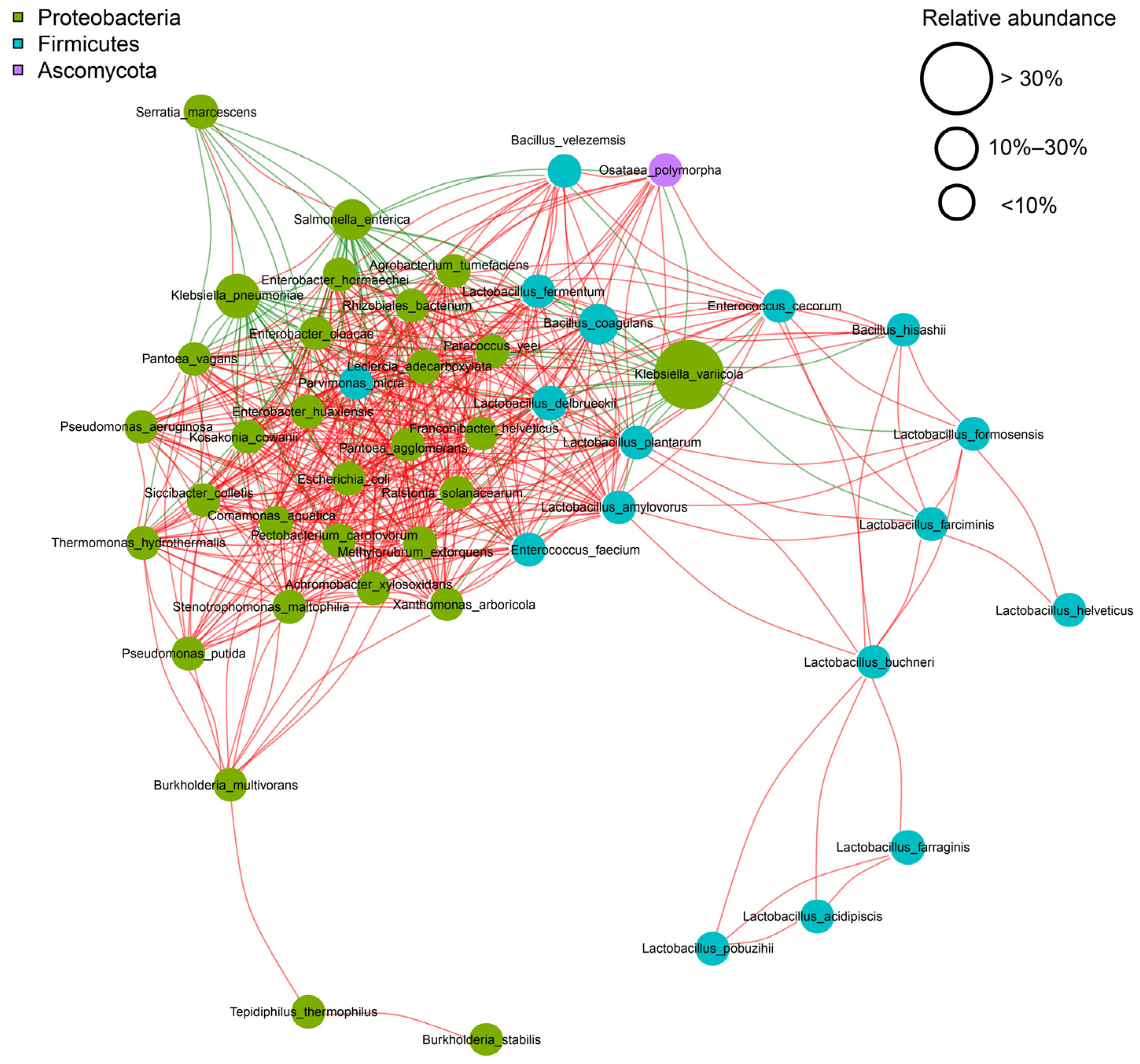
3.1.1. Microbial Interactions and Fermentation Dynamics
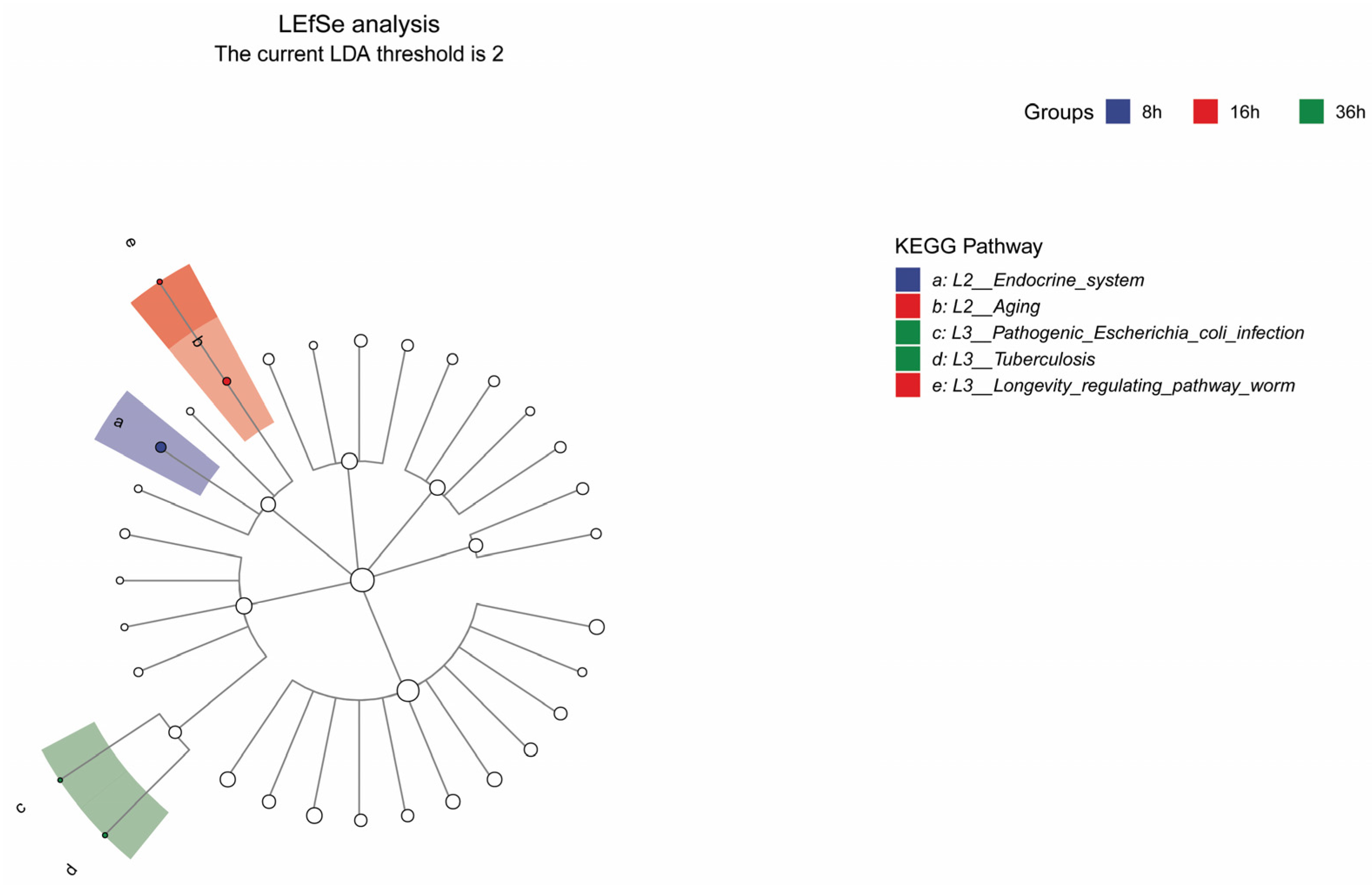
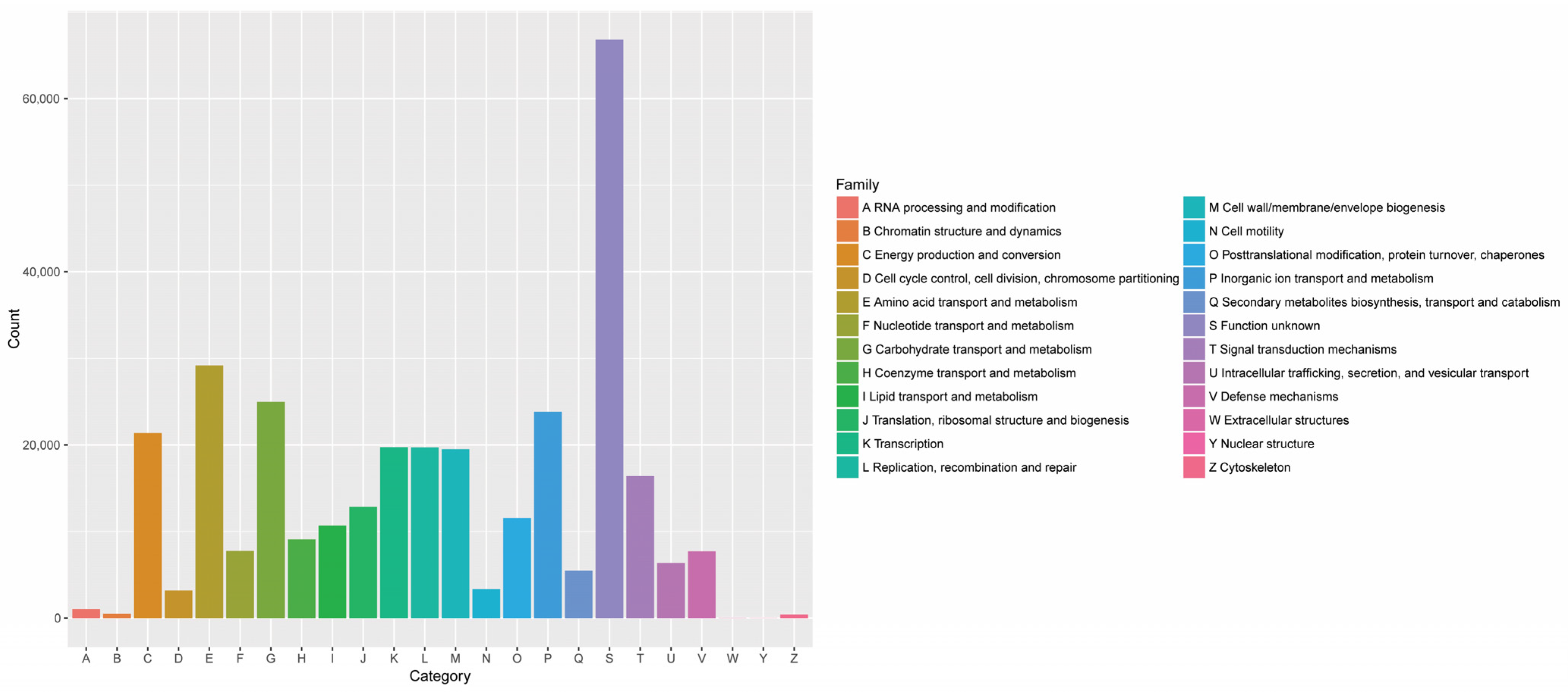
3.1.2. Analysis of FRTLC Microbial Gene Function
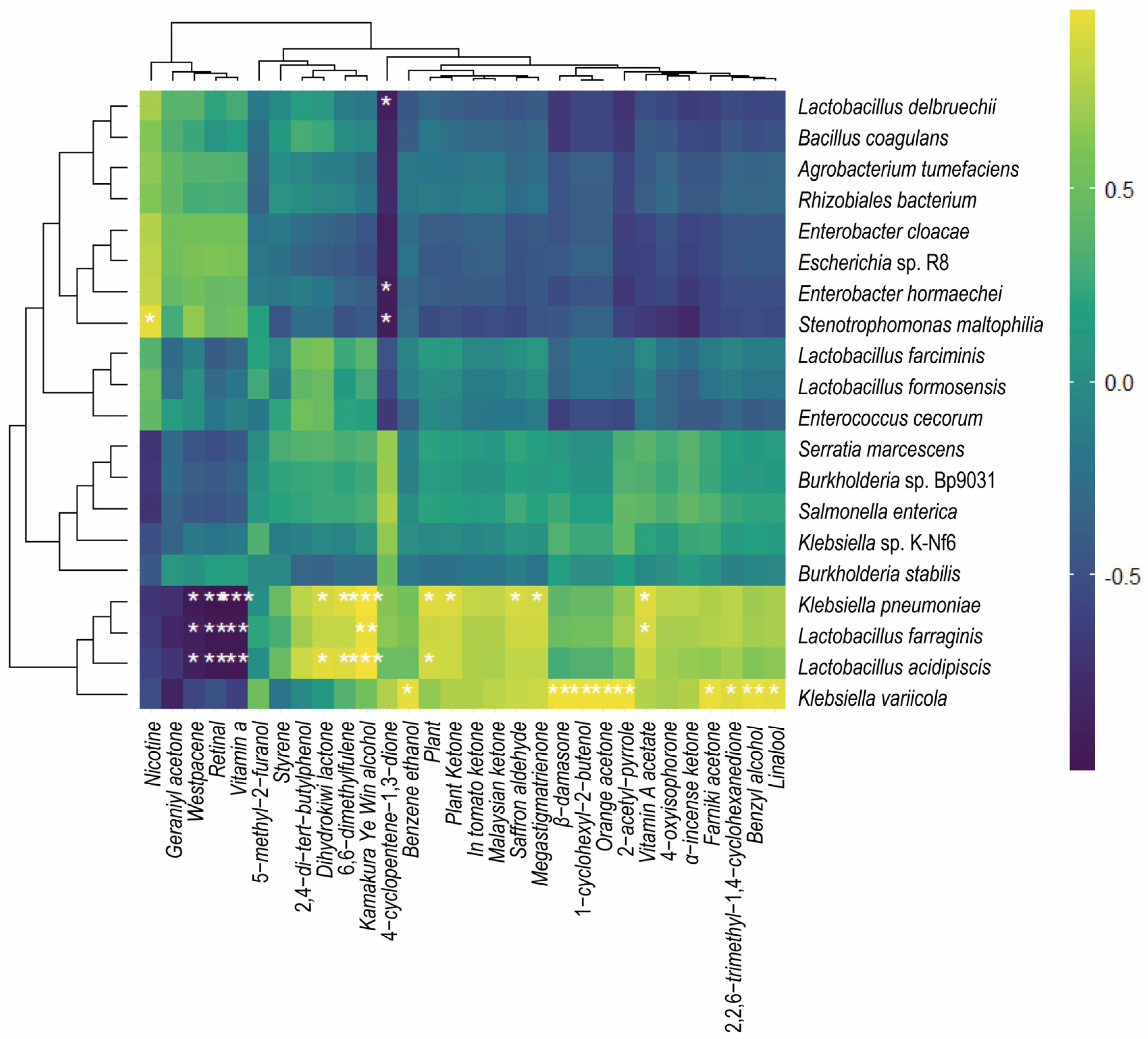
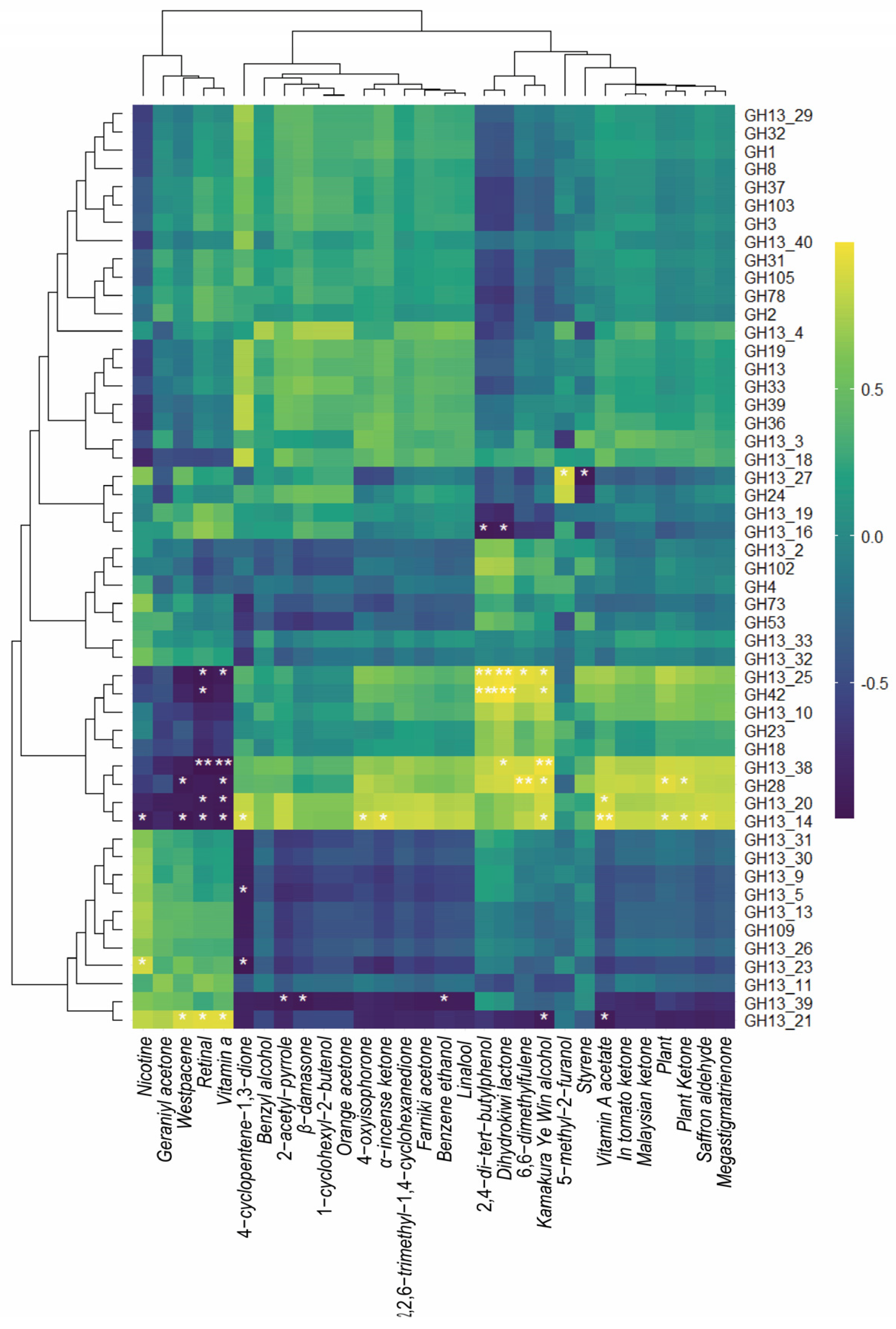
3.2. Correlation between Microbial Growth, NAECs, and Nicotine Level in FRTLC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Lu, J.J.; Xue, A.Q.; Cao, Z.Y.; Yang, S.J.; Hu, X.F. Diversity of Plant Growth-Promoting Paenibacillus mucilaginosus Isolated from Vegetable Fields in Zhejiang, China. Ann. Microbiol. 2014, 64, 1745–1756. [Google Scholar] [CrossRef]
- Li, H.; Qu, Y.; Cao, K.; Xiong, J.; Hua, K.; Liu, Z.; Wang, H.; Yue, L.; Tang, Y.; Ding, G. Cigarette Flavouring Regulation by Using Aroma-Producing Microorganism Isolated from Maotai Daqu. E3S Web Conf. 2021, 271, 04031. [Google Scholar] [CrossRef]
- Huang, S.; Wang, M.; Mao, D.B.; Rasool, A.; Jia, C.; Yang, P.; Han, L.; Yan, M. Isolation, Identification and Characterization of Growth Parameters of Pseudomonas putida HSM-C2 with Coumarin-Degrading Bacteria. Molecules 2022, 27, 6007. [Google Scholar] [CrossRef]
- Potts, R.J.; Bombick, B.R.; Meckley, D.R.; Ayres, P.H.; Pence, D.H. A Summary of Toxicological and Chemical Data Relevant to the Evaluation of Cast Sheet Tobacco. Exp. Toxicol. Pathol. 2010, 62, 117–126. [Google Scholar] [CrossRef]
- Wang, W.S.; Wang, Y.; Yang, L.J.; Liu, B.; Lan, M.; Sun, W. Studies on Thermal Behavior of Reconstituted Tobacco Sheet. Thermochim. Acta 2005, 437, 7–11. [Google Scholar] [CrossRef]
- Brown, C.J.; Cheng, J.M. Electronic Cigarettes: Product Characterization and Design Considerations. Tob. Control 2014, 23, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Liu, Y.X.; Jiang, Y.F.; Zhang, L.; Zhang, Y.; Zhao, L.; Jiang, X.; Zhang, Q. Advanced Materials Design for Adsorption of Toxic Substances in Cigarette Smoke. Adv. Sci. 2023, 10, e2301834. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.H.; Fan, H.; Liu, J.L.; Yang, Y.; Xu, S.; Gao, Y.; Zhou, G. High-Throughput Bacterial Analysis on Aqueous Extract of Waste Tobacco Scrap. Tob. Sci. Technol. 2016, 49, 1–7. [Google Scholar] [CrossRef]
- Liu, H.G.; He, H.L.; Cheng, C.H.; Liu, J.; Shu, M.; Jiao, Y.; Tao, F.; Zhong, W. Diversity Analysis of the Bacterial Community in Tobacco Waste Extract during Reconstituted Tobacco Process. Appl. Microbiol. Biotechnol. 2015, 99, 469–476. [Google Scholar] [CrossRef]
- Ruan, A.; Min, H. Studies on Microbiological Degradation of Tobacco Tar. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2005, 40, 2073–2083. [Google Scholar] [CrossRef]
- Zhang, Z.L.; Mei, X.T.; He, Z.L.; Xie, X.; Yang, Y.; Mei, C.; Xue, D.; Hu, T.; Shu, M.; Zhong, W. Nicotine Metabolism Pathway in Bacteria: Mechanism, Modification, and Application. Appl. Microbiol. Biotechnol. 2022, 106, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.F.; Zhang, Q.Y.; Wu, Q.Y.; Li, D.; Wu, X.; Li, P.; Zhou, Q.; Cai, W.; Zhang, J.; Du, G. Effects of Inoculation with Acinetobacter on Fermentation of Cigar Tobacco Leaves. Front. Microbiol. 2022, 13, 911791. [Google Scholar] [CrossRef] [PubMed]
- Gravely, L.E.; Geiss, V.L. Microbial digestion of tobacco materials using mixed cultures. US04476881A, 9 May 1983. [Google Scholar]
- Huang, Y.G.; Wu, Q.; Xu, Y. Isolation and Identification of a Black Aspergillus Strain and the Effect of Its Novel Protease on the Aroma of Moutai-Flavoured Liquor. J. Inst. Brew. 2014, 120, 268–276. [Google Scholar] [CrossRef]
- Wang, L. Research Trends in Jiang-Flavor Baijiu Fermentation: From Fermentation Microecology to Environmental Ecology. J. Food Sci. 2022, 87, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.X.; Xu, L.Y.; Zhu, Y.P.; Li, J.; Li, X. Optimization of Synthesis Technology of Galacto-Oligosaccharides by Heterologous Expression of β-Galactosidase from Klebsiella. J. Chin. Inst. Food Sci. Technol. 2021, 21, 245–251. [Google Scholar] [CrossRef]
- Ansari, S.A.; Satar, R. Recombinant β-Galactosidases—Past, Present and Future: A Mini Review. J. Mol. Catal. B Enzym. 2012, 81, 1–6. [Google Scholar] [CrossRef]
- Lu, L.L.; Guo, L.C.; Wang, K.; Liu, Y.; Xiao, M. β-Galactosidases: A Great Tool for Synthesizing Galactose-Containing Carbohydrates. Biotechnol. Adv. 2020, 39, 107465. [Google Scholar] [CrossRef]
- Oliveira, C.; Guimarães, P.M.R.; Domingues, L. Recombinant Microbial Systems for Improved β-Galactosidase Production and Biotechnological Applications. Biotechnol. Adv. 2011, 29, 600–609. [Google Scholar] [CrossRef]
- Wöhner, G.; Wöber, G. Pullulanase, an Enzyme of Starch Catabolism, Is Associated with the Outer Membrane of Klebsiella. Arch. Microbiol. 1978, 116, 303–310. [Google Scholar] [CrossRef]
- Boyd, J.; Turvey, J.R. Isolation of a Poly-α-l-Guluronate Lyase from Klebsiella Aerogenes. Carbohydr. Res. 1977, 57, 163–171. [Google Scholar] [CrossRef]
- Caffrey, A.; Ebeler, S.E. The Occurrence of Glycosylated Aroma Precursors in Vitis vinifera Fruit and Humulus lupulus Hop Cones and Their Roles in Wine and Beer Volatile Aroma Production. Foods 2021, 10, 935. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.J. Characteristic Analysis of Carbapenem-Resistant Klebsiella pneumoniae and Study on the Mechanism of Tigecycline Resistance. Master’s Thesis, Fujian Medical University, Fuzhou, China, 2020; p. 1. [Google Scholar]
- Zhong, Y.M.; Luo, X.Y.; Li, Y.M.; Li, H.L.; Jian, Z.J.; Yan, Q.; Liu, W.E. Clinical characteristics of patients with Klebsiella variicola infection. Chin. J. Infect. Control 2023, 22, 38–43. [Google Scholar]
- Huang, H.; Huang, Y.L.; Gu, D.X.; Zhang, R.; Zhou, H.W. Analysis of drug resistance genes and molecular typing of carbapenem-resistant Klebsiella variicola strains. Chin. J. Microbiol. Immunol. 2019, 39, 197–201. [Google Scholar] [CrossRef]
- Moreno, F.L.; Quintanilla-Carvajal, M.X.; Sotelo, L.I.; Osorio, C.; Raventós, M.; Hernández, E.; Ruiz, Y. Volatile Compounds, Sensory Quality and Ice Morphology in Falling-Film and Block Freeze Concentration of Coffee Extract. J. Food Eng. 2015, 166, 64–71. [Google Scholar] [CrossRef]
- Fan, X.; Zi, W.H.; Ao, J.; Li, B.; Qiao, J.; Wang, Y.; Nong, Y. Analysis and Application Evaluation of the Flavour-Precursor and Volatile-Aroma-Component Differences between Waste Tobacco Stems. Heliyon 2022, 8, e10658. [Google Scholar] [CrossRef] [PubMed]
- Zi, W.H.; Zhang, X.L.; Peng, J.H.; Zhang, L.; Long, M.; Zuo, J. Optimization of Microwave Drying Biomass Material of Stem Granules from Waste Tobacco Using Response Surface Methodology. Dry Technol. 2013, 31, 1234–1244. [Google Scholar] [CrossRef]
- Pang, X.N.; Han, B.Z.; Huang, X.N.; Zhang, X.; Hou, L.F.; Cao, M.; Gao, L.J.; Hu, G.H.; Chen, J.Y. Effect of the Environment Microbiota on the Flavour of Light-Flavour Baijiu during Spontaneous Fermentation. Sci. Rep. 2018, 8, 3396. [Google Scholar] [CrossRef]
- Su, C.; Zhang, K.Z.; Cao, X.Z.; Yang, J.G. Effects of Saccharomycopsis fibuligera and Saccharomyces cerevisiae Inoculation on Small Fermentation Starters in Sichuan-Style Xiaoqu Liquor. Food Res. Int. 2020, 137, 109425. [Google Scholar] [CrossRef]
- Neuman, H.; Debelius, J.W.; Knight, R.; Koren, O. Microbial Endocrinology: The Interplay between the Microbiota and the Endocrine System. FEMS Microbiol. Rev. 2015, 39, 509–521. [Google Scholar] [CrossRef]
- Williams, C.L.; Garcia-Reyero, N.; Martyniuk, C.J.; Tubbs, C.W.; Bisesi, J.H. Regulation of Endocrine Systems by the Microbiome: Perspectives from Comparative Animal Models. Gen. Comp. Endocrinol. 2020, 292, 113437. [Google Scholar] [CrossRef]
- Liu, A.M.; Yuan, K.L.; Li, Q.; Liu, S.; Li, Y.; Tao, M.; Xu, H.; Tian, J.; Guan, S.; Zhu, W. Metabolomics and Proteomics Revealed the Synthesis Difference of Aroma Precursors in Tobacco Leaves at Various Growth Stages. Plant Physiol. Biochem. 2022, 192, 308–319. [Google Scholar] [CrossRef]
- Fan, J.H.; Kong, G.H.; Yao, H.; Wu, Y.; Zhao, G.; Li, F.; Zhang, G. Widely Targeted Metabolomic Analysis Reveals That Volatile Metabolites in Cigar Tobacco Leaves Dynamically Change during Fermentation. Biochem. Biophys. Rep. 2023, 35, 101532. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Hu, W.R.; Li, P.H.; Liu, J.; Zhang, Q.; Zhou, Q.; Luo, C.; Li, D. Effects of Fermentation Medium on Cigar Filler. Front. Bioeng. Biotechnol. 2022, 10, 1069796. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hu, Y.Q.; Wang, Q.; Liu, J.; Fang, S.; Huang, D.; Pang, X.; Cao, J.; Gao, Y.; Ning, Y. Study on the Correlation between the Dominant Microflora and the Main Flavor Substances in the Fermentation Process of Cigar Tobacco Leaves. Front. Microbiol. 2023, 14, 1267447. [Google Scholar] [CrossRef]
- Chen, J.; He, X.; Zhang, X.Y.; Chen, Y.; Zhao, L.; Su, J.; Qu, S.; Ji, X.; Wang, T.; Li, Z.; et al. The Applicability of Different Tobacco Types to Heated Tobacco Products. Ind. Crops Prod. 2021, 168, 113579. [Google Scholar] [CrossRef]
- Tang, Z.X.; Chen, L.L.; Chen, Z.B.; Fu, Y.; Sun, X.; Wang, B.; Xia, T. Climatic Factors Determine the Yield and Quality of Honghe Flue-Cured Tobacco. Sci. Rep. 2020, 10, 19868. [Google Scholar] [CrossRef] [PubMed]
- Kurt, D. Impacts of Environmental Variations on Quality and Chemical Contents of Oriental Tobacco. Beiträge zur Tab. Int./Con. to Tob. Res. 2021, 30, 50–62. [Google Scholar] [CrossRef]
- Li, J.J.; Zhao, Y.Y.; Qin, Y.Q.; Shi, H. Influence of Microbiota and Metabolites on the Quality of Tobacco during Fermentation. BMC Microbiol. 2020, 20, 356. [Google Scholar] [CrossRef]
- Huang, W.J.; Yang, J.K.; Duan, Y.Q.; Gu, W.; Gong, X.; Zhe, W.; Su, C.; Zhang, K.Q. Bacterial Diversities on Unaged and Aging Flue-Cured Tobacco Leaves Estimated by 16S RRNA Sequence Analysis. Appl. Microbiol. Biotechnol. 2010, 88, 553–562. [Google Scholar] [CrossRef]
- Gong, X.W.; Yang, J.K.; Duan, Y.Q.; Dong, J.Y.; Zhe, W.; Wang, L.; Li, Q.H.; Zhang, K.Q. Isolation and Characterization of Rhodococcus sp. Y22 and Its Potential Application to Tobacco Processing. Res. Microbiol. 2009, 160, 200–204. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Zheng, T.F.; Yang, Z.; Yang, S.; Cai, W.; Li, P.; Huang, Y.; Zhang, J.; Li, D. Analysis of the Structure and Metabolic Function of Microbial Community in Cigar Tobacco Leaves in Agricultural Processing Stage. Front. Microbiol. 2023, 14, 1230547. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wu, Z.Y.; Zhang, X.P.; Xi, G.; Zhao, Z.; Lai, M.; Zhao, M. Microbial Community and Metabolic Function Analysis of Cigar Tobacco Leaves during Fermentation. Microbiologyopen 2021, 10, e1171. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Liu, Y.F.; Hu, W.R.; Cai, W.; Zheng, Z.; Luo, C.; Li, D. Development of Candida Autochthonous Starter for Cigar Fermentation via Dissecting the Microbiome. Front. Microbiol. 2023, 14, 1138877. [Google Scholar] [CrossRef]
- Huang, X.L.; Lei, L.P.; Xai, Z.Y.; Wu, Y.P.; Yang, K.J. Preliminary study on the mechanism of degradation of nicotine and increasing scent of Arthrobacter sp. Southwest Chin. J. Agric. Sci. 2010, 23, 1867–1869. [Google Scholar] [CrossRef]
- Ardö, Y. Flavour Formation by Amino Acid Catabolism. Biotechnol. Adv. 2006, 24, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Bustamante, E.; Sánchez, S. Microbial Production of C13-Norisoprenoids and Other Aroma Compounds via Carotenoid Cleavage. Crit. Rev. Microbiol. 2007, 33, 211–230. [Google Scholar] [CrossRef]
- Johnson, R.R.; Nicholson, J.A. The Structure, Chemistry, and Synthesis of Solanone. A New Anomalous Terpenoid Ketone from Tobacco. J. Org. Chem. 1965, 30, 2918–2921. [Google Scholar] [CrossRef]
- Demole, E.; Berthet, D. A Chemical Study of Burley Tobacco Flavour (Nicotiana tabacum L.). I. Volatile to Medium-volatile Constituents (b.p. ≦ 84°/0.001 Torr. Helv. Chim. Acta 1972, 55, 1866–1882. [Google Scholar] [CrossRef]
- Ahmed, A.; Aslam, M.; Ashraf, M.; ul-Hassan Nasim, F.; Batool, K.; Bibi, A. Microbial β-Glucosidases: Screening, Characterization, Cloning and Applications. J. Appl. Environ. Microbiol. 2017, 5, 57–73. [Google Scholar] [CrossRef]
- Wahlberg, I.; Karlsson, K.; Austin, D.J.; Junker, N.; Roeraade, J.; Enzell, C.R.; Johnson, W.H. Effects of Flue-Curing and Ageing on the Volatile, Neutral and Acidic Constituents of Virginia Tobacco. Phytochemistry 1977, 16, 1217–1231. [Google Scholar] [CrossRef]
- Sathya, T.A.; Khan, M. Diversity of Glycosyl Hydrolase Enzymes from Metagenome and Their Application in Food Industry. J. Food Sci. 2014, 79, R2149–R2156. [Google Scholar] [CrossRef]
- Abdul Manas, N.H.; Md. Illias, R.; Mahadi, N.M. Strategy in Manipulating Transglycosylation Activity of Glycosyl Hydrolase for Oligosaccharide Production. Crit. Rev. Biotechnol. 2018, 38, 272–293. [Google Scholar] [CrossRef]
- Amin, K.; Tranchimand, S.; Benvegnu, T.; Abdel-Razzak, Z.; Chamieh, H. Glycoside Hydrolases and Glycosyltransferases from Hyperthermophilic Archaea: Insights on Their Characteristics and Applications in Biotechnology. Biomolecules 2021, 11, 1557. [Google Scholar] [CrossRef]
- Sun, W.; Yang, J.; Zhou, J.; Xu, D.; Bai, R.; Zhang, G.; Ma, Y.; Shi, H. Effect of Different Nitrogen Forms on Nitrate Nitrogen Content and TSNAs Formation in Tobacco. Acta Tabacaria Sin. 2015, 21, 1557. [Google Scholar] [CrossRef]
- Schwab, W.; Fischer, T.C.; Giri, A.; Wüst, M. Potential Applications of Glucosyltransferases in Terpene Glucoside Production: Impacts on the Use of Aroma and Fragrance. Appl. Microbiol. Biotechnol. 2015, 99, 165–174. [Google Scholar] [CrossRef]
- Liang, Z.J.; Fang, Z.X.; Pai, A.; Luo, J.; Gan, R.; Gao, Y.; Lu, J.; Zhang, P. Glycosidically Bound Aroma Precursors in Fruits: A Comprehensive Review. Crit. Rev. Food Sci. Nutr. 2022, 62, 215–243. [Google Scholar] [CrossRef]
- Zhu, F.M.; Du, B.; Ma, Y.G.; Li, J. The Glycosidic Aroma Precursors in Wine: Occurrence, Characterization and Potential Biological Applications. Phytochem. Rev. 2017, 16, 565–571. [Google Scholar] [CrossRef]
- Zhang, Q.; Luo, C.; Li, D.; Cai, W. Research Progress in Curing and Fermentation Technology for Cigar Tobacco Leaf Production. Acta Tabacaria Sin. 2020, 26, 692–697. [Google Scholar]
- Yang, Y.; Peng, Q.R.; Ou, M.Y.; Wu, Y.; Fang, J. Research Progress in Tobacco Fermentation. J. Biosci. Med. 2018, 6, 105–114. [Google Scholar] [CrossRef]
- Xia, Y.N.; Shuang, Q. Metagenomic Analysis of Microbial Diversity and Key Flavor-Related Genes in Solid-State Fermented Jujube Mash for Jujube Wine. Shipin Kexue/Food Sci. 2022, 43, 192–198. [Google Scholar] [CrossRef]

| Number | CAS No. | Compound Name | Concentration (mg/mL) | Increase (%) | ||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 8 h | 16 h | 24 h | 36 h | ||||
| 1 | 100-42-5 | Styrene | 0.28 ± 0.014 | 0.37 ± 0.015 | 0.40 ± 0.018 | 0.32 ± 0.016 | 0.47 ± 0.017 | 67.85 |
| 2 | 3857-25-8 | 5-Methyl-2-furanol | 0.03 ± 0.002 | 0.02 ± 0.001 | 0.02 ± 0.002 | 0.03 ± 0.001 | 0.02 ± 0.002 | NA |
| 3 | 100-52-7 | Benzaldehyde | 0.03 ± 0.003 | 0.05 ± 0.002 | 0.04 ± 0.002 | 0.04 ± 0.001 | 0.04 ± 0.002 | 66.66 |
| 4 | 100-51-6 | Benzyl alcohol | 0.24 ± 0.016 | 0.27 ± 0.018 | 0.27 ± 0.021 | 0.38 ± 0.023 | 0.32 ± 0.001 | 58.33 |
| 5 | 122-78-1 | Phenylacetaldehyde | 0.67 ± 0.031 | 0.9 ± 0.044 | 0.84 ± 0.058 | 1.07 ± 0.062 | 0.98 ± 0.064 | 59.70 |
| 6 | 1072-83-9 | 2-Acetylpyrrole | 0.17 ± 0.015 | 0.16 ± 0.014 | 0.20 ± 0.02 | 0.31 ± 0.019 | 0.21 ± 0.021 | 82.35 |
| 7 | 78-70-6 | Linalool | 0.08 ± 0.002 | 0.09 ± 0.003 | 0.09 ± 0.002 | 0.12 ± 0.019 | 0.10 ± 0.001 | 50 |
| 8 | 60-12-8 | Phenylethanol | 0.19 ± 0.008 | 0.21 ± 0.011 | 0.19 ± 0.009 | 0.28 ± 0.017 | 0.21 ± 0.016 | 47.36 |
| 9 | 1125-21-9 | 4-oxoisofolkone | 0.06 ± 0.003 | 0.09 ± 0.004 | 0.10 ± 0.011 | 0.13 ± 0.015 | 0.12 ± 0.009 | 116.66 |
| 10 | 432-25-7 | β-Cyclocitral | 0.22 ± 0.012 | 0.27 ± 0.014 | 0.26 ± 0.008 | 0.36 ± 0.037 | 0.35 ± 0.025 | 63.63 |
| 11 | 56797-40-1 | (Z)-7-Cetylenal | 0.07 ± 0.004 | 0.09 ± 0.008 | 0.09 ± 0.011 | 0.12 ± 0.013 | 0.12 ± 0.009 | 71.42 |
| 12 | 116-26-7 | Crocin aldehyde | 0.08 ± 0.002 | 0.09 ± 0.004 | 0.09 ± 0.004 | 0.12 ± 0.009 | 0.11 ± 0.007 | 50 |
| 13 | 475-03-6 | 1,2,3,4-tetrahydro-1,1,6-trimethylnaphthalene | 0.09 ± 0.004 | 0.09 ± 0.005 | 0.08 ± 0.008 | 0.25 ± 0.021 | 0.23 ± 0.022 | 177.77 |
| 14 | 54-11-5 | Nicotine | 0.08 ± 0.005 | 0.08 ± 0.003 | 0.06 ± 0.004 | 0.06 ± 0.004 | 0.06 ± 0.005 | 25 |
| 15 | 21852-80-2 | 1,9-Heptadecadiene-4,6-diyn-3-ol | 0.06 ± 0.004 | 0.06 ± 0.004 | 0.06 ± 0.003 | 0.13 ± 0.009 | 0.16 ± 0.014 | 116.67 |
| 16 | 23726-93-4 | β-Damastrone | 0.06 ± 0.003 | 0.06 ± 0.002 | 0.07 ± 0.005 | 0.13 ± 0.008 | 0.06 ± 0.002 | 116.67 |
| 17 | 1883-13-2 | (±)-3-hydroxylauric acid | - | - | 0.05 ± 0.001 | - | - | NA |
| 18 | 54868-48-3 | Solanone | 0.91 ± 0.056 | 1.11 ± 0.079 | 1.07 ± 0.047 | 1.34 ± 0.102 | 1.26 ± 0.084 | 47.25 |
| 19 | 35044-68-9 | α-Damarone | 0.63 ± 0.032 | 0.75 ± 0.049 | 0.72 ± 0.036 | 0.91 ± 0.051 | 0.84 ± 0.047 | 44.44 |
| 20 | 3796-70-1 | Geranyl acetone | 0.26 ± 0.021 | 0.32 ± 0.025 | 0.31 ± 0.024 | 0.17 ± 0.019 | 0.24 ± 0.023 | D |
| 21 | 3879-26-3 | Neroli acetone | - | - | - | 0.07 ± 0.004 | - | NA |
| 22 | 96-76-4 | 2,4-di-tert-butylphenol | 0.04 ± 0.003 | 0.05 ± 0.004 | 0.05 ± 0.003 | 0.09 ± 0.005 | 0.52 ± 0.004 | 125 |
| 23 | 17092-92-1 | Dihydrokiwi lactone | 0.42 ± 0.044 | 0.34 ± 0.041 | 0.33 ± 0.03 | 1.23 ± 0.098 | 0.58 ± 0.054 | 192.86 |
| 24 | 13215-88-8 | Megastigmatrienone a | 0.18 ± 0.009 | 0.20 ± 0.011 | 0.17 ± 0.01 | 0.39 ± 0.042 | 0.28 ± 0.03 | 116.67 |
| 25 | 13215-88-8 | Megastigmatrienone b | 1.03 ± 0.057 | 1.03 ± 0.061 | 0.98 ± 0.044 | 1.24 ± 0.081 | 1.09 ± 0.093 | 20.39 |
| 26 | 13215-88-8 | Megastigmatrienone c | 0.82 ± 0.052 | 0.96 ± 0.074 | 0.91 ± 0.063 | 1.13 ± 0.089 | 0.99 ± 0.076 | 37.80 |
| 27 | 13215-88-8 | Megastigmatrienone d | 0.18 ± 0.008 | 0.19 ± 0.012 | 0.25 ± 0.026 | 0.34 ± 0.015 | 0.28 ± 0.024 | 88.89 |
| 28 | 473-08-5 | α-Cyperone | 0.28 ± 0.014 | 0.33 ± 0.021 | 0.37 ± 0.027 | 0.43 ± 0.032 | 0.40 ± 0.028 | 53.57 |
| 29 | 102608-53-7 | Phytol | 1.67 ± 0.075 | 1.99 ± 0.09 | 1.93 ± 0.074 | 2.47 ± 0.095 | 2.50 ± 0.112 | 49.70 |
| 30 | 502-69-2 | Phytone | 0.43 ± 0.025 | 0.50 ± 0.034 | 0.48 ± 0.03 | 0.64 ± 0.051 | 0.61 ± 0.058 | 48.837 |
| 31 | 65646-68-6 | 4-hydroxyphenyl retinamide | 0.36 ± 0.035 | 0.38 ± 0.031 | 0.34 ± 0.029 | 0.94 ± 0.084 | 0.37 ± 0.026 | 161.11 |
| 32 | 25360-09-2 | potassium,2,6-ditert-butylphenolate | 0.05 ± 0.002 | - | 0.05 ± 0.003 | 0.08 ± 0.005 | - | NA |
| 33 | 1117-52-8 | farnesyl acetone | 0.87 ± 0.084 | 0.92 ± 0.075 | 0.97 ± 0.082 | 1.27 ± 0.1 | 1.06 ± 0.087 | 45.98 |
| 34 | 112-39-0 | Methyl hexadecanoate | 1.05 ± 0.095 | 1.13 ± 0.102 | 1.15 ± 0.098 | 1.52 ± 0.137 | 1.31 ± 0.124 | 44.76 |
| 35 | 68-26-8 | Vitamin a | 0.89 ± 0.074 | 0.9 ± 0.085 | 0.85 ± 0.073 | 0.55 ± 0.042 | 0.47 ± 0.037 | D |
| 36 | 1898-13-1 | CEMBRENE (Westpac ene) | 0.25 ± 0.018 | 0.24 ± 0.02 | 0.23 ± 0.019 | 0.21 ± 0.207 | 0.20 ± 0.013 | D |
| 37 | 84-74-2 | dibutyl phthalate | 0.29 ± 0.018 | 0.31 ± 0.026 | 0.42 ± 0.037 | 0.65 ± 0.051 | 0.54 ± 0.048 | 124.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Zhu, L.; Wang, K.; Zhang, X.; Mao, D.; Rasool, A. Unravel the Supremacy of Klebsiella variicola over Native Microbial Strains for Aroma-Enhancing Compound Production in Reconstituted Tobacco Concentrate through Metagenomic Analysis. Metabolites 2024, 14, 158. https://doi.org/10.3390/metabo14030158
Huang S, Zhu L, Wang K, Zhang X, Mao D, Rasool A. Unravel the Supremacy of Klebsiella variicola over Native Microbial Strains for Aroma-Enhancing Compound Production in Reconstituted Tobacco Concentrate through Metagenomic Analysis. Metabolites. 2024; 14(3):158. https://doi.org/10.3390/metabo14030158
Chicago/Turabian StyleHuang, Shen, Li Zhu, Ke Wang, Xinlong Zhang, Duobin Mao, and Aamir Rasool. 2024. "Unravel the Supremacy of Klebsiella variicola over Native Microbial Strains for Aroma-Enhancing Compound Production in Reconstituted Tobacco Concentrate through Metagenomic Analysis" Metabolites 14, no. 3: 158. https://doi.org/10.3390/metabo14030158
APA StyleHuang, S., Zhu, L., Wang, K., Zhang, X., Mao, D., & Rasool, A. (2024). Unravel the Supremacy of Klebsiella variicola over Native Microbial Strains for Aroma-Enhancing Compound Production in Reconstituted Tobacco Concentrate through Metagenomic Analysis. Metabolites, 14(3), 158. https://doi.org/10.3390/metabo14030158






